Abstract
Food intake induces synthesis of N-acylphosphatidylethanolamines (NAPEs) in the intestinal tract. While NAPEs exert leptin-like (leptogenic) effects, including reduced weight gain and food intake, the mechanisms by which NAPEs induce these leptogenic effects remain unclear. One key question is whether intestinal NAPEs act directly on cognate receptors or first require conversion to N-acylethanolamides (NAEs) by NAPE-hydrolyzing phospholipase D (NAPE-PLD). Previous studies using Nape-pld−/− mice were equivocal because intraperitoneal injection of NAPEs led to nonspecific aversive effects. To avoid the aversive effects of injection, we delivered NAPEs and NAEs intestinally using gut bacteria synthesizing these compounds. Unlike in wild-type mice, increasing intestinal levels of NAPE using NAPE-synthesizing bacteria in Nape-pld−/− mice failed to reduce food intake and weight gain or alter gene expression. In contrast, increasing intestinal NAE levels in Nape-pld−/− mice using NAE-synthesizing bacteria induced all of these effects. These NAE-synthesizing bacteria also markedly increased NAE levels and decreased inflammatory gene expression in omental adipose tissue. These results demonstrate that intestinal NAPEs require conversion to NAEs by the action of NAPE-PLD to exert their various leptogenic effects, so that the reduced intestinal NAPE-PLD activity found in obese subjects may directly contribute to excess food intake and obesity.
Keywords: obesity; liver; adipose; phosphatidylethanolamine; diet effects/lipid metabolism; N-acylphosphatidylethanolamine,; N-acylethanolamides; feeding behavior; phospholipases
Energy homeostasis is regulated by complex feedback loops, dysregulation of which leads to positive energy balance and obesity. Recent studies have identified N-acylphosphatidylethanolamines (NAPEs), a family of lipid mediators, as potentially important in satiety regulation and, thereby, inhibition of obesity (1–4). However, whether NAPEs act directly on cognate receptors or are simply precursors to the active signaling lipid is unclear (5). A better understanding of the mechanism of action for NAPEs should allow for identification of subjects at risk for obesity and appropriate molecular targets for potential pharmacotherapy.
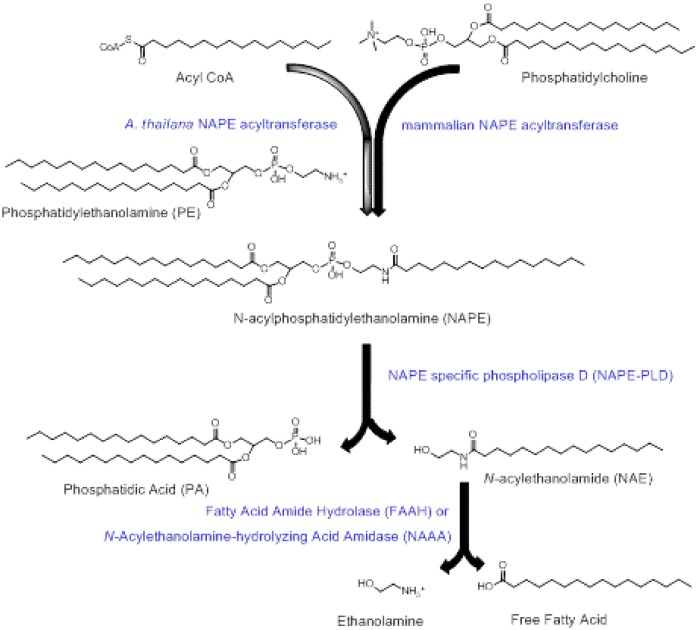
In response to feeding, intestinal cells synthesize NAPEs by the transfer of acyl chains to phosphatidylethanolamine (PE) using NAPE-acyltransferases (ATs) (Fig. 1) (6, 7). NAPEs can then be directly converted to N-acylethanolamides (NAEs) by the action of a NAPE-hydrolyzing phospholipase D (NAPE-PLD) (8). NAPE-PLD is a zinc metallo-hydrolase enzyme of the β-lactamase fold family with no known homology to other PLD enzymes (9) and is ubiquitously expressed in eukaryotic organisms, including yeast, worms, and mammals (10). In mammals, NAPE-PLD is expressed in many tissues, including the brain, various sections of the intestinal tract, the liver, and adipose tissue (8). Nape-pld−/− mice, created by removal of exon 4 of the Nape-pld gene, have significant reductions in levels of NAEs such as N-palmitoyl-ethanolamide (PEA) and N-oleoylethanolamide (OEA), but not N-arachidonylethanolamide (AEA) (11), suggesting that endogenous synthesis of these saturated and monounsaturated NAEs particularly requires NAPE-PLD. NAPEs can also be indirectly converted to NAEs by two alternative two-step processes: first to glycerophospho-N-acylethanolamines by α/beta-hydroxylase 4 (ABHD4) and then to NAEs by glycerophosphodiesterase 1 or first to phospho-NAEs by phospholipase C (PLC) and then to NAEs by protein phosphatase 22 (12, 13). NAEs are hydrolyzed to free fatty acids and ethanolamine by fatty acid amide hydrolase (FAAH) (14) and NAE-hydrolyzing acid amidase (NAAA) (15). To date, no cognate receptor has been identified for NAPEs, while a number of receptors have been identified for various NAEs. AEA is a potent agonist for endocannabinoid receptor 1 (CB1). In contrast to AEA, OEA and PEA are not agonists for CB1 (16, 17), but are agonists for PPARα (18, 19), TRPV1 (18, 19), GPR119 (20, 21), and GPR55 (22, 23) and the activation of these receptors by OEA and PEA (16, 19, 24, 25) or other agonists reduces food intake and increases fatty acid oxidation (26–30).
Fig. 1.
Formation of NAPE and conversion to NAE by NAPE-PLD. Transfer of an acyl group to PE by either mammalian NAPE-ATs from the sn-1 group of phosphatidylcholine or from acyl-CoA by the A. thaliana NAPE-AT generates NAPEs. NAPEs can then be converted to NAEs and phosphatidic acid (PA) by the action of NAPE-PLD. NAEs can be hydrolyzed to ethanolamine and free fatty acid by either FAAH or by NAE-hydrolyzing acid amidase.
Interventions that increase NAPEs exert leptin-like (i.e., leptogenic) effects, including reduced food intake and increased lipolysis and fatty acid oxidation (1–4), but whether these are direct effects of NAPEs on cognate receptors or require conversion of NAPEs to NAEs has been unclear. Intraperitoneal administration of NAPEs causes acute reductions in food intake and inhibits obesity induced by feeding a high fat diet (1–3). Administration of saturated or monounsaturated NAEs, such as PEA and OEA, respectively, also reduces food intake and increases fatty acid oxidation (16, 19, 25), while AEA (also known as anandamide) increases food intake, most likely via its activation of CB1 (31–34). About 20-fold higher concentrations of NAPE compared with OEA are required to reduce food intake when these compounds are administered intraperitoneally (1–3). It has been unclear if the leptogenic effects of NAPEs and NAEs result from a single integrated pathway where NAPEs are converted to NAEs and then act on cognate receptors for NAEs or if, instead, NAPEs and NAEs activate completely independent pathways (5). In a number of lipid signaling pathways, the substrate and the product of phospholipase action have separate bioactivities (e.g., phosphatidylinositol 4,5 bisphosphate has separate activities than its products inositol 1,4,5-trisphosphate and diacylglycerol that form by the action of phosphatidylinositol-specific PLCs).
Several studies have suggested that intact NAPE exerts leptogenic effects that are independent of its conversion to NAEs. Gillum et al. (1) found that intracerebroventricular injection of NAPE dramatically reduced food intake, while injection of NAEs or PE did not. They also showed that intraperitoneal injection of NAPE increased systemic circulating levels of NAPE and that intravenous injection of NAPE increased brain levels of NAPE, demonstrating that NAPE had the capacity to transfer intact from portal circulation to the brain. Wellner et al. (3) demonstrated that intraperitoneal injection of NAPE reduced food intake in Nape-pld−/− mice, as well as in wild-type mice. While these findings in their Nape-pld−/− mice could support either NAPEs acting directly on cognate receptors or after conversion to NAEs by non-NAPE-PLD-dependent pathways, Wellner et al. (3) also found that intraperitoneal injection of similar concentrations of PE or phosphatidylserine reduced food intake. Therefore, these investigators concluded that intraperitoneal injection of highly concentrated phospholipids resulted in a nonspecific aversive reduction in food intake and not that intact NAPE necessarily exerted a specific anorexigenic effect via cognate receptors. Studies by Geurts et al. (35) using adipose-specific deletion of NAPE-PLD showed that these mice had modestly increased body weight and body fat compared with wild-type mice, but not reductions in food intake, suggesting either that the satiety-inducing effects were exerted by intact NAPE or that the action of NAPE-PLD in other tissues, such as the intestinal tract, were required.
