Abstract
Fungi can produce jasmonic acid (JA) and its isoleucine conjugate in large quantities, but little is known about the biosynthesis. Plants form JA from 18:3n-3 by 13S-lipoxygenase (LOX), allene oxide synthase, and allene oxide cyclase. Shaking cultures of Fusarium oxysporum f. sp. tulipae released over 200 mg of jasmonates per liter. Nitrogen powder of the mycelia expressed 10R-dioxygenase-epoxy alcohol synthase activities, which was confirmed by comparison with the recombinant enzyme. The 13S-LOX of F. oxysporum could not be detected in the cell-free preparations. Incubation of mycelia in phosphate buffer with [17,17,18,18,18-2H5]18:3n-3 led to biosynthesis of a [2H5]12-oxo-13-hydroxy-9Z,15Z-octadecadienoic acid (α-ketol), [2H5]12-oxo-10,15Z-phytodienoic acid (12-OPDA), and [2H5]13-keto- and [2H5]13S-hydroxyoctadecatrienoic acids. The α-ketol consisted of 90% of the 13R stereoisomer, suggesting its formation by nonenzymatic hydrolysis of an allene oxide with 13S configuration. Labeled and unlabeled 12-OPDA were observed following incubation with 0.1 mM [2H5]18:3n-3 in a ratio from 0.4:1 up to 47:1 by mycelia of liquid cultures of different ages, whereas 10 times higher concentration of [2H5]13S-hydroperoxyoctadecatrienoic acid was required to detect biosynthesis of [2H5]12-OPDA. The allene oxide is likely formed by a cytochrome P450 or catalase-related hydroperoxidase. We conclude that F. oxysporum, like plants, forms jasmonates with an allene oxide and 12-OPDA as intermediates.
Keywords: cytochrome P450, epoxy alcohol synthase, fatty acid dioxygenase, jasmonates, lipids/oxidation, lipoxygenase, mass spectrometry, methods/high-performance liquid chromatography
Fusarium oxysporum is a plant pathogenic fungus, which infects the root tips and causes wilt disease (1, 2). F. oxysporum can infect virtually all plants except grasses. A special form [forma specialis (f. sp.)] of F. oxysporum is so designated from its capacity to infect a specific host. F. oxysporum f. sp. cubense is one of most infamous strains, as it causes Panama disease of banana, a constant threat to this commodity (3).
The methyl ester of (−)-jasmonic acid [(−)-JA] was isolated and structurally characterized by Demole in 1962 (4). The (+)-7-iso-jasmonic acid [(+)-JA], which is the biologically relevant form of JA, was conclusively identified more than two decades later in lemon (5) and in media from two fungi, Lasiodiplodia theobromae (6) and Fusarium fujikoroi (7).
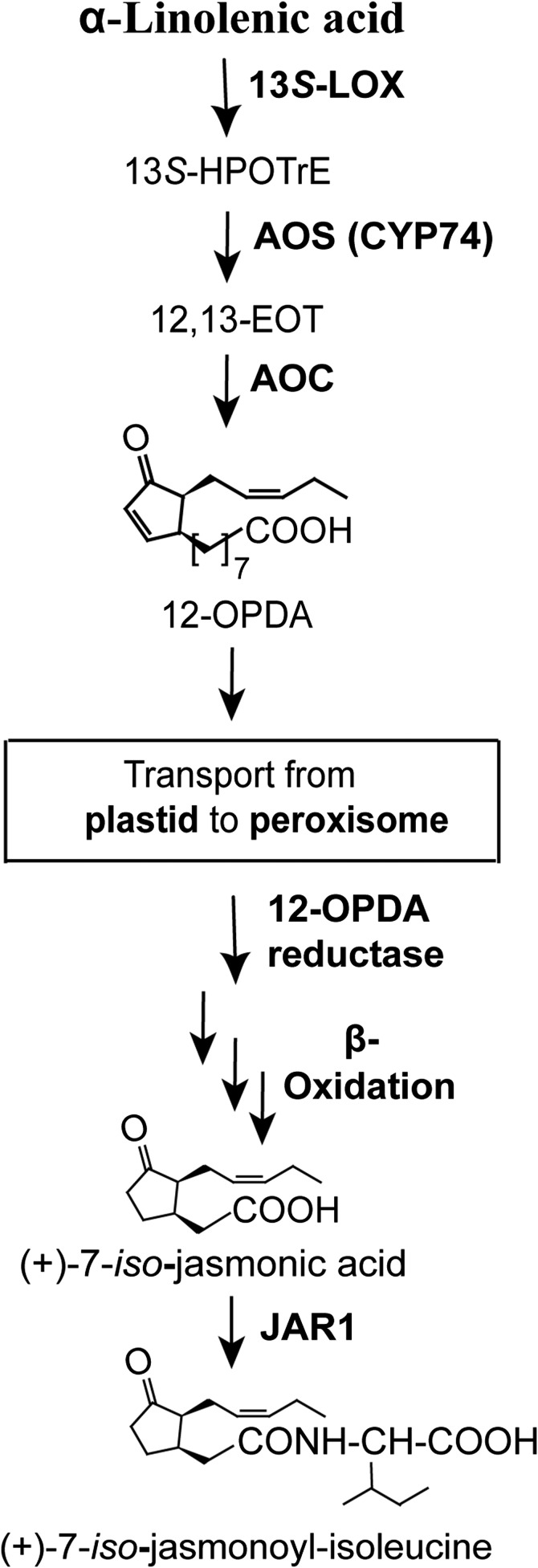
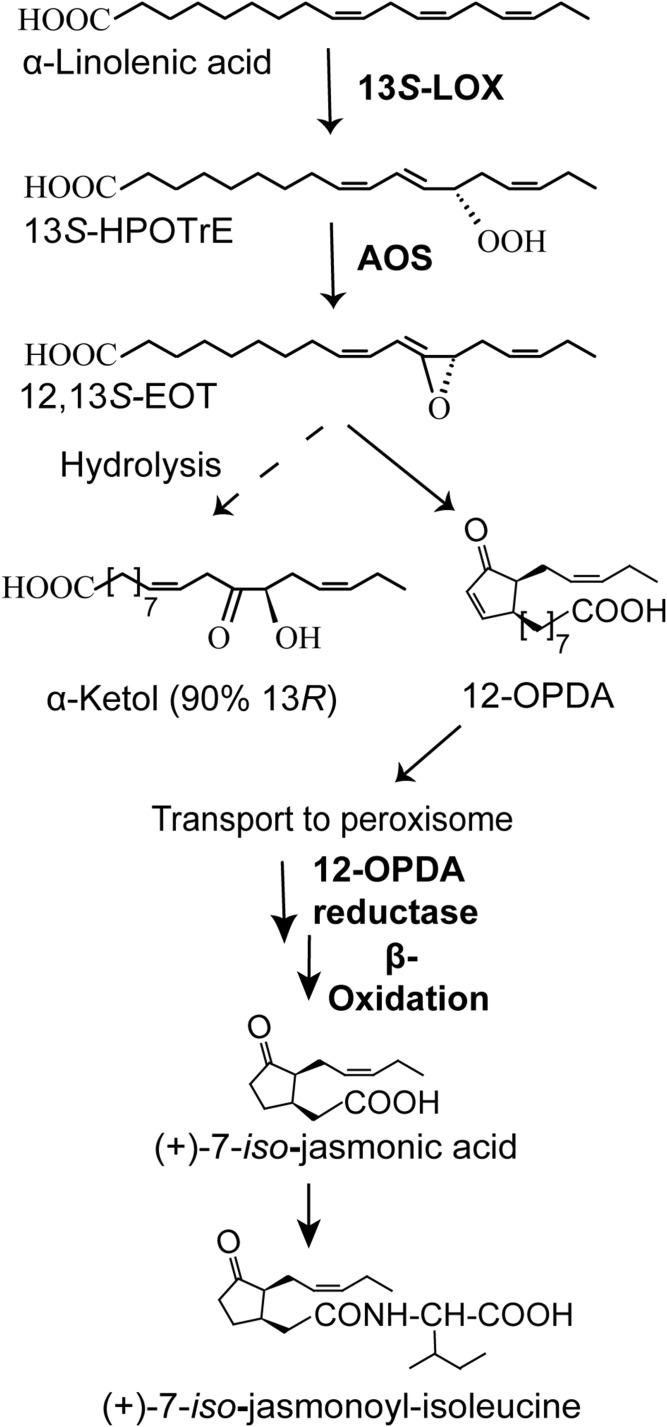
The plant biosynthesis of JA has been investigated in detail. Many of the enzymes have been crystallized, and the biological importance of jasmonates as growth hormones and physiological regulators is well-documented (8–14). The 13S-lipoxygenase (LOX) of plants oxidizes 18:3n-3 to 13S-hydroperoxy-9Z,11E,15Z-octadecatrienoic acid [13S-hydroperoxyoctadecatrienoic acid (HPOTrE)], which is dehydrated by allene oxide synthase (AOS) to an allene oxide, which in turn is converted by allene oxide cyclase (AOC) to 12-oxo-10,15Z-phytodienoic acid (12-OPDA) in plastids. The 12-OPDA is reduced and subject to β-oxidation with formation of (+)-JA in the peroxisomes and then conjugated with Ile (Fig. 1).