To investigate specific signaling by NAPE requires delivery of NAPEs to the intestine without adverse effects, so we had previously developed a method to deliver NAPEs intestinally using the commensal gut bacteria, Escherichia coli Nissle 1917 (EcN), engineered to overproduce NAPE (4). Unlike intraperitoneal NAPE administration that circumvents the intestine to deliver compounds to portal and systemic circulation, our bacterial NAPE system delivers them to the intestine to more closely mimic endogenous intestinal NAPE/NAE biosynthesis in response to food intake. Administering approximately 5 mg of NAPE per kilogram body weight per day by this bacterial method significantly reduced food intake and adiposity induced by a high fat diet, even though this amounts to less than 5% of the minimal NAPE dose required by intraperitoneal injection (4). Furthermore, the effect of NAPE-producing gut bacteria persisted up to 4 weeks after ending active administration, suggesting that even the lesser amounts of NAPE produced by the remaining colonized pNAPE-EcN was sufficient to sustain ongoing leptogenic signaling (4). As importantly, administration of control EcN transformed with the empty expression vector did not significantly alter either food intake or adiposity compared with untreated mice (4). Because expression of Arabidopsis thaliana NAPE-AT converts less than 0.1% of bacterial PE to NAPE, these empty expression vector EcNs controlled for any adverse effect of delivering PE or EcN to the intestinal lumen. While these previous studies clearly demonstrated that NAPEs absorbed from the intestinal tract reduced food intake, weight, and fat gain, and increased resting energy expenditure, they did not address whether these effects of intestinal NAPE required the action of NAPE-PLD. If so, then preventing the reduction in NAPE-PLD activity and NAE levels reported to occur in obesity (36–40) would be critical in the treatment of obesity. We therefore used Nape-pld−/− mice to test which of the various leptogenic effects (reduced body weight, reduced body fat, reduced food intake, increased fatty acid oxidation gene expression, reduced inflammatory gene expression) induced by intestinal NAPEs required conversion to NAEs by NAPE-PLD and found that NAPE-PLD is required for all of these processes.
MATERIALS AND METHODS
Plasmids
Generation of pQE80L1 (supplemental Fig. S1A) was previously described (4). In brief, commercially available plasmid pQE-80L (Qiagen) was modified to remove one lac operator to allow basal expression of NAPE-AT while maintaining the ability to further induce gene expression using the synthetic inducer, IPTG, or natural inducer, allolactose, that might be present in the gut during fermentation. The plasmid, pQE80L1-NAPE (supplemental Fig. S1B), was also previously generated by inserting the gene sequence for A. thaliana NAPE-AT into the multiple cloning site of pQE80L1 plasmid (4). Because previous reports raised the possibility that isolation of N-terminal tagged NAPE-PLD resulted isolation of truncated nonfunctional NAPE-PLD, we desired to create a plasmid that allowed inducible expression of NAPE-PLD with a C-terminal 6×His tag. To do this, we used the two synthetic oligos to create an insertion sequence encoding for a ribosomal binding site, start codon, and multiple cloning site followed by C-terminal 6×His tag and stop codon flanked by a 5′ EcoRI site and a 3′ HindIII site. Forward oligo: TCAATCACACAGAATTCATTAAAGAGGAGAAATTAACTATGAGAGGATCGCATCACCATCACCATCACGGATCCGCATG; reverse oligo: CATGCGGATCCGTGATGGTGATGGTGATGCGATCCTCTCATAGTTAATTTCTCCTCTTTAATGAATTCTGTGTGATTGA. These oligos were annealed together, digested with EcoRI and HindIII, and then ligated into pQE80L1 plasmid that had also been digested with EcoRI and HindIII. This insertion removes the original start codon and N-terminal RGS-6×His tag region, while leaving the T5 promoter and one lac operator sequence intact to create pQE80L1His (supplemental Fig. S1C). We then used PCR to amplify the murine Nape-pld using the plasmid pYX-ASC-Nape-pld purchased from Thermo Scientific Open Biosystems (clone ID, 5702354; catalog number MMM1013-9497346) as template and forward primer CGC GGATCC GAT GAG TAT GAG GAC AGC CAG and reverse primer GGG TGTTTCTTCAAAAGCTCTATCATCGG. This generated an amplicon with a BamHI site 5′ of the Nape-pld sequence. After digestion with BamHI, this amplicon was ligated into pQE80L1His plasmid that had been digested with BamHI and SmaI (a blunt end restriction enzyme) to create pQE80L1PLDHis (supplemental Fig. S1D). To create a plasmid for inducible coexpression of both NAPE-AT and NAPE-PLD, we first digested pQE80L1PLDHis with XhoI and XbaI under partial digestion conditions and filled in the resulting overhangs using Klenow fragment DNA Polymerase I to create blunt ended fragments. Of note, pQE80L1PLDHis contains three XbaI sites, one near the 3′ end of the Nape-pld gene, one immediately following the rrnB transcriptional terminator sequence, and one after the LacI gene, so the resulting linear fragments were separated on agarose gel to isolate the fragment containing the full-length Nape-pld gene, but not the additional LacI gene. The pQE80L1NAPE plasmid was also digested with BamHI and resulting fragments treated with Klenow fragment DNA polymerase to create blunt ends. This plasmid also contains two XbaI sites, with the LacI gene being located between these XbaI sites, so the fragment of appropriate size was also isolated by agarose gel. The two appropriate fragments were then ligated together to generate pQE80L1NAE (supplemental Fig. S1E). To confirm that pQE80L1NAE allowed for coexpression of both NAPE-AT and NAPE-PLD, we transformed pQE80L1NAE into E. coli DH5a and resulting single clones inoculated into LB broth with 1 mg/ml ampicillin, overnight cultures diluted and induced with 1 mM IPTG, incubated overnight at 30°C, then pelleted by centrifugation, lysed under native conditions with 1 mg/ml lysozyme, and supernatants mixed with 200 μl Ni-NTA-beads on ice. After three washes, the captured protein was eluted with 3× LDS/15% β-mercaptoethanol and subjected to SDS-PAGE followed by Coomassie G250 staining for visualization of protein (supplemental Fig. S2).
To determine whether expression of these proteins altered NAPE and NAE levels in our bacteria, we transformed EcN into which we had previously inserted a luciferase operon into the RecA gene (Lux-EcN) (4) with either the empty expression vector plasmid pQE80L1 (pEcN), with pQE80L1NAPE (pNAPE-EcN), or with pQE80L1NAE (pNAE-EcN). Bacteria were grown on LB agar plates with 1 mg/ml ampicillin added and single clones chosen for further amplification in liquid LB broth/ampicillin culture and for subsequent experimentation.
NAPE and NAE measurement
NAPE and NAE concentrations were quantified by LC/MS/MS using a revised version of our previously published protocol (41). For bacteria, the concentration of bacteria in liquid culture was determined by absorption at 600 nm. Bacteria were then pelleted by centrifugation, the supernatant discarded, and the bacterial pellet mixed with 4 ml of water, 8 ml of prechilled chloroform/methanol (2:1) solution, 1 nmol of C17:0NAPE (10 μl in chloroform), and 2 nmol of [2H4]C17:0NAE (8 μl in ethanol). For tissue, frozen sections were placed in 1 ml RIPA buffer and homogenized with a Polytron homogenizer and an aliquot removed for protein measurement. Then 3 ml water, 8 ml of prechilled chloroform/methanol (2:1) solution, 1 nmol of C17:0NAPE (10 μl in CHCl3) and 2 nmol of [2H4]C17:0NAE (8 μl in ethanol) were added. For either bacteria or tissue, the solvent mixture was then vortexed for 5 min and centrifuged at 4,000 g for 5 min. The resulting lower (chloroform) layer was carefully collected and dried under nitrogen gas. The extracted lipids were reconstituted in 1 ml of chloroform and loaded onto a Sep-Pak silica cartridge that had been preequilibrated with 4 ml methanol followed by 8 ml chloroform. The cartridge was washed with 1 ml of chloroform, and NAEs eluted with 8 ml chloroform/methanol (9:1) solution and eluant collected. NAPEs were eluted with 8 ml of chloroform/methanol (2:1) solution and eluant collected separately. NAEs were dried under nitrogen gas, redissolved in 200 ml of chloroform/ethanol (1:9) and analyzed by high-performance LC/MS/MS. NAPEs were dried under nitrogen gas, and then hydrolyzed to glycerophospho-NAEs by addition of 200 μl methylamine working solution (1-butanol/methanol/40% methylamine, 1/4/4, v/v/v) and incubation at 53°C for 1 h. After cooling, solutions were analyzed by LC/MS/MS. For analysis of NAEs, an Agilent XDB-C8 2.1 × 50 mm column was used with initial HPLC conditions of 99% solvent 1 (0.1% acetic acid in water) and 1% solvent 2 (0.1% acetic acid in methanol) and constant flow rate of 0.4 ml/min, then gradient ramp to 99% solvent 2 over 3 min, hold at 99% solvent 2 for 3 min, then return to initial conditions over 0.5 min, and hold for 1.5 min prior to initiating injection of next sample. HPLC eluant was coupled directly to a triple quadrupole mass spectrometer using electrospray ionization and operating in positive ion multiple reaction monitoring mode. The following transitions were monitored: m/z 298.3 → 62.2 at 35 eV, C16:1NAE; m/z 300.3 → 62.2 at 35 eV, C16:0NAE; m/z 312.3 → 62.2 at 35 eV, C17cyclopropaneNAE; m/z 318.3 → 66.2 at 35 eV, [2H4]C17:0NAE; and m/z 326.3 → 62.2 at 35eV, C18:1NAE. Quantitation was performed using the ratio of the peak area for [2H4]C17:0NAE versus each of the other NAE peaks in the sample. For analysis of NAPEs, a Phenomenex Kinetex 2.6 μ C18 100A column was used with initial HPLC conditions of 99% solvent 1 (1 mM triethylammonium acetate in water) and 1% solvent 2 (1 mM triethylammonium acetate in acetonitrile) and constant flow rate of 0.25 ml/min. A 0.5 min hold at the initial condition was followed by a gradient ramp to 99% solvent 2 over 4.5 min, then a hold at 99% solvent 2 for an additional 1.5 min, before returning to initial conditions over 0.5 min and holding for 2 min prior to initiating injection of next sample. HPLC eluant was coupled directly to a triple quadrupole mass spectrometer using electrospray ionization and operating in negative ion multiple reaction monitoring mode. The following transitions were monitored: m/z 450.3 → 79.1 at 50 eV, C16:1GP-NAE; m/z 452.3 → 79.1 at 50 eV, C16:0GP-NAE; m/z 464.3 → 79.1 at 50 eV, C17cyclopropaneGP-NAE; m/z 466.3 → 79.1 at 50 eV, C17:0NAE; and m/z 478.3 → 79.1 at 50 eV, C18:1GP-NAE. Quantitation was performed using the ratio of the peak area for C17:0NAPE versus each of the other NAPE peaks in the sample.