Fig. 1.
Overview of the plant pathway to jasmonates. The first steps to 12-OPDA are catalyzed in plastid. The 12-OPDA is then transferred to peroxisome for further transformations to (+)-JA. The end product, (+)-JA-Ile, has potent biological effects.
The fungal biosynthesis of (+)-JA is intriguing in many aspects. First, fungi can form remarkably large amounts of (+)-JA in the laboratory, as discussed below. Second, in spite of the large production of (+)-JA, the mechanism of the fungal biosynthesis is not firmly established. Third, (+)-JA produced by fungi can augment plant disease (15, 16); yet many plants form jasmonates as defense reactions to fungal infections (11, 12). Fourth, the initial steps of (+)-JA biosynthesis in plants occur in plastids, which are absent in fungi.
L. theobromae and members of the Fusarium family remain prototypes for studies of (+)-JA biosynthesis. L. theobromae (strain 2334) can routinely produce 300–500 mg (+)-JA per liter of growth medium and occasionally much more (17–19). In the fields, L. theobromae causes excessive growth of tropical fruit trees, possibly due to (+)-JA secretion (15, 18). Several subspecies of F. oxysporum, which infect garden stock, cabbage, and tulips, are also known to produce (+)-JA, 9,10-dihydro-JA, and their conjugates with isoleucine (15, 20). The most prominent biosynthesis occurs in still cultures of F. oxysporum f. sp. tulipae (Fot). Fot can produce milligram amounts of jasmonates per liter and induce gum disease in tulips (15, 16).
The mechanism of (+)-JA biosynthesis by L. theobromae was investigated by Tsukada, Takahashi, and Nabeta (21), who found that L. theobromae produced [2H4](−)-JA following feeding with [9,10,12,13,14,15-2H6]18:3n-3 (21). Traces of labeled 12-OPDA were detected (21); however, additional compounds indicative of the plant pathway [13S-HPOTrE and 12-oxo-13-hydroxy-9Z,15Z-octadecadienoic acid (α-ketol)] were not observed. The fungal biosynthesis of jasmonates from 18:3n-3 therefore remains an enigma.
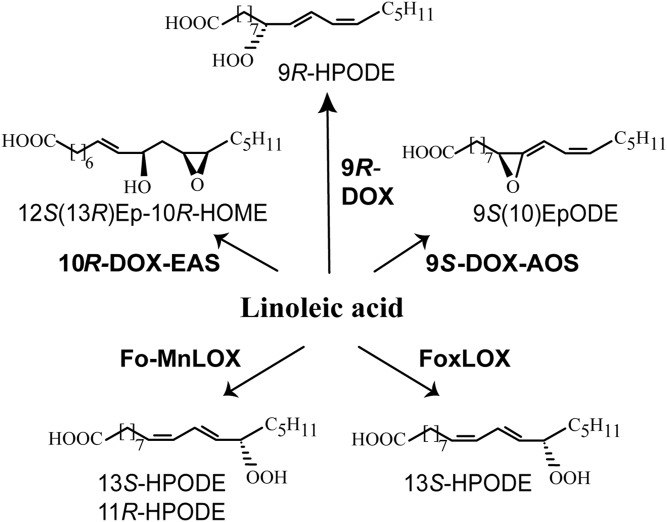
Dioxygenases (DOXs) of C18 fatty acids, which belong to the cyclooxygenase gene family, and LOXs have been investigated by genome analysis of F. oxysporum and recombinant protein expression (22–25). The genome codes for two LOXs and three DOX-cytochrome P450 (CYP) fusion proteins (Fig. 2). The LOXs contain either catalytic manganese (Fo-MnLOX) or catalytic iron (FoxLOX). Both enzymes oxidize 18:2n-6 and 18:3n-3 to 13S-hydroperoxy metabolites, and Fo-MnLOX also forms 11-hydroperoxy metabolites (22, 26). These LOXs could be candidates for the first fungal enzyme in a plant pathway to jasmonates (26). The 9S-DOX-AOS oxidizes 18:2n-6 to 9S-HPODE and the 9S-AOS transforms this hydroperoxide to an allene oxide (Fig. 2), but it does not transform 13-HPOTrE (24, 27). This 9S-AOS is therefore not involved in fungal biosynthesis of (+)-JA. This also applies to 9R-DOX, which does not form 13-hydroperoxy C18 fatty acids (23). The 10R-DOX-epoxy alcohol synthase (EAS) of F. oxysporum has, so far, only been characterized as a recombinant homolog to 10R-DOX-EAS of the rice blast fungus, Magnaporthe oryzae, but only with respect to oxidation of 18:2n-6 (25). Expression of 10R-DOX-EAS in mycelia of F. oxysporum has not been reported, but expressed sequence tags have been found.
Fig. 2.
Overview of LOXs and DOX-CYPs of F. oxysporum. Recombinant Fo-MnLOX (FOXB_09004) and FoxLOX (FOMG_06104) oxidize 18:2n-6 to 13S-HPODE, whereas 18:2n-6 is oxidized by recombinant 10R-DOX-EAS (FOXB_03425), 9R-DOX (FOXB_09952), and 9S-DOX-AOS (FOXB_01332) to the illustrated end products. HOME, hydroxyoctadecenoic acid. EpODE, epoxyoctadecadienoic acid.
The present investigation had three goals. The first goal was to optimize the growth conditions of F. oxysporum with respect to biosynthesis of jasmonates, and we chose to investigate Fot (15). The second goal was to determine the transformation of unsaturated C18 fatty acids by nitrogen powder of these mycelia. This led us to investigate the oxidation of 18:3n-3 and 18:1n-9 by recombinant 10R-DOX-EAS of F. oxysporum. The third goal was to detect possible intermediates in jasmonate biosynthesis, which could be formed by the intact mycelium.
MATERIALS AND METHODS
Materials
Cartridges, silicic acid, and C18 silica (SepPak) were from Waters. Fatty acids were dissolved in ethanol and stored in stock solutions (50–100 mM) at −20°C. The 18:2n-6 (99%) was from Sigma. The 18:1n-9 (99%), 18:3n-3 (99%), [13C18]18:2n-6 (98%), [17,17,18,18,18-2H5]18:3n-3, (+)-JA, 12-OPDA, and α-ketol were from Larodan (Stockholm, Sweden). When required, fatty acids were purified by SiO2 chromatography (SepPak) and eluted with 30 ml diethyl ether/hexane/acetic acid, 5/95/0.2, in one step. The [13C18]10S/R-hydroperoxy-8E,12Z-octadecadienoic acid (HPODE) was prepared by photo oxidation of [13C18]18:2n-6 (24). The [2H5]13S-HPOTrE was prepared with soybean LOX-1 (Sigma) and 13R-HPOTrE with 13R-MnLOX. The (−)-JA, RNaseA, potato dextrose broth (PDB), and ampicillin were from Sigma. Czapec-Dox broth (CDB) was prepared with reagents from Sigma and VWR [per liter: saccharose ( 30 g), NaNO3 (3 g), K2HPO4 (1 g), KCl (0.5 g), MgSO4·7H2O (0.5 g), and FeSO4·7H2O (0.01 g)]. JA-Ile was synthesized as described (28). The Ile conjugate of 18:3n-3 was prepared as earlier described (29) and purified by reversed phase (RP)-HPLC using a solvent system of methanol-water-acetic acid (85:15:0.01, v/v/v). Fot (NRRL 26954) was from ARS Culture Collection (Peoria, IL), imported with due permission (Jordbruksverket), and stored on potato dextrose agar (PDA) plates at +4°C. The fungus was recultivated for 7–10 days at room temperature (PDA plates). The 10R-DOX-EAS of F. oxysporum (EGU86021; FOXB_03425) was expressed in Escherichia coli BL21 Star cells (Invitrogen), as described (25).
N-([5-2H2,7-2H](−)-jasmonoyl)-(S)-isoleucine O-methyloxime {[2H3](−)-JA-Ile-MO} was prepared as follows. N-[(−)-jasmonoyl]-(S)-isoleucine (95 mg), prepared as described in (28), was added to 2H2O (10 ml) and stirred with 300 mg of K2CO3 at 23°C for 46 h. A solution of 30 mM O-methyl-hydroxylamine hydrochloride in methanol was added, and the mixture was kept at 23°C for 18 h. Extraction with ethyl acetate gave a residue that was subjected to RP-HPLC using a 250 × 10 mm column of Nucleosil 100-7 C18 eluted with methanol/water/acetic acid (60:40:0.015, v/v/v) at a flow rate of 4 ml/min. This afforded pure [2H3](−)-JA-Ile-MO (68 mg) as a white solid. An aliquot was treated with diazomethane and analyzed by GC-MS. Two peaks (ratio, 6:1), due to the MO syn-anti isomers, appeared. The mass spectrum of the major isomer showed the following prominent ions [deuterium isotope shifts relative to the corresponding unlabeled derivative (30) are given in parenthesis]: m/z 338 (+3), 307 (+2), 270 (+3), 183 (+3), 150 and 151 (+2 and +3, respectively), and 86 (+0). The isotopic composition was 93.07% ([2H3]), 4.85% (2H2), 1.30% (2H1), and 0.78% (2H0).
Fungal cultures
In initial experiments, 50–100 ml CDB or PDB in 250–500 ml flasks were inoculated from PDA plates (0.5–1 cm2) and grown under various conditions [dark or light, 22°C or 28°C, with or without moderate shaking (100 rpm)]. These incubations were terminated after 18–21 days.
In subsequent studies, PDB was inoculated with growth medium (2.5–50 ml PDB) from a growing culture and incubated at 28°C (100 rpm) in the dark (shaking incubator SI-300R; Jeio Tech). After 4–5 days, the culture was colored deep red due to bikaverin, and they were harvested after 10–15 days. The reddish mycelia were collected by filtration, washed with saline, frozen in liquid N2, and ground to a fine powder in a mortar with liquid N2. The powder was stored at −80°C. An aliquot was added to 0.1 M KHPO4 buffer (pH 7.4)/2 mM EDTA/0.04% Tween-20 and homogenized with a Potter-Elvehjem glass homogenizer with Teflon pestle (10 passes) and then incubated with fatty acids.
The growth media were centrifuged (3,100 g) and the supernatant was then assayed for jasmonates. An aliquot (7–10 ml) was extracted on a cartridge of C18 silica (SepPak/C18), washed with water (2 ml), and lipids were eluted with ethyl acetate (4 ml) (31). The organic extract was evaporated under a stream of N2, dissolved in ethanol (1 ml), and 1–10 μl were then analyzed for JA-conjugates by LC-MS/MS. For detection of JA, the organic extract of 10 ml medium was evaporated to dryness and purified by semi-preparative RP-HPLC (65% methanol) with subsequent analysis of the evaporated polar fractions by RP-HPLC-MS/MS (60% methanol).
For studies of oxidation of fatty acid by mycelia and release of intermediates in JA biosynthesis in small scale, 2.5 ml of the F. oxysporum liquid culture was centrifuged (3,100 g; 20 min, +4°C) and the supernatant was discarded. The mycelia were suspended in 2.5 ml 0.1 NaBO3 buffer (pH 8.2) or 2.5 ml 0.1 M KHPO4 buffer (pH 7.4)/2 mM EDTA/0.04% Tween-20 in 15 ml tubes at 22°C. The 18:3n-3 (2.3 mM) and 30, 100, 300, or 2,300 μM [17,17,18,18,18-2H5]18:3n-3 were added in some experiments, and 100 μM N-[9Z,12Z,15Z-octadecatrienoyl]-S-isoleucine (18:3n-3-Ile) or 100 μM 13S-HPOTrE-Ile in other experiments. The tubes were slowly tilted for 1 h (Duomax 1030; Heidolph; 22°C). The mycelia were then precipitated by centrifugation (3,100 g) and the supernatant was diluted with water and extracted with C18 silica (SepPak/C18). For large-scale experiments, we used the same protocol with mycelia from 25 ml of the liquid culture, 25 ml of suspension KHPO4 buffer in 50 ml Falcon tubes (1 h, 22°C), and several rounds of extraction on C18 silica (SepPak/C18). The extracts were reduced to dryness under a stream of N2 and dissolved in ethanol for analysis.
Enzyme assay
The N2 powder was added to 100 μM [13C18]18:2n-6 or 100 μM other C18 fatty acids in 0.1 M KHPO4 (pH 7.4), 2 mM EDTA, and 0.04% Tween-20. After homogenization, the crude homogenate was incubated for 30–40 min on ice. After centrifugation (17,000 g; 10 min, +4°C), we extracted the supernatant with C18 silica, as above.