Animal experiments
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee at Vanderbilt University. Nape-pld−/− mice were a generous gift from Dr. Benjamin Cravatt at Scripps Institute in La Jolla, CA and a breeding colony was established at Vanderbilt University. These Nape-pld−/− mice were created by deletion of exon 4 of the Nape-pld gene (11). Mice were individually housed in the Vanderbilt University animal facility in a 12 h-light/12 h-dark cycle. All mice were initially fed a standard chow diet (Lab Diet 5001; 13.5% kcal from fat, 60% kcal from carbohydrate, 28.5% kcal from protein) until start of the bacterial treatment. For studies with Nape-pld−/− mice, 18 male mice (6–10 weeks old) were used. Prior to start of bacterial treatment, mice were pretreated with 0.5 g/l ampicillin in drinking water for 7 days. During this time, body weight and body composition were determined for each mouse. Body weight was measured using a portable electronic scale. For body composition, mice were scanned by magnetic resonance imaging using a Bruker Minispec MQ10 NMR analyzer to determine fat mass, lean mass, and free fluid. The mice were then divided into three groups of six mice each, with mice grouped to ensure that each group had the same number of mice with identical birthdates and a similar mean and variation in body weight and body fat. All mice continued to be individually housed throughout the treatment and follow-up periods. Each group of mice was then randomly assigned to receive one of three bacterial treatments: pEcN, pNAPE-EcN, and pNAE-EcN. On treatment day 0, ampicillin treatment was stopped and standard drinking water was replaced with water containing 0.125% gelatin and 5 × 109 cfu/ml of the appropriate bacteria. The chow diet was also replaced with the high fat diet (TestDiet 58Y1; containing 60% fat by kilocalories) for the remainder of the study. Cumulative food intake, body weight, and body fat were measured at regular intervals. Daily food intake was measured by adding preweighed food pellets to each cage and then reweighing these pellets at the next time point. On treatment day 56, the administration of bacteria was stopped. On treatment day 72, mice were euthanized and tissue collected and stored at −80°C until analysis. Omental fat was collected from around the intestinal tract. Retroperitoneal fat was collected from the flanks on the side of the peritoneal cavity. For the brain sample, the entire hemisphere was collected. For analysis of cecum, the sample was cut lengthwise to expose luminal content; the luminal content was collected to measure fecal NAPE and NAE levels; any remaining content was then washed away from cecum with PBS. The set of tissues harvested from one mouse in the pNAE-EcN group was lost during storage.
To verify that pNAE-EcN did not exert a markedly greater effect than pNAPE-EcN in wild-type mice, an identical study to that carried out with Nape-pld−/− mice was performed with male wild-type C57BL/6J mice purchased from the Jackson Laboratory, as described in (4). For this study, the pNAE-EcN was an additional (previously unpublished observations) arm of the published study and 10 mice were used for each group.
For studies examining the effect of oral administration of purified C18:1NAPE, a fine particle dispersion in water containing either 3.75% C18:1NAPE with vehicle or vehicle only (soy lecithin, caprylic triglycerides, polyglycerol ester of fatty acids, citric acid, and potassium sorbate, where additional soy lecithin equal to the amount of added C18:1NAPE was substituted in vehicle) was generated by 3i Solutions (Wooster, OH) in collaboration with Chemi SpA (Patrica, Italy). The 3.75% C18:1NAPE was added to drinking water at a final concentration of 1 mg/ml C18:1NAPE and an identical dilution of vehicle was used as control. C57BL6 mice were switched to the 60% high fat diet at the same time that administration of C18:1NAPE began, and mice were treated for 8 weeks with either vehicle (n = 6) or C18:1NAPE (n = 7). Drinking water was changed every 2–3 days.
Measurement of liver and omental adipose gene expression
For measurement of gene expression in tissue, total RNA was extracted from liver tissue using an RNeasy Micro kit (Qiagen Sciences) and the concentration was measured spectrophotometrically. The extracted RNA was reverse transcribed into cDNA using a cDNA reverse transcription kit (Applied Biosystems) and the RNA expression level was quantified by a quantitative (q)real-time-PCR using SYBR Green PCR Master Mix (Qiagen Sciences) and the 7500 Real-Time PCR system (Applied Biosystems) in the Molecular Cell Biology Resource Core of Vanderbilt University. For Cpt1a, the primers were GTGACTGGTGGGAGGAATAC and GAGCATCTCCATGGCGTAG. For AOX, the primers were GTGCAGCTCAGAGTCTGTCCAA and TACTGCTGCGTCTGAAAATCCA. For Ppara, the primer pair used was GTACGGTGTGTATGAAGCCATCTT and GCCGTACGCGATCAGCAT. For Ppard, the primers were GCCATATTCCCAGGCTGTC and CAGCACAAGGGTCATCTGTG. For Scd1, the primers were TTCTTACACGACCACCACCA and CCGAAGAGGCAGGTGTAGAG. For Cd36, the primers were CCTTAAAGGAATCCCCGTGT and TGCATTTGCCAATGTCTAGC. For Tnfa, the primers were CCATTCCTGAGTTCTGCAAAG and GCAAATATAAATAGAGGGGGGC. For Ccl2, the primers were ACTGAAGCCAGCTCTCTCTTCCTC and TTCCTTCTTGGGGTCAGCACAGAC. For β-actin (Actb), the primers were GAGCGCAAGTACTCTGTGTG and CGGACTCATCGTACTCCTG. Relative quantification of gene expression with real-time PCR data was calculated relative to β-actin.
RESULTS
To assess the requirement for NAPE-PLD hydrolysis to the leptogenic effects of NAPE, we needed to be able to deliver NAPE to the intestinal tract of Nape-pld−/− mice at sufficient concentrations to induce a robust leptogenic effect without inducing the adverse effects reported to occur with intraperitoneal injection (3). While we had previously found that administration in drinking water of EcN engineered to express NAPE (pNAPE-EcN) could effectively deliver NAPE intestinally and inhibit weight gain without adverse effect (4), we recognized that, for mechanistic studies, interpretation of outcomes might be simplified if it was feasible to orally administer purified compound directly to the mice rather than relying on living bacteria for delivery. We therefore tested to determine whether we could achieve robust NAPE-induced alterations in body weight gain by administering drinking water with 1 g/l C18:1NAPE as a fine particle dispersion in order to deliver approximately 200 mg/kg body weight of C18:1NAPE per day. Weight gain tended to be reduced by this treatment with C18:1NAPE, but this change was not sufficiently robust to be useful for our studies, as it did not reach statistical significance during the 8 weeks of treatment (supplemental Fig. S3). Additional optimization of the C18:1NAPE dose, duration of treatment, and/or formulation would therefore be required to achieve the necessary robust effect. For this reason, we chose instead to continue using our previously established recombinant bacteria strategy in order to deliver NAPE to test the requirement for NAPE-PLD for NAPE action.