GC- and LC-MS/MS analysis
An Agilent mass selective detector model 5977E connected to an Agilent model 7820A gas chromatograph were used for GC-MS. A capillary column of 5% phenylmethylsiloxane (12 m, 0.33 μm film thickness) with helium as the carrier gas was used. The temperature was raised from 80°C to 300°C at a rate or 10°C/min. The scan and selected ion monitoring modes were used for data acquisition. For quantitative determination of (−)-JA-Ile and (+)-7-iso-jasmonoyl-(S)-isoleucine [(+)-JA-Ile] in media from cultures of F. oxysporum, samples of medium were treated with 30 mM O-methyl hydroxylamine hydrochloride in methanol and 0.5 M sodium acetate (proportions, 1:1:10, v/v/v) at room temperature overnight. After addition of 2.2 μg of [2H3](−)-JA-Ile-MO, the mixture was extracted with ethyl acetate. The product was dissolved in CHCl3/2-propanol, 2:1 (v/v) and applied to an aminopropyl cartridge (Supelcoclean LC-NH2). The CHCl3/2-propanol eluate was discarded and the conjugates were obtained by elution with methanol/acetic acid, 98:2 (v/v). The product was treated with diazomethane and subjected to GC-MS using selected monitoring of the ions m/z 335 (unlabeled conjugates) and m/z 338 ([2H3] standard). The peaks of the methyl ester/MO derivatives of (−)-JA-Ile, (+)-JA-Ile, and [2H3](−)-JA-Ile were integrated and the amounts of the two unlabeled conjugates were calculated from the area ratios (following, in the case of (−)-JA-Ile, subtraction of the 0.78% of unlabeled material present in the [2H3]standard). The (+)- and (−)-JA were analyzed in an analogous way using [2H3](−)-JA-MO as an internal standard (M. Hamberg, unpublished observations).
RP-HPLC with MS/MS analysis was performed with a Surveyor MS pump (ThermoFisher) and an analytical or semi preparative octadecyl silica column (5 μm; 2.0 × 150 mm, Phenomenex; 5 μm; 4.6 × 150 mm, Dr. Maisch). The columns were eluted at 0.25–0.3 ml/min or 1 ml/min, respectively, with methanol/water/acetic acid, 600:400:0.05, 650:350:0.05, 700:300:0.05, 750:250:0.05, or 800:200:0.05 (v/v/v). The effluent was subject to ESI in a linear ion trap mass spectrometer (LTQ; ThermoFisher). The heated transfer capillary was set at 315°C, the ion isolation width at 1.5 amu (5 amu for analysis of hydroperoxides), the collision energy at 35 (arbitrary scale), and the tube lens at −112 V. Samples were injected manually (Rheodyne 7510) or by an autosampler (Surveyor Autosampler Plus; ThermoFisher).
Chiral phase (CP)-HPLC-MS/MS was performed with Reprosil Chiral AM (5 μ; 2 × 300 mm; Dr. Maisch), which was eluted (0.2 ml/min) with hexane/methanol/acetic acid, 95:5:0.02 (v/v/v). The eluate was mixed in-line with 2-propanol/water [3:2 (v/v); 0.15 ml/min] from a second pump (Constametric 3200, LDC/MiltonRoy). The combined effluents were introduced by ESI into the ion trap mass spectrometer above. Normal phase-HPLC-MS/MS was performed in the same way and the silica column (5 μm; 2 × 250 mm; Reprosil 100 SI, Dr. Maisch) was eluted with 2-propanol/hexane/acetic acid, 3:97:0.05 (v/v/v). Hydrogenation of JA-Ile was performed with H2 and catalytic Pd/C, and hydroperoxides were reduced to alcohols with triphenylphosphine.
RESULTS
Analysis of jasmonates in the growth medium
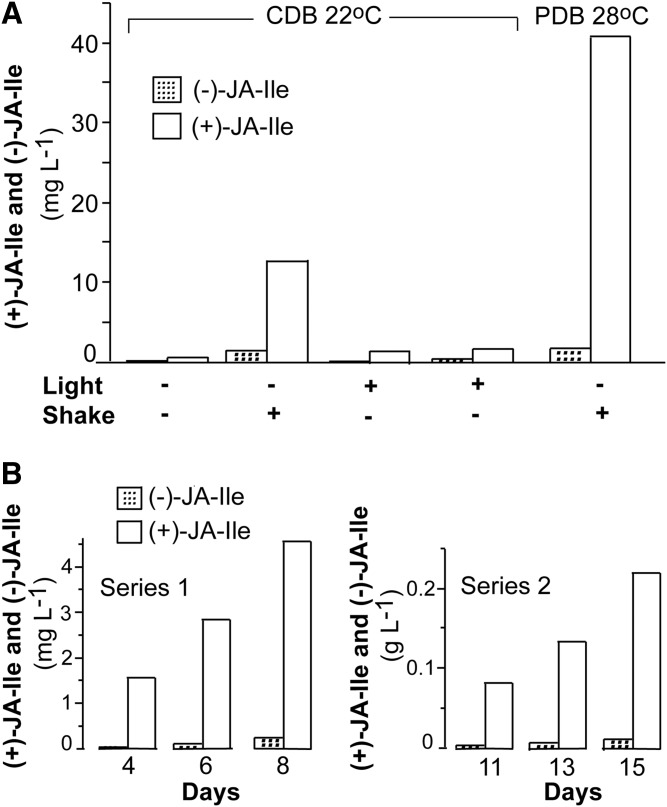
Cultures were inoculated from agar plates. We first examined the secretion of JA and JA-conjugates by cultures for 3 weeks (22°C) in flasks with CDB with and without shaking (100 rpm) and in the dark or under fluorescent light. Cultures in CDB, which were grown with shaking in the dark, released the largest amounts of (−)-JA-Ile and (+)-JA-Ile, as shown in Fig. 3A. Cultures grown in PDB in the dark (or in subdued light) at 28°C with shaking (100 rpm) released the highest amounts of jasmonates after 3 weeks (Fig. 3A). The amount of (+)- and (−)-JA was only 0.2% of the amount of (+)-JA-Ile in this sample, which is lower than in other strains of F. oxysporum (15, 20). Growth in PDB with shaking at 28°C and in the dark yielded the highest levels and these conditions were, therefore, used routinely.
Fig. 3.
Formation of JA-Ile conjugates after 3 weeks under different conditions and two time curves. A: Different growth conditions in CDB and a comparison with PDB. Light reduced biosynthesis and it was augmented in shaking flasks (100 rpm). Changing from CDB to PDB and increasing the temperature to 28°C yielded the largest amounts of jasmonates and this growth condition was used routinely. B: Partial time curves for biosynthesis of (−)-JA-Ile and (+)-JA-Ile by two different cultures in PDB, which were inoculated from liquid cultures in PDB. Left: The biosynthesis of (+)-JA-Ile and (−)-JA-Ile reached 4.5 and 0.25 mg l−1, respectively, after 8 days. Right: The biosynthesis of (+)-JA-Ile and (−)-JA-Ile reached over 0.22 and 0.01 g l−1, respectively, after 15 days.
GC-MS analysis showed that the secretion of jasmonates to the PDB was low for the first 4 days, but caught momentum after about a week and reached about 4 mg l−1 (+)-JA-Ile (Fig. 3B). The yield varied between incubations (15). Inoculation of PDB with an aliquot of a liquid culture led to accumulation of over 0.23 g l−1 (+)-JA-Ile after 15 days (Fig. 3C). The largest amounts obtained were 0.31 g l−1 (+)-JA-Ile (almost 1 mM).
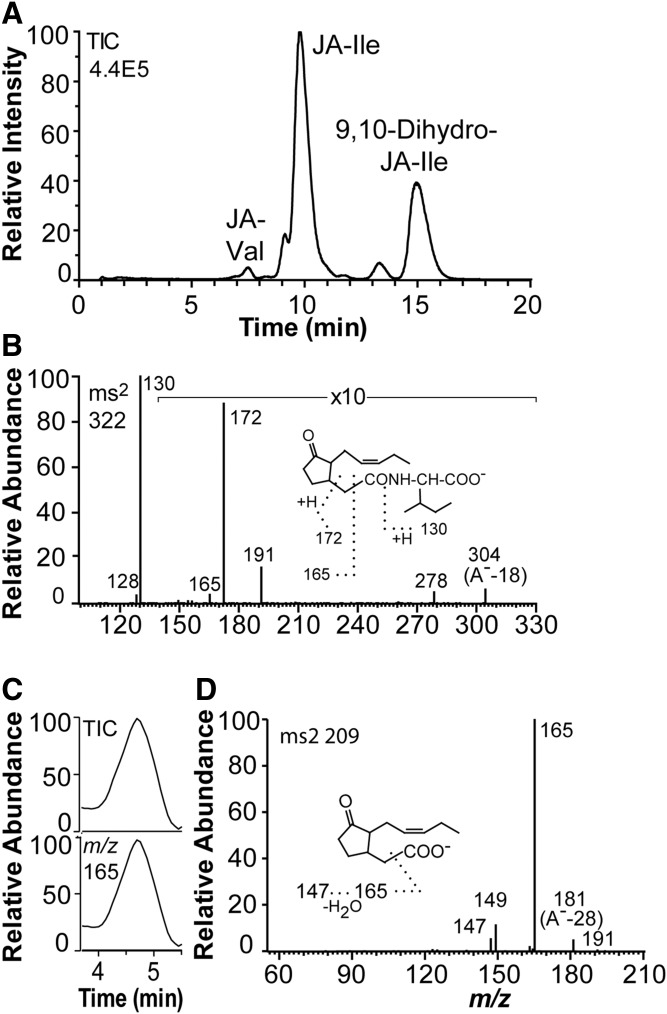
The elution of jasmonates on RP-HPLC is shown in Fig. 4A. JA-Ile was the main product along with variable relative amounts of 9,10-dihydro-JA-Ile. In addition, small amounts of Ile-Val and 9,10-dihydro-JA-Val were also detected. The MS2 spectrum of JA-Ile is shown in Fig. 4B, and dominated by the signal at m/z 130 (Ile carboxylate anion; compare inset in Fig. 4B). The MS2 spectra of 9,10-dihydro-JA-Ile, JA-Val, and 9,10-dihydro-JA-Val showed a similar fragmentation pattern (supplemental Fig. S1).
Fig. 4.
LC-MS/MS analysis of JA-Ile, 9,10-dihydro-JA-Ile, and JA in the growth medium. A: LC-MS/MS analysis of JA and 9,10-dihydro-JA conjugates with Ile and Val (total ion current). The column was eluted with methanol/water/acetic acid, 650:350:0.5 (0.25 ml/min). Small amounts of JA-Val were also detected. B: MS2 spectrum of JA-Ile. Major fragment ions are indicated by the inset. C: LC-MS/MS analysis of JA (partial chromatograms). Top: Total ion current. Bottom: Single ion monitoring of m/z 165. D: MS2 spectrum of JA. Authentic (−)-JA and (+)-JA showed identical mass spectra.
JA was detectable by LC-MS/MS after two rounds of RP-HPLC purification (semi-preparative RP-HPLC, 65% methanol; RP-HPLC-MS/MS, 60% methanol) (Fig. 4C). The MS2 spectrum of JA (Fig. 4D) lacked a signal at m/z 59, which is abundant in the mass spectra of JA recorded with triple quadrupole instruments (15, 18).
Oxidation of C18 fatty acids by N2 powder of mycelia
We first evaluated mycelia grown in CDB. N2 powder, prepared from nonshaking cultures under light or dark conditions for 3 weeks (22°C) did not oxidize [13C18]18:2n-6, whereas significant oxidation to [13C18]10-HPODE was detected in N2 powder of mycelia from shaking dark cultures at 100 rpm (22°C). This oxidation appeared to be further increased in N2 powder of mycelia grown with shaking at 28°C (dark). The highest activities were noted of N2 powder of mycelia, which was grown under these conditions in PDB. The PDB medium also contained the largest amounts of jasmonates, as discussed above (Fig. 3). We conclude that large secretion of jasmonates occurred by mycelia, which also oxidized 18:2n-6 to 10-HPODE. Mycelia from these cultures in PDB were intensely dark red due to bikaverin expression [confirmed by CHCl3 extraction and UV analysis, compare (32)].
The N2 powder oxidized [13C18]18:2n-6 to [13C18]10-HPODE and [13C18]10-HODE as main products (Fig. 5A). Relatively small amounts of [13C18]8-HPODE were also detected. In addition, 10-HODE was formed from endogenous 18:2n-6 (Fig. 5A). We expected to detect signs of 13S-LOX or 9-DOX activities in the N2 powder, but neither [13C18]9-HPODE nor [13C18]13-HPODE could be detected.
Fig. 5.
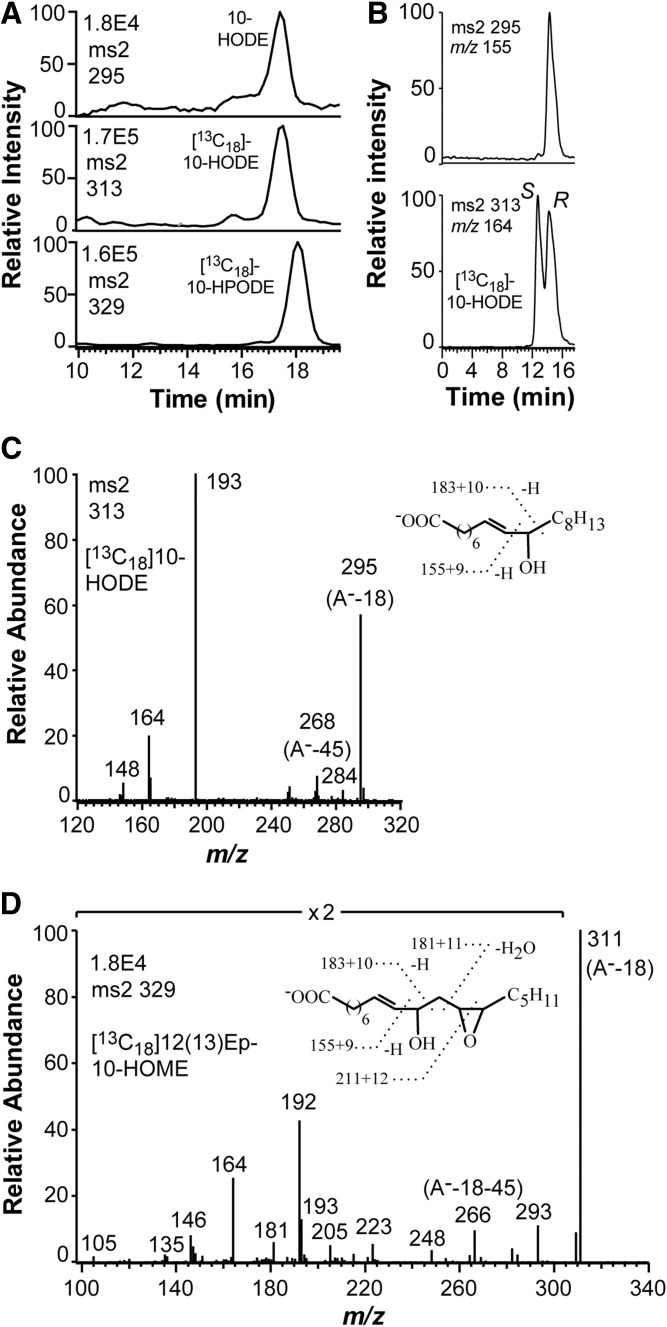
LC-MS/MS analysis of HPODE formed from endogenous and [13C18]18:2n-6 by nitrogen powder of F. oxysporum. A: Chromatograms from LC-MS/MS analysis of HODE (top), [13C18]HODE (middle), and [13C18]HPODE (bottom). The 10-HODE, [13C18]10-HODE, and [13C18]10-HPODE could be detected along with small amounts of 8-HPODE. B: Steric analysis of 10-HODE formed from 18:2n-6 by the N2 powder. The [13C18]10R/S-HODE was added, and the 10-HODE (top) eluted at the same time as the R stereoisomer (bottom). The elution order on this chiral column was established with10R-HODE formed by recombinant 10R-DOX-EAS (25). C: MS2 spectrum of [13C18]10-HODE formed by N2 powder. The insert and the labels describe the corresponding ions in this and the unlabeled spectrum. D: MS2 spectrum of [13C18]12(13)Ep-10-HOME. The insert illustrates the ions of the labeled and unlabeled compound.