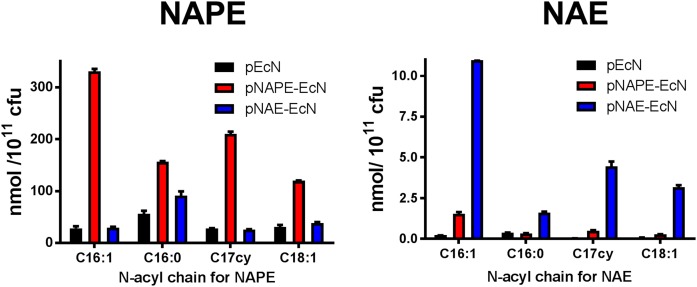
We previously showed that transformation of EcN with a plasmid encoding for expression of A. thaliana NAPE-AT (pQE80L1-NAPE) results in significant synthesis of NAPEs compared with EcN that was transformed with a plasmid encoding only empty expression vector (pQE80L1) (4). To generate EcN-synthesizing NAEs, we created a plasmid for coexpressing both NAPE-AT and NAPE-PLD. To do this, we inserted the mouse Nape-pld gene with an additional sequence for a C-terminal 6×His tag into the plasmid, pQE80L1NAPE, to create the plasmid, pQE80L1NAE (supplemental Fig. S1). Transformation with pQE80L1-NAE resulted in strong expression of both NAPE-AT and NAPE-PLD (supplemental Fig. S2). We then transformed EcN with pQE80L1 (pEcN), pQE80L1NAPE (pNAPE-EcN), or pQE80L1NAE (pNAE-EcN), and measured the resulting biosynthesis of NAPEs and NAEs in these bacteria using MS. Total NAPEs were only 1.3-fold higher in pNAE-EcN compared with pEcN (Fig. 2A), while total NAEs were 27.1-fold higher in pNAE-EcN compared with pEcN (Fig. 2B). An inverse pattern for total NAPEs and NAEs was seen with pNAPE-EcN. Administration of pNAE-EcN in drinking water to wild-type mice resulted in reduced weight and fat gain very similar to what we had previously published for administration of pNAPE-EcN (supplemental Fig. S4), which is consistent with NAPE requiring conversion to NAE for its actions.
Fig. 2.
Coexpression of NAPE-PLD and NAPE-AT in EcN increases bacterial levels of NAE, but not NAPE. EcN was transformed either with empty expression plasmid pQE80L1 (pEcN), with plasmid pQE80L1NAPE for expression of A. thaliana NAPE-AT only (pNAPE-EcN), or with plasmid pQEL1NAE for coexpression of A. thaliana NAPE-AT and murine Nape-pld (pNAE-EcN). Gene expression was induced with IPTG for 4 h in duplicate batches of transformed EcN and levels of NAPE and NAE measured after solid phase extraction by LC/MS/MS using C17:0NAPE and [2H4]C17:0NAE as internal standards.
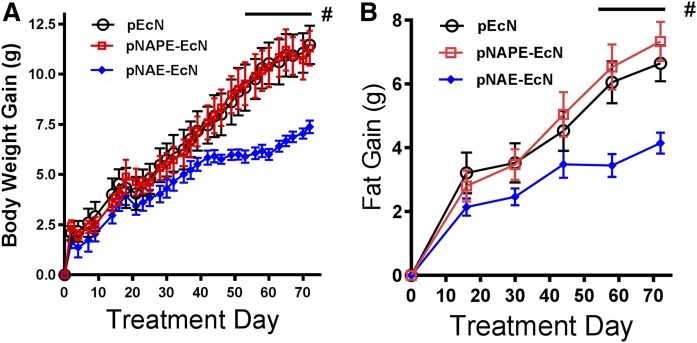
To determine whether NAPE required the action of NAPE-PLD to exert its leptogenic effects, we used mice with whole-body genetic deletion of exon 4 of the Nape-pld gene (Nape-pld−/−), that had previously been shown to have markedly decreased levels of saturated and monounsaturated NAEs in brain (11). We found that peripheral tissues of these Nape-pld−/− mice also had reduced NAEs levels compared with wild-type mice (supplemental Fig. S5). We then fed Nape-pld−/− mice a high fat diet (60% kcal fat) and simultaneously administered pNAPE-EcN, pNAE-EcN, or pEcN for 8 weeks (to treatment day 56). At the end of the 8 weeks, bacterial treatments were withdrawn and mice continued on the high fat diet for another 2 weeks of follow-up before being euthanized. Unlike wild-type mice, Nape-pld−/− mice that received pNAPE-EcN did not differ in weight gain (Fig. 3A) or fat gain (Fig. 3B) compared with those that received control bacteria (pEcN). In contrast, Nape-pld−/− mice that received pNAE-EcN gained significantly less body weight and body fat than those receiving pEcN.
Fig. 3.
Treatment with pNAE-EcN, but not pNAPE-EcN, is effective at inhibiting high fat diet-induced gain in body weight and body fat in Nape-pld−/− mice. Male Nape-pld−/− mice that had initially been maintained on a chow diet were switched to a 60% fat diet at day 0 and administered either pEcN, pNAPE-EcN, or pNAE-EcN through day 56, after which treatment was stopped and the mice continued on a 60% fat diet to day 70. A: Effect of bacterial treatment on body weight gain (two-way ANOVA: P < 0.0001 for time; P < 0.0356 for treatment; P < 0.0001 for interaction; Bonferroni post hoc analysis for individual treatment days: #P < 0.05 pNAE-EcN vs. pEcN; P = not significant (n.s.) for pNAPE-EcN vs. pEcN). B: The effect of treatment on body fat gain (two-way ANOVA: P < 0.0001 for time; P < 0.0253 for treatment; P < 0.0001 for interaction; Bonferroni post hoc analysis for individual treatment days: #P < 0.05 pNAE-EcN vs. pEcN; P = n.s. for pNAPE-EcN vs. pEcN).
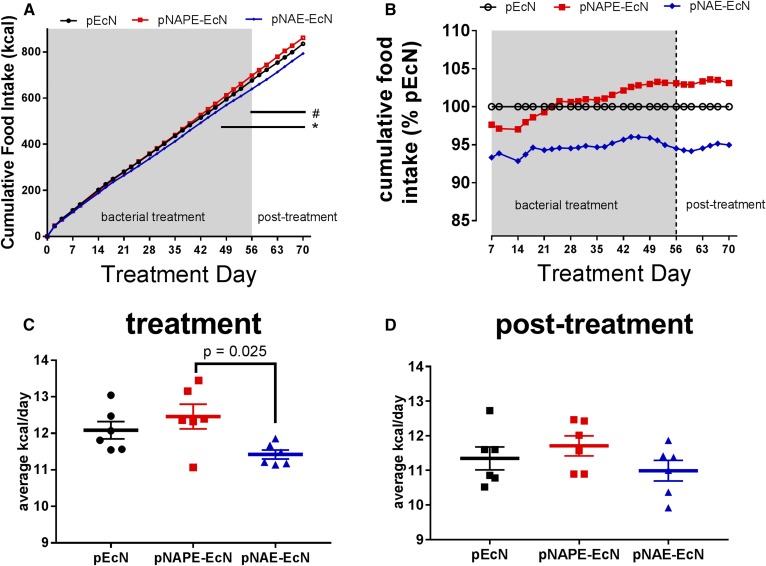
To assess whether the reduction in weight gain and adiposity induced by administration of pNAE-EcN was mediated, in part, by increased satiation leading to reduced overeating, we calculated cumulative food intake throughout the treatment and post-treatment period. By day 56 of bacterial administration, mice given pNAE-EcN, but not those given pNAPE-EcN, had statistically reduced cumulative food intake compared with those given pEcN (Fig. 4A), although a tendency toward lower relative cumulative food intake (about 5% lower) than mice given pEcN or pNAPE-EcN was manifest from day 7 of treatment and throughout the follow-up period (Fig. 4B). Average daily food intake during the 56 days of bacterial treatment was 11.42 kcal/day for mice given pNAE-EcN compared with 12.08 kcal/day for mice given pEcN (Fig. 4C). During the 2 week posttreatment period, average daily food intake for mice given pNAE-EcN was 10.99 kcal/day compared with 11.35 kcal/day for mice given pEcN (Fig. 4D).
Fig. 4.
Treatment with pNAE-EcN, but not pNAPE-EcN, inhibits food intake. A: Cumulative food intake in Nape-pld−/− mice [two-way ANOVA: P < 0.0001 for time; P = 0.062 for treatment; P ≤ 0.0001 for interaction; Tukey’s post hoc multiple comparison testing for individual treatments at each time point: #P < 0.05 pNAE-EcN vs. pEcN (days 56–70), *P < 0.05 pNAE-EcN vs. pNAPE-EcN (days 44–70)]. B: Cumulative food intake expressed as percentage of the pEcN treatment group. C: Average daily food intake during treatment period (days 0–56) (one-way ANOVA: P = 0.030; Tukey’s multiple comparisons test: P = 0.025 for pNAE-EcN vs. pNAPE-EcN). D: Average daily food intake during posttreatment period (day 57–70) (one-way ANOVA: P = 0.284).
Our previous studies had shown that fecal levels of pNAPE-EcN dropped dramatically in the first few days after cessation of bacterial administration, but then remained fairly stable, and that the leptogenic effects of pNAPE-EcN in wild-type mice persisted for at least 4 weeks after cessation of bacterial administration, despite the much lower fecal pNAPE-EcN levels (4). During the bacterial administration period, fecal EcN likely consisted primarily of the planktonic EcN, which passed through the intestinal tract without adhering; whereas within a few days of withdrawing EcN from drinking water, fecal EcN likely consisted primarily of EcN shed by stable intestinal colonies. Thus, the persistence of the leptogenic effect of pNAPE-EcN in the wild-type mice suggested that the amount of NAPE/NAE produced by the colonized pNAPE-EcN was sufficient to produce the leptogenic effect without any contribution from planktonic pNAPE-EcN. We therefore followed our Nape-pld−/− mice for an additional 2 weeks after cessation of pNAE-EcN administration to confirm that the leptogenic effects persisted and then euthanized the mice to determine the tissue levels of NAE produced in the colonized mice. Fecal NAE levels were elevated in mice previously administered pNAE-EcN compared with those previously administered pEcN at this 2 week posttreatment time point, consistent with continued colonization and production (supplemental Fig. S6). The differences in weight and fat gain (Fig. 3) and reduced food intake (Fig. 4) were also maintained during the 2 week follow-up period, consistent with the colonized pNAE-EcN producing sufficient NAE to induce leptogenic effects. When we then measured NAPE and NAE in the large intestine (cecum) and plasma, we found that treatment with pNAPE-EcN significantly increased intestinal and plasma levels of NAPE (Fig. 5A, B) without increasing NAE levels (Fig. 5C, D) compared with treatment with pEcN. In contrast, treatment with pNAE-EcN significantly increased intestinal and plasma NAE levels without increasing NAPE levels (Fig. 5A–D). These results are consistent with absorption from the intestinal tract of the expected product from each type of colonized bacteria, and a markedly reduced capacity of the Nape-pld−/− mice to convert NAPEs to NAEs.
Fig. 5.
Colonization with pNAPE-EcN or pNAE-EcN increases NAPE and NAE levels, respectively, in intestine and plasma of Nape-pld−/− mice. Nape-pld−/− mice were administered pEcN, pNAPE-EcN, or pNAE-EcN in drinking water for 8 weeks while on high fat diet, and then kept on high fat diet for another 2 weeks prior to tissue collection. Cecum was homogenized and NAPE (A) and NAE (B) levels were measured by LC/MS/MS (n = 5–6 cecum per group). Treatment significantly altered NAPE levels (one-way ANOVA: P = 0.0446; Tukey’s multiple comparison test: pNAPE-EcN vs. pEcN, P = 0.048; pNAE-EcN vs. pEcN, P = 0.896) and NAE levels (one-way ANOVA:P = 0.0296; Tukey’s multiple comparison test: pNAPE-EcN vs. pEcN, P = 0.818; pNAE-EcN vs. pEcN, P = 0.030). Plasma was also collected and NAPE (C) and NAE (D) measured (n = 6 per group). Treatment significantly altered NAPE levels (one-way ANOVA: P = 0.0036; Tukey’s multiple comparison test: pNAPE-EcN vs. pEcN, P = 0.003; pNAE-EcN vs. pEcN, P = 0.075) and NAE levels (one-way ANOVA: P = 0.0104; Tukey’s multiple comparison test: pNAPE-EcN vs. pEcN, P = 0.204; pNAE-EcN vs. pEcN, P = 0.008).
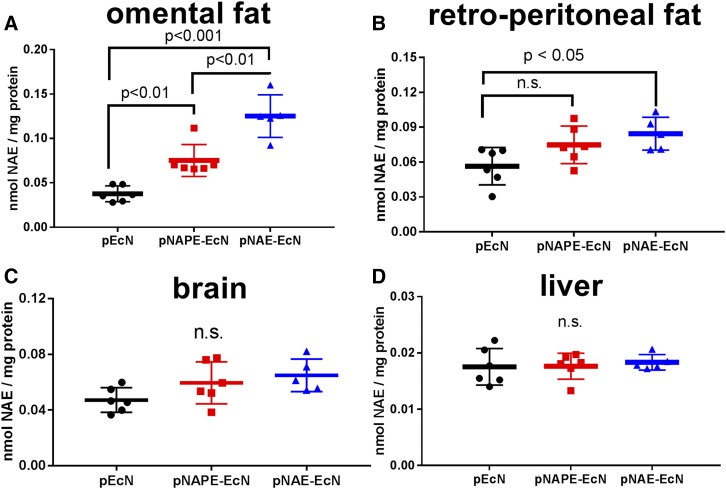
We then assessed whether intestinally absorbed NAE elevated NAE levels in target tissues. We found that Nape-pld−/− mice colonized with pNAE-EcN also had elevated NAE levels in omental adipose tissue (Fig. 6A) and retroperitoneal adipose tissue (Fig. 6B) compared with those colonized with pNAPE-EcN or pEcN. Non-statistically significant trends toward elevation in whole brain NAE levels were found in pNAE-EcN-colonized mice (Fig. 6C), while no elevation in liver NAEs were detected (Fig. 6D).
Fig. 6.
Colonization with pNAE-EcN increases NAE levels in extra-intestinal tissue of Nape-pld−/− mice. Total tissue NAE levels 2 weeks post-administration were measured by LC/MS/MS. A: Omental adipose tissue (one-way ANOVA: P < 0.0001; Tukey’s multiple comparisons test: pNAPE-EcN vs. pEcN, P < 0.01; pNAE-EcN vs. pEcN, P < 0.001; pNAE-EcN vs. pNAPE-EcN, P < 0.01). B: Retroperitoneal adipose tissue (one-way ANOVA: P = 0.0283; Tukey’s multiple comparisons test: pNAPE-EcN vs. pEcN, P = not significant (n.s.); pNAE-EcN vs. pEcN, P < 0.05). C: Brain (one-way ANOVA: P = 0.0741). D: Liver (one-way ANOVA: P = 0.8519).
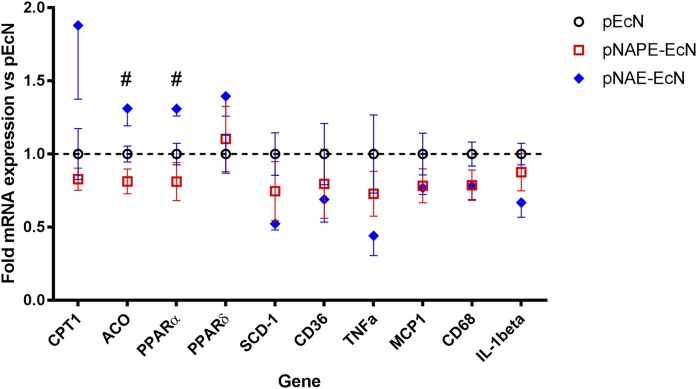
We previously showed that in wild-type mice, pNAPE-EcN treatment increased hepatic expression of genes related to fatty acid oxidation, including Cpt1, Acox1, Ppara, and Ppard, in addition to its effects on food intake and proposed that these changes might contribute to the increased basal metabolic rate induced by NAPE. Geurts et al. (35) had also shown that specifically deleting NAPE-PLD in adipose tissue resulted in altered hepatic gene expression. To test whether NAPE-PLD was required for the hepatic response to intestinally absorbed NAPE, we examined the effect of pNAPE-EcN treatment on hepatic gene expression in Nape-pld−/− mice. In Nape-pld−/− mice, colonization with pNAPE-EcN failed to increase hepatic expression of fatty acid oxidation genes, while colonization with pNAE-EcN increased their expression (Fig. 7). Colonization with pNAE-EcN also tended to reduce hepatic expression of inflammatory genes, such as Tnfa and IL-1b, but these differences were not statistically significant.
Fig. 7.
Colonization of Nape-pld−/− mice with pNAE-EcN, but not with pNAPE-EcN, induces changes in hepatic expression of genes related to fatty acid oxidation. Gene expression was measured by qPCR relative to β-actin and normalized to expression in pEcN treated group. #One-way ANOVA, P < 0.05.