Steric analysis of 10-HODE showed that it consisted of the 10R stereoisomer (90%; Fig. 5B). The mass spectrum of [13C18]10-HODE is shown in Fig. 5C; the insert compares the mass spectrum with that of unlabeled 10-HODE. A polar metabolite, which was formed from [13C18]18:2n-6, was identified as [13C18]12(13)epoxy-10-hydroxy-8E-octadecenoic acid [12(13)Ep-10-HOME] by the mass spectra illustrated in Fig. 5D; the insert compares the spectra of the [13C18]12(13)Ep-10-HOME and 12(13)Ep-10-HOME.
The 18:3n-3 was oxidized in analogy with 18:2n-6 to 10-HPOTrE and 12(13)epoxy-10-hydroxy-8E,15Z-octadecadienoic acid [12(13)Ep-10-HODE], but 18:1n-9 was mainly oxidized to 8-HOME (data not shown). The oxidation of 18:2n-6 was consistent with expression of 10R-DOX-EAS of F. oxysporum (25), but the transformation of 18:1n-9 and 18:3n-3 by this enzyme has not been established.
Recombinant 10R-DOX-EAS oxidized 18:1n-9 to 8-H(P)OME and to small amounts of 10-H(P)OME (supplemental Fig. S2A). The 18:3n-3 was oxidized in analogy with 18:2n-6 to 10-HPOTrE and 12(13)Ep-10-HODE (supplemental Fig. S2B, C). In addition, 8- and 16-hydroxyoctadecatrienoic acids (HOTrEs) were also detected. Steric analysis by CP-HPLC-MS/MS showed that the latter was racemic (data not shown). We conclude that the transformations of C18 fatty acids by N2 powder of mycelia and by recombinant 10R-DOX-EAS are strikingly similar.
Metabolism of 18:3n-3, [2H5]18:3n-3, and [2H5]13S-HPOTrE by mycelia in buffer
Mycelia, in 2.5 ml of suspended cultures, were harvested by centrifugation and resuspended (2.5 ml) in 0.1 M NaBO3 (pH 8.2) or 0.1 M KHPO4 (pH 7.4)/2 mM EDTA/0.02% Tween-20 with or without added 18:3n-3 (2.3 mM) or [2H5]18:3n-3 (from 30 μM to 2.3 mM).
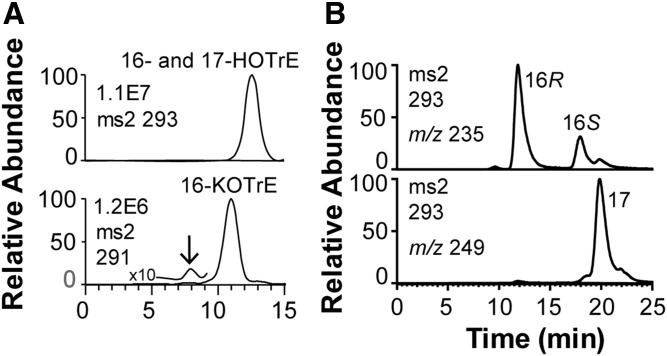
The 2.3 mM 18:3n-3 was extensively metabolized in 1 h to 16-HOTrE, 16-keto-9Z,12Z,13E-octadecatrienoic acid (KOTrE), 16-HPOTrE, and 17-KOTrE and to the ω3 epoxide of 18:3n-3 in both buffers within 1 h. The elution of 16- and 17-HOTrE and 16-KOTrE is shown in Fig. 6A. The 16- and 17-KOTrE were separated on Reprosil-Chiral AM, as illustrated by the ion chromatograms for 16-HOTrE (m/z 235; A−-58; loss of HCO-CH2-CH3) and 17-HOTrE (m/z 249; A−-44; loss of HCO-CH3 or CO2) in Fig. 6B. R stereoisomers of cis-trans conjugated hydroxy fatty acids elute before S stereoisomers on columns with amylose tris-3,5-dimethylphenylcarbamate modified silica (33). The 16R-HOTrE thus appeared to be the main stereoisomer. The MS2 spectrum of 16-KOTrE (supplemental Fig. S3) and the MS3 spectrum of 16-HPOTrE (data not shown) appeared to be identical. In addition, a series of polar products were formed, including tentative epoxy alcohols formed from 16-HPOTrE. Incubation with 2.3 mM 18:3n-3 overnight yielded only traces of ω2 and ω3 hydroxy metabolites. The prominent ω3 and ω2 oxidations are presumably due the catalytic activities of P450foxy (34).
Fig. 6.
Oxidation of 18:3n-3 by mycelia of Fot during short-time incubation in 0.1 M NaBO3 (pH 8.2). A: MS/MS analysis of products. Top: MS/MS analysis of HOTrE (m/z 293). Bottom: MS/MS analysis of KOTrE (m/z 291). The insets mark the elution of the identified products, 16- and 17-HOTrE, and 16-KOTrE, respectively. Small amounts of 12-OPDA (marked by arrow) eluted 3 min before 16-KOTrE. B: Chiral HPLC-MS/MS analysis (m/z 293 → full scan) of 16-HOTrE [m/z 235 (A−-58); top] and separation of 17-HOTrE [m/z 249 (A−-44); bottom].