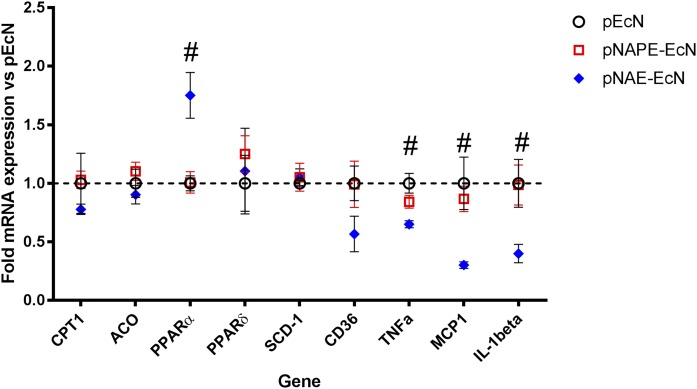
Because we had found that pNAE-EcN colonization markedly increased NAE levels in omental adipose tissue, we also looked at its effect on gene expression in this tissue. We found that colonization of Nape-pld−/− with pNAPE-EcN did not alter adipose gene expression compared with treatment with pEcN. In contrast, pNAE-EcN colonization significantly increased expression of Ppara and decreased expression of inflammatory cytokines, including Tnfa, Ccl2, and Il1b (Fig. 8).
Fig. 8.
Colonization of Nape-pld−/− mice with pNAE-EcN, but not pNAPE-EcN, induces changes in omental adipose gene expression. Gene expression was measured by qPCR relative to β-actin and normalized to expression in pEcN treated group. #One-way ANOVA P < 0.05.
DISCUSSION
Increasing levels of NAPE, either via intestinal synthesis in response to feeding or by pharmacological administration of NAPE, was previously shown to exert leptogenic effects, such as reduced food intake, body weight, and body fat, with increased expression of fat oxidation genes. However, previous studies did not resolve whether NAPE arising in the intestinal tract exerts these various leptogenic effects directly as intact NAPE and independent of conversion to NAE or, instead, requires hydrolysis to NAE by NAPE-PLD (5). Increasing intestinal NAPE levels using engineered gut bacteria reduced food intake, weight gain, and fat gain and modulated gene expression in wild-type mice, but not Nape-pld−/− mice. In contrast, increasing intestinal NAEs levels using engineered bacteria did induce these beneficial changes in Nape-pld−/− mice. Together these results support the proposition that the leptogenic effects of intestinal NAPEs require their conversion to NAEs by NAPE-PLD.
Geurts et al.’s (35) previous demonstration that adipose-specific deletion of NAPE-PLD increased body weight and body fat compared with wild-type mice clearly implicated conversion of NAPEs to NAEs in adipose as an important mediator of adiposity and adipose inflammation. However, because they found no changes in food intake in this model, whether NAPE regulation of feeding behavior was independent of NAPE-PLD activity or simply required NAPE-PLD in non-adipose tissue was unclear. Furthermore, whether activity of NAPE-PLD in non-adipose tissue was also required for other metabolic effects of NAPE, such as changes in hepatic fat oxidation genes, was also unclear. Our previous studies with pair-feeding wild-type untreated mice to the caloric intake of NAPE-treated mice demonstrated that about two-thirds of the overall effects of NAPE on inhibiting weight gain on a high fat diet resulted from reduced food intake. The remaining one-third of weight gain inhibition appeared to stem from alteration in basal metabolic rate through upregulation of genes related to fatty acid oxidation, including Cpt1a, Acox1, Ppara, and Ppard (4). The results of our current study demonstrate that the effects of NAPE on food intake require its conversion to NAEs by NAPE-PLD. We found that, unlike in wild-type mice, increasing intestinal NAPE levels by administering pNAPE-EcN failed to reduced food intake in Nape-pld−/− mice. In contrast, increasing intestinal NAE levels by administering pNAE-EcN reduced food intake in both wild-type and Nape-pld−/− mice. Our findings that the leptogenic effects of intestinal NAPE require metabolism to NAE is consistent with studies demonstrating that oral or intraperitoneal administration of PEA or OEA is sufficient to reduce food intake and inhibit weight gain (16, 19, 24, 25). Our findings also suggest that future studies should focus on elucidating how NAEs, rather than the initially synthesized NAPEs, act at various receptors and tissues to induce satiety, increase metabolism, and inhibit inflammation and also on the extent to which reduced metabolism of NAPEs to NAEs accounts for dysregulation of these pathways in obesity.
A number of signaling pathways that may be relevant to satiety, increased metabolism, and inhibition of inflammation have already been identified for saturated and monounsaturated NAEs. Previous reports that direct injection of these NAEs into the brain do not induce decreased food intake (1, 16) and our finding that pNAE-EcN colonization did not significantly elevate brain NAE levels suggest that intestinally absorbed NAEs primarily act on peripheral sites, including adipose tissue and the enteric nervous system, which then generate downstream signals that reduce food intake and altered metabolic rate. Known receptors for saturated and monounsaturated NAEs include PPARα (18, 19), TRPV1 (42, 43), GPR119 (20, 21), and GPR55 (22, 23). Unrelated agonists of these receptors induce leptogenic effects (26–30). Potential downstream pathways from these receptors, including induction of stimulation of lipolysis in adipocytes by PPARα activation (44), inhibition of SCD-1 by activation of PPARα (45, 46), activation of kinases by PPARα (47), repression of nitric oxide signaling by PPARα (18), induction of white adipose remodeling to beige adipose by PPARα activation (48), induction of hypothalamic oxytocin secretion by PPARα-mediated stimulation of vagal fibers (49, 50), GLP-1 secretion from enteroendocrine L cells induced by GPR119 activation (20, 51), enhancement of GLP1 receptor activation by direct binding of OEA to GLP-1 (52), inhibition of CB1-mediated activation of gustatory insula by GPR119 activation (53), and induction of vagal nerve currents via TRPV1 activation (54). Despite these significant advances in our understanding of how the NAEs can exert the broad range of leptogenic effects attributed to the NAPE/NAE pathway, additional studies are clearly still needed.
For instance, our finding that NAPE-PLD activity was required for increases in hepatic gene expression of Acox1 and Ppara even though hepatic NAE levels were not elevated warrants future studies. Because treatment with pNAE-EcN markedly elevated NAE levels in other tissues, such as adipose tissue and the intestinal tract, changes in hepatic gene expression are likely to be induced indirectly by signals released from these tissues. This would be consistent with Geurts et al.’s (35) finding that deleting Nape-pld specifically in adipose tissue reduced hepatic expression of Acox1 and Ppara. The extent to which NAPE-PLD in intestinal cells versus adipose tissue contributes to appropriate regulation of hepatic fat oxidation gene expression will require future studies where NAPE-PLD activity is abrogated in specific intestinal cell types.
Our finding that adipose tissue beds appeared to undergo the most marked elevation in NAE levels in comparison to other tissues after pNAE-EcN treatment also warrants further investigation because of the therapeutic implications of the apparent inverse association between adipose NAE levels and adipose inflammatory gene expression. Geurts et al. (35) found that reducing adipose NAE levels by specifically deleting adipose Nape-pld markedly increased expression of inflammatory genes in adipose tissue, including Ccl2 and Il1b, while we found that increasing adipose NAE levels in Nape-pld−/− mice by administering pNAE-EcN significantly downregulated inflammatory genes, Tnfa, Ccl2, and Il1b, in adipose tissue. Inflammation in visceral adipose tissue has been implicated as a key factor driving the markedly increased risk for cardiovascular disease seen with obesity (55), so that reducing adipose inflammation by increasing adipose NAE level may be a powerful therapeutic intervention for obese subjects.
A related therapeutic implication of our findings is that they clearly suggest that any condition that reduces peripheral NAPE-PLD activity will likely make individuals vulnerable to obesity (through reduced satiety and lower basal metabolic rate) and to obesity-driven inflammatory diseases. Currently, the factors that regulate peripheral expression and activity of NAPE-PLD are poorly defined. Short-term feeding of a high fat diet to rodents (1–7 days) is sufficient to markedly decrease NAPE-PLD mRNA expression in the small intestine (36), while longer term feeding downregulates NAPE-PLD in adipose tissue (40). Other factors that have been shown to modulate NAPE-PLD activity are exposure to LPS (56) and the presence of bile acids (57), although whether either of these alters NAPE-PLD activity in the intestinal tract or adipose tissue remains to be determined. Therefore, our current studies highlight the need for future studies seeking to understand how NAPE-PLD expression and activity is regulated, especially in humans exposed to obesogenic conditions.
Supplementary Material
Acknowledgments
The authors thank Dr. Benjamin Cravatt of the Scripps Institute for his generous gift of Nape-pld−/− mice, Dr. Linda Zhang of Vanderbilt University for discussion and comments on the manuscript, and Dr. Wade Calcutt of Vanderbilt University for helpful suggestions with the mass spectrometry analysis.