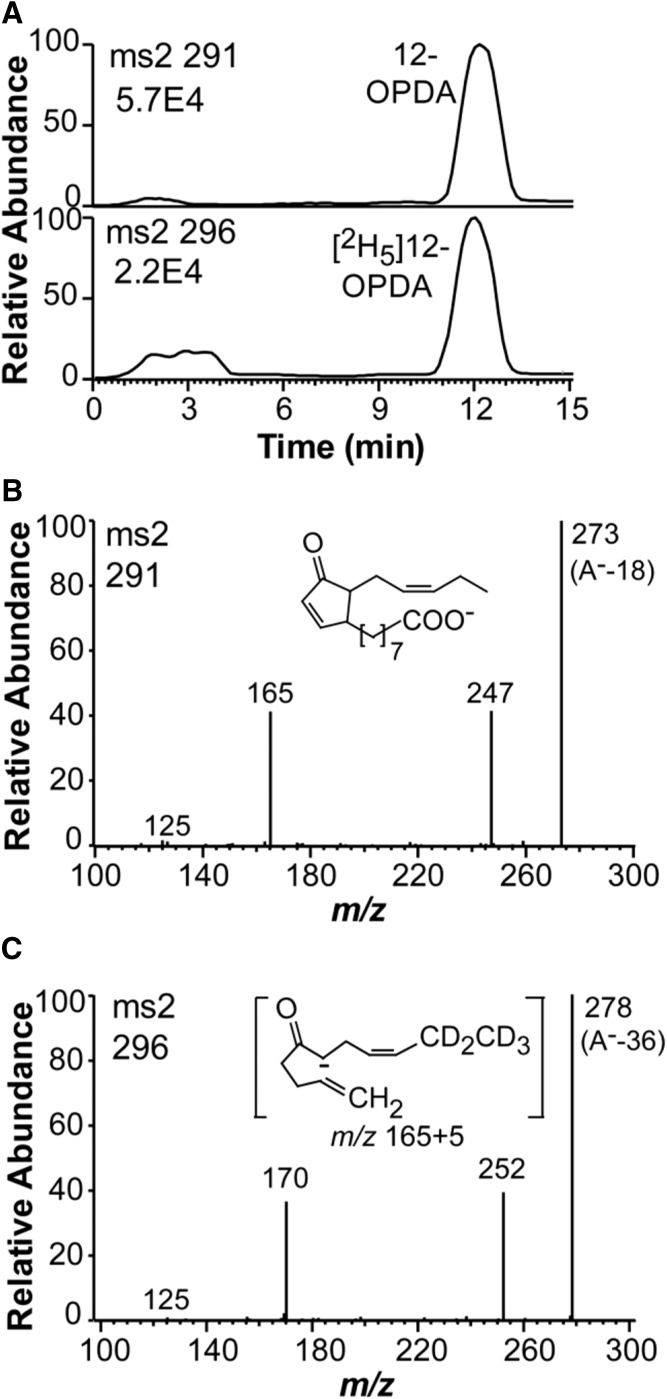
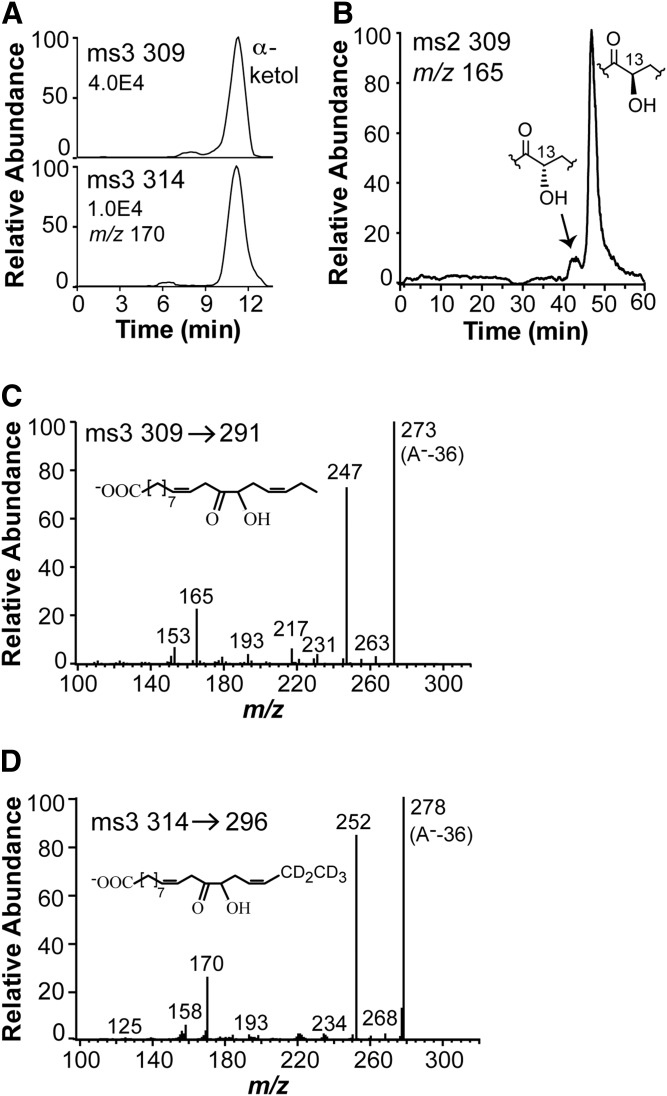
A minor metabolite eluted before 16-KOTrE (marked by arrow in the chromatogram, Fig. 6A). The MS2 spectrum and the retention time of this product were both identical to that of authentic 12-OPDA. We next examined its formation with [2H5]18:3n-3 as a substrate.
Incubation of mycelia with 300 μM [2H5]18:3n-3 in the KHPO4 buffer yielded [2H5]10-OPDA and 12-OPDA in a ratio of 0.4:1 (Fig. 7A), but only small amounts were formed (about 10 and 25 ng, respectively). Incubation with 100 μM [2H5]18:3n-3 led to almost the same ratio, but it was lower with 30 μM [2H5]18:3n-3 (0.1:1). This clearly showed that [2H5]12-OPDA was formed from [2H5]18:3n-3 in a concentration-dependent manner. We repeated the experiments with mycelia from a young culture and obtained the [2H5]12-OPDA and 12-OPDA in a ratio of 47:1 (peak heights 7.56E5 and 1.6E4, respectively).
Fig. 7.
LC-MS/MS analysis of the transformation of 300 μM [2H5]18:3n-3 by mycelium of Fot for 1 h. A: The mycelium released endogenous 12-OPDA and newly formed [2H5]12-OPDA to the KHPO4 buffer, as shown by the top and bottom chromatograms, respectively. B: LC-MS/MS analysis of 12-OPDA. C: LC-MS/MS analysis of [2H5]12-OPDA. The inset in (C) shows the formation of the fragmentation ion at m/z 165 and 170, respectively, in (A, C) (35).
The mass spectra of 12-OPDA and [2H5]12-OPDA are shown in Fig. 7B, C. The signal at m/z 165 in the spectrum of 12-OPDA is due to charge-directed fragmentation by the carboxyl group and the fragment is illustrated in the insert in Fig. 7C (35).