Footnotes
Abbreviations:
- AEA
- N-arachidonylethanolamide
- CB1
- endocannabinoid receptor 1
- EcN
- Escherichia coli Nissle 1917
- FAAH
- fatty acid amide hydrolase
- NAAA
- NAE-hydrolyzing acid amidase
- NAE
- N-acylethanolamide
- NAPE
- N-acylphosphatidylethanolamine
- NAPE-AT
- N-acylphosphatidylethanolamine-acyltransferase
- NAPE-PLD
- N-acylphosphatidylethanolamine-hydrolyzing phospholipase D
- OEA
- N-oleoylethanolamide
- PE
- phosphatidylethanolamine
- PEA
- N-palmitoyl-ethanolamide
- PLC
- phospholipase C
- pEcN
- Escherichia coli Nissle 1917 transformed with empty expression vector
- pNAPE-EcN
- Escherichia coli Nissle 1917 transformed with expression plasmid for N-acylphosphatidylethanolamine-acyltransferase
- pNAE-EcN
- Escherichia coli Nissle 1917 transformed with expression plasmid for N-acylphosphatidylethanolamine-acyltransferase and N-acylphosphatidylethanolamine-hydrolyzing phospholipase D
This work was supported, in part, by National Institutes of Health/National Center for Complementary and Integrative Health Grant AT007830 and Vanderbilt University Department of Pharmacology and Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Gillum M. P., Zhang D., Zhang X. M., Erion D. M., Jamison R. A., Choi C., Dong J., Shanabrough M., Duenas H. R., Frederick D. W., et al. . 2008. N-acylphosphatidylethanolamine, a gut- derived circulating factor induced by fat ingestion, inhibits food intake. Cell. 135: 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srisai D., Gillum M. P., Panaro B. L., Zhang X. M., Kotchabhakdi N., Shulman G. I., Ellacott K. L., and Cone R. D.. 2011. Characterization of the hyperphagic response to dietary fat in the MC4R knockout mouse. Endocrinology. 152: 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellner N., Tsuboi K., Madsen A. N., Holst B., Diep T. A., Nakao M., Tokumura A., Burns M. P., Deutsch D. G., Ueda N., et al. . 2011. Studies on the anorectic effect of N-acylphosphatidylethanolamine and phosphatidylethanolamine in mice. Biochim. Biophys. Acta. 1811: 508–512. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z., Guo L., Zhang Y., Walzem R. L., Pendergast J. S., Printz R. L., Morris L. C., Matafonova E., Stien X., Kang L., et al. . 2014. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J. Clin. Invest. 124: 3391–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romano A., Tempesta B., Provensi G., Passani M. B., and Gaetani S.. 2015. Central mechanisms mediating the hypophagic effects of oleoylethanolamide and N-acylphosphatidylethanolamines: different lipid signals? Front. Pharmacol. 6: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen H. S., and Diep T. A.. 2009. N-acylethanolamines, anandamide and food intake. Biochem. Pharmacol. 78: 553–560. [DOI] [PubMed] [Google Scholar]
- 7.Fu J., Astarita G., Gaetani S., Kim J., Cravatt B. F., Mackie K., and Piomelli D.. 2007. Food intake regulates oleoylethanolamide formation and degradation in the proximal small intestine. J. Biol. Chem. 282: 1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto Y., Morishita J., Tsuboi K., Tonai T., and Ueda N.. 2004. Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem. 279: 5298–5305. [DOI] [PubMed] [Google Scholar]
- 9.Wang J., and Ueda N.. 2009. Biology of endocannabinoid synthesis system. Prostaglandins Other Lipid Mediat. 89: 112–119. [DOI] [PubMed] [Google Scholar]
- 10.McPartland J. M., Matias I., Di Marzo V., and Glass M.. 2006. Evolutionary origins of the endocannabinoid system. Gene. 370: 64–74. [DOI] [PubMed] [Google Scholar]
- 11.Leung D., Saghatelian A., Simon G. M., and Cravatt B. F.. 2006. Inactivation of N-acyl phosphatidylethanolamine phospholipase D reveals multiple mechanisms for the biosynthesis of endocannabinoids. Biochemistry. 45: 4720–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon G. M., and Cravatt B. F.. 2010. Characterization of mice lacking candidate N-acyl ethanolamine biosynthetic enzymes provides evidence for multiple pathways that contribute to endocannabinoid production in vivo. Mol. Biosyst. 6: 1411–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Wang L., Harvey-White J., Huang B. X., Kim H. Y., Luquet S., Palmiter R. D., Krystal G., Rai R., Mahadevan A., et al. . 2008. Multiple pathways involved in the biosynthesis of anandamide. Neuropharmacology. 54: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinney M. K., and Cravatt B. F.. 2005. Structure and function of fatty acid amide hydrolase. Annu. Rev. Biochem. 74: 411–432. [DOI] [PubMed] [Google Scholar]
- 15.Ueda N., Tsuboi K., and Uyama T.. 2010. N-acylethanolamine metabolism with special reference to N-acylethanolamine-hydrolyzing acid amidase (NAAA). Prog. Lipid Res. 49: 299–315. [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez de Fonseca F., Navarro M., Gómez R., Escuredo L., Nava F., Fu J., Murillo-Rodríguez E., Giuffrida A., LoVerme J., Gaetani S., et al. . 2001. An anorexic lipid mediator regulated by feeding. Nature. 414: 209–212. [DOI] [PubMed] [Google Scholar]
- 17.Lambert D. M., and Di Marzo V.. 1999. The palmitoylethanolamide and oleamide enigmas: are these two fatty acid amides cannabimimetic? Curr. Med. Chem. 6: 757–773. [PubMed] [Google Scholar]
- 18.Fu J., Gaetani S., Oveisi F., Lo Verme J., Serrano A., Rodriguez De Fonseca F., Rosengarth A., Luecke H., Di Giacomo B., Tarzia G., et al. . 2003. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 425: 90–93. [DOI] [PubMed] [Google Scholar]
- 19.Fu J., Oveisi F., Gaetani S., Lin E., and Piomelli D.. 2005. Oleoylethanolamide, an endogenous PPAR-alpha agonist, lowers body weight and hyperlipidemia in obese rats. Neuropharmacology. 48: 1147–1153. [DOI] [PubMed] [Google Scholar]
- 20.Overton H. A., Babbs A. J., Doel S. M., Fyfe M. C., Gardner L. S., Griffin G., Jackson H. C., Procter M. J., Rasamison C. M., Tang-Christensen M., et al. . 2006. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 3: 167–175. [DOI] [PubMed] [Google Scholar]
- 21.Moran B. M., Abdel-Wahab Y. H., Flatt P. R., and McKillop A. M.. 2014. Activation of GPR119 by fatty acid agonists augments insulin release from clonal beta-cells and isolated pancreatic islets and improves glucose tolerance in mice. Biol. Chem. 395: 453–464. [DOI] [PubMed] [Google Scholar]
- 22.McKillop A. M., Moran B. M., Abdel-Wahab Y. H., and Flatt P. R.. 2013. Evaluation of the insulin releasing and antihyperglycaemic activities of GPR55 lipid agonists using clonal beta-cells, isolated pancreatic islets and mice. Br. J. Pharmacol. 170: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryberg E., Larsson N., Sjogren S., Hjorth S., Hermansson N. O., Leonova J., Elebring T., Nilsson K., Drmota T., and Greasley P. J.. 2007. The orphan receptor GPR55 is a novel cannabinoid receptor. Br. J. Pharmacol. 152: 1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen H. S. 2014. Role of anorectic N-acylethanolamines in intestinal physiology and satiety control with respect to dietary fat. Pharmacol. Res. 86: 18–25. [DOI] [PubMed] [Google Scholar]
- 25.Mattace Raso G., Santoro A., Russo R., Simeoli R., Paciello O., Di Carlo C., Diano S., Calignano A., and Meli R.. 2014. Palmitoylethanolamide prevents metabolic alterations and restores leptin sensitivity in ovariectomized rats. Endocrinology. 155: 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington W. W., Britt C. S., Wilson J. G., Milliken N. O., Binz J. G., Lobe D. C., Oliver W. R., Lewis M. C., and Ignar D. M.. 2007. The effect of PPARalpha, PPARdelta, PPARgamma, and PPARpan agonists on body weight, body mass, and serum lipid profiles in diet-induced obese AKR/J mice. PPAR Res. 2007: 97125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergeron R., Yao J., Woods J. W., Zycband E. I., Liu C., Li Z., Adams A., Berger J. P., Zhang B. B., Moller D. E., et al. . 2006. Peroxisome proliferator-activated receptor (PPAR)-alpha agonism prevents the onset of type 2 diabetes in Zucker diabetic fatty rats: a comparison with PPAR gamma agonism. Endocrinology. 147: 4252–4262. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L. L., Yan Liu D., Ma L. Q., Luo Z. D., Cao T. B., Zhong J., Yan Z. C., Wang L. J., Zhao Z. G., Zhu S. J., et al. . 2007. Activation of transient receptor potential vanilloid type-1 channel prevents adipogenesis and obesity. Circ. Res. 100: 1063–1070. [DOI] [PubMed] [Google Scholar]
- 29.Saito M. 2015. Capsaicin and related food ingredients reducing body fat through the activation of TRP and brown fat thermogenesis. Adv. Food Nutr. Res. 76: 1–28. [DOI] [PubMed] [Google Scholar]
- 30.Al-Barazanji K., McNulty J., Binz J., Generaux C., Benson W., Young A., and Chen L.. 2015. Synergistic effects of a GPR119 agonist with metformin on weight loss in diet-induced obese mice. J. Pharmacol. Exp. Ther. 353: 496–504. [DOI] [PubMed] [Google Scholar]
- 31.Williams C. M., and Kirkham T. C.. 1999. Anandamide induces overeating: mediation by central cannabinoid (CB1) receptors. Psychopharmacology (Berl.). 143: 315–317. [DOI] [PubMed] [Google Scholar]
- 32.Hao S., Avraham Y., Mechoulam R., and Berry E. M.. 2000. Low dose anandamide affects food intake, cognitive function, neurotransmitter and corticosterone levels in diet-restricted mice. Eur. J. Pharmacol. 392: 147–156. [DOI] [PubMed] [Google Scholar]
- 33.Di Marzo V., Goparaju S. K., Wang L., Liu J., Batkai S., Jarai Z., Fezza F., Miura G. I., Palmiter R. D., Sugiura T., et al. . 2001. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature. 410: 822–825. [DOI] [PubMed] [Google Scholar]
- 34.Jamshidi N., and Taylor D. A.. 2001. Anandamide administration into the ventromedial hypothalamus stimulates appetite in rats. Br. J. Pharmacol. 134: 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geurts L., Everard A., Van Hul M., Essaghir A., Duparc T., Matamoros S., Plovier H., Castel J., Denis R. G., Bergiers M., et al. . 2015. Adipose tissue NAPE-PLD controls fat mass development by altering the browning process and gut microbiota. Nat. Commun. 6: 6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diep T. A., Madsen A. N., Holst B., Kristiansen M. M., Wellner N., Hansen S. H., and Hansen H. S.. 2011. Dietary fat decreases intestinal levels of the anorectic lipids through a fat sensor. FASEB J. 25: 765–774. [DOI] [PubMed] [Google Scholar]
- 37.Rivera P., Luque-Rojas M. J., Pastor A., Blanco E., Pavon F. J., Serrano A., Crespillo A., Vida M., Grondona J. M., Cifuentes M., et al. . 2013. Diet-dependent modulation of hippocampal expression of endocannabinoid signaling-related proteins in cannabinoid antagonist-treated obese rats. Eur. J. Neurosci. 37: 105–117. [DOI] [PubMed] [Google Scholar]
- 38.Starowicz K. M., Cristino L., Matias I., Capasso R., Racioppi A., Izzo A. A., and Di Marzo V.. 2008. Endocannabinoid dysregulation in the pancreas and adipose tissue of mice fed with a high-fat diet. Obesity (Silver Spring). 16: 553–565. [DOI] [PubMed] [Google Scholar]
- 39.Artmann A., Petersen G., Hellgren L. I., Boberg J., Skonberg C., Nellemann C., Hansen S. H., and Hansen H. S.. 2008. Influence of dietary fatty acids on endocannabinoid and N-acylethanolamine levels in rat brain, liver and small intestine. Biochim. Biophys. Acta. 1781: 200–212. [DOI] [PubMed] [Google Scholar]
- 40.Igarashi M., DiPatrizio N. V., Narayanaswami V., and Piomelli D.. 2015. Feeding-induced oleoylethanolamide mobilization is disrupted in the gut of diet-induced obese rodents. Biochim. Biophys. Acta. 1851: 1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo L., Amarnath V., and Davies S. S.. 2010. A liquid chromatography-tandem mass spectrometry method for measurement of N-modified phosphatidylethanolamines. Anal. Biochem. 405: 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahern G. P. 2003. Activation of TRPV1 by the satiety factor oleoylethanolamide. J. Biol. Chem. 278: 30429–30434. [DOI] [PubMed] [Google Scholar]
- 43.Ambrosino P., Soldovieri M. V., Russo C., and Taglialatela M.. 2013. Activation and desensitization of TRPV1 channels in sensory neurons by the PPARalpha agonist palmitoylethanolamide. Br. J. Pharmacol. 168: 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guzmán M., Lo Verme J., Fu J., Oveisi F., Blázquez C., and Piomelli D.. 2004. Oleoylethanolamide stimulates lipolysis by activating the nuclear receptor peroxisome proliferator-activated receptor alpha (PPAR-alpha). J. Biol. Chem. 279: 27849–27854. [DOI] [PubMed] [Google Scholar]
- 45.Serrano A., Del Arco I., Javier Pavon F., Macias M., Perez-Valero V., and Rodriguez de Fonseca F.. 2008. The cannabinoid CB1 receptor antagonist SR141716A (Rimonabant) enhances the metabolic benefits of long-term treatment with oleoylethanolamide in Zucker rats. Neuropharmacology. 54: 226–234. [DOI] [PubMed] [Google Scholar]
- 46.Terrazzino S., Berto F., Dalle Carbonare M., Fabris M., Guiotto A., Bernardini D., and Leon A.. 2004. Stearoylethanolamide exerts anorexic effects in mice via down-regulation of liver stearoyl-coenzyme A desaturase-1 mRNA expression. FASEB J. 18: 1580–1582. [DOI] [PubMed] [Google Scholar]
- 47.Martínez de Ubago M., García-Oya I., Pérez-Pérez A., Cánfran-Duque A., Quintana-Portillo R., Rodríguez de Fonseca F., González-Yanes C., and Sánchez-Margalet V.. 2009. Oleoylethanolamide, a natural ligand for PPAR-alpha, inhibits insulin receptor signalling in HTC rat hepatoma cells. Biochim. Biophys. Acta. 1791: 740–745. [DOI] [PubMed] [Google Scholar]
- 48.Suárez J., Rivera P., Arrabal S., Crespillo A., Serrano A., Baixeras E., Pavón F. J., Cifuentes M., Nogueiras R., Ballesteros J., et al. . 2014. Oleoylethanolamide enhances beta-adrenergic-mediated thermogenesis and white-to-brown adipocyte phenotype in epididymal white adipose tissue in rat. Dis. Model. Mech. 7: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romano A., Cassano T., Tempesta B., Cianci S., Dipasquale P., Coccurello R., Cuomo V., and Gaetani S.. 2013. The satiety signal oleoylethanolamide stimulates oxytocin neurosecretion from rat hypothalamic neurons. Peptides. 49: 21–26. [DOI] [PubMed] [Google Scholar]
- 50.Gaetani S., Fu J., Cassano T., Dipasquale P., Romano A., Righetti L., Cianci S., Laconca L., Giannini E., Scaccianoce S., et al. . 2010. The fat-induced satiety factor oleoylethanolamide suppresses feeding through central release of oxytocin. J. Neurosci. 30: 8096–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lauffer L. M., Iakoubov R., and Brubaker P. L.. 2009. GPR119 is essential for oleoylethanolamide-induced glucagon-like peptide-1 secretion from the intestinal enteroendocrine L-cell. Diabetes. 58: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Y. H., Ho M. S., Huang W. T., Chou Y. T., and King K.. 2015. Modulation of glucagon-like peptide-1 (GLP-1) potency by endocannabinoid-like lipids represents a novel mode of regulating GLP-1 receptor signaling. J. Biol. Chem. 290: 14302–14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang Y., Sato H., Saito M., Yin D. X., Park S. K., Oh S. B., Bae Y. C., and Toyoda H.. 2016. A role of CB1R in inducing theta-rhythm coordination between the gustatory and gastrointestinal insula. Sci. Rep. 6: 32529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thabuis C., Tissot-Favre D., Bezelgues J. B., Martin J. C., Cruz-Hernandez C., Dionisi F., and Destaillats F.. 2008. Biological functions and metabolism of oleoylethanolamide. Lipids. 43: 887–894. [DOI] [PubMed] [Google Scholar]
- 55.Autieri M. V. 2016. Adipose inflammation at the heart of vascular disease. Clin. Sci. 130: 2101–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu C., Solorzano C., Sahar S., Realini N., Fung E., Sassone-Corsi P., and Piomelli D.. 2011. Proinflammatory stimuli control N-acylphosphatidylethanolamine-specific phospholipase D expression in macrophages. Mol. Pharmacol. 79: 786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magotti P., Bauer I., Igarashi M., Babagoli M., Marotta R., Piomelli D., and Garau G.. 2015. Structure of human N-acylphosphatidylethanolamine-hydrolyzing phospholipase D: regulation of fatty acid ethanolamide biosynthesis by bile acids. Structure. 23: 598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.