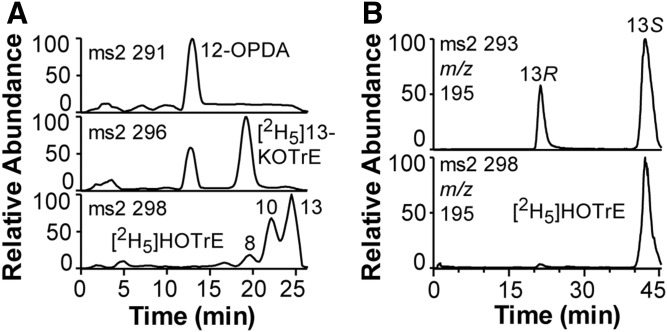
The transformation of endogenous 18:3n-3 and 100 μM [2H5]18:3n-3 by mycelia appeared to differ. The latter was transformed to [2H5]12-OPDA and also to [2H5]13-KOTrE and [2H5]-labeled 8-, 10-, and 13-HOTrE (Fig. 8A). The [2H5]11-HOTrE could not be detected, which is a major product of Fo-MnLOX (22). Steric analysis by CP-HPLC showed that over 95% of [2H5]13-HOTrE coeluted with 13S-HOTrE (Fig. 8B).
Fig. 8.
Mycelia transform endogenous and [2H5]18:3n-3 to partly different products. A: RP-HPLC-MS analysis of products. The top chromatogram shows that endogenous 18:3n-3 is transformed to 12-OPDA, but not to significant amounts of 13-KOTrE. The middle and bottom chromatograms show that [2H5]18:3n-3 is transformed to [2H5]12-OPDA and also to [2H5]13-KOTrE and [2H5]-labeled 8-, 10-, and 13-HOTrE. B: Steric analysis by CP-HPLC-MS/MS of [2H5]13-HOTrE (bottom) showed that over 95% co-chromatographed with 13S-HOTrE (top chromatogram).
Products formed in a large-scale incubation of mycelia with the KHPO4 buffer were extracted and purified by semi-preparative RP-HPLC, and 10 μl of 1 ml fractions were analyzed by direct injection into the mass spectrometer. The fraction with 12-OPDA was reduced to dryness and then analyzed by GC-MS, which separated the cis and trans side chain isomers of 12-OPDA (supplemental Fig. S4A). The electron impact mass spectrum is shown in supplemental Fig. S4B. We conclude that 12-OPDA was identified by both LC- and GC-MS analyses.
The α-ketol was detected when mycelia were incubated with buffer or buffer with [2H5]18:3n-3 (Fig. 9A). It is well-known that the allene oxide [12(13S)epoxy-9Z,11,15Z-octadecatrienoic acid (12,13S-EOT)], which is formed from 13S-HPOTrE, is hydrolyzed to the α-ketol by inversion of configuration at C-13 (36). Steric analysis showed that the fungal α-ketol consisted mainly of the 13R stereoisomer (90%; Fig. 9B), and thus likely originated from 13S-HPOTrE via 12,13S-EOT.
Fig. 9.
Metabolism of 18:3n-3 by the mycelium of Fot. A: RP-HPLC-MS/MS analysis of the α-ketol, 12-oxo-13R-hydroxy-9Z,15Z-octadecadienoic acid, which is released in labeled and unlabeled form by the mycelium after incubation with 300 μM [2H5]18:3n-3. B: CP-HPLC-MS/MS analysis of the unlabeled α-ketol showing a large excess of the 13R enantiomer. C: MS3 spectrum of the unlabeled α-ketol. D: MS3 spectrum of the deuterated α-ketol.
The MS2 spectrum of the α-ketol was noncharacteristic with weak signals except for the trivial signals at m/z 291 (loss of water), 273 (291-18), and 247 (291-44; loss of CO2) (supplemental Fig. S5). The MS3 spectrum of the α-ketol was more characteristic with strong signals at m/z 165 and 153 (Fig. 9B). These mass spectra were identical with those of the authentic standard. We next purified the [2H5]-labeled α-ketol by normal phase-HPLC. The MS3 spectrum of [2H5]α-ketol showed signals, among other things, at m/z 158 (153+5) and 170 (165+5) (Fig. 9C). We also prepared the [2H5]-labeled α-ketol and [2H5]12-OPDA from [2H5]13S-HPOTrE using AOS in acetone powder of flaxseed (37), and their MS2 and MS3 spectra were identical with the fungal metabolites.
We expected exogenous 13S-HPOTrE to be transformed to 12-OPDA. Incubation of mycelia with 100 μM [2H5]13S-HPOTrE for 1 h did not lead to significant amounts of [2H5]12-OPDA, but [2H5]13-KOTrE was formed. This is illustrated in supplemental Fig. S6A. The ratio of signal intensity of m/z 170 (possibly due to [2H5]12-OPDA) and the signal intensity of m/z 165 (12-OPDA) was only 0.002:1 (supplemental Fig. S6B). We repeated the experiments with 10 times higher concentration of [2H5]13S-HPOTrE. The [2H5]13S-HPOTrE (1 mM) was transformed to [2H5]12-OPDA and the signal ratio [m/z 170/(m/z 165)] increased to 0.035:1 (supplemental Fig. S6C). In spite of the low conversion, we obtained the MS2 spectrum of [2H5]12-OPDA (supplemental Fig. S6D; compare Fig. 7C). We conclude that mycelia of Fot form 12-OPDA by the same pathway as in plants.
Metabolism of 18:3n-3-Ile by mycelia of Fot
Standards were prepared with flaxseed AOS. The 13S-HPOTrE-Ile was obtained by oxidation with sLOX-1 and further transformed by acetone powder of flaxseed (AOS) to two major products, identified as N-[12-oxo-13-hydroxy-9Z,15Z-octadecadienoyl]-S-isoleucine (α-ketol-Ile; 90%) and 12-OPDA-Ile (10%) (supplemental Fig. S7A). The MS2 spectra of 12-OPDA-Ile and the MS3 spectrum of the α-ketol-Ile are shown in supplemental Fig. S7B, C. The spectrum of the former was identical to the reported MS2 spectrum of 12-OPDA-Ile (38). Small amounts of 13-HOTrE-Ile were also detected. In contrast, mycelia of Fot oxidized 100 μM 18:3n-3-Ile to three major products, likely formed by the prominent subterminal ω hydroxylase activities. The 12-OPDA-Ile or α-ketol-Ile could not be detected.
Mycelia of Fot did not release detectable amounts of 12-OPDA-Ile or the α-ketol-Ile conjugate. We conclude that conjugation of (+)-JA with Ile likely occurs as a final step.
DISCUSSION
We report a prominent biosynthesis of the JA conjugate (+)-JA-Ile by the fungus Fot, about 0.2 g l−1. This is of the same magnitude as that reported for the biosynthesis of (+)-JA by L. theobromae, which commonly produces 0.3–0.5 g l−1 (17, 19). The prominent (+)-JA biosynthesis by Fot enabled our main finding, the detection of two key intermediates in plant biosynthesis of (+)-JA. The [2H5]18:3n-3 was converted by mycelia of Fot to [2H5]12-OPDA and to an allene oxide-derived α-ketol, [2H5]12-oxo-13R-hydroxy-9Z,15Z-octadecadienoic acid. In addition, 13S-HOTrE and 13-KOTrE could be detected, and [2H5]13S-HPOTrE was also converted to 12-OPDA, albeit in low yields. This suggests that the initial steps of the fungal biosynthetic pathway to jasmonates are analogous to those operating in plants (Fig. 10).
Fig. 10.
Proposed pathway for the formation in F. oxysporum of 12-OPDA and its further conversion to (+)-JA-Ile. Involvement of F. oxysporum iron 13S-LOX (26) in the initial step was deduced from the parallel formations of 13S-HOTrE, 13-KOTrE, and a 13R-configured α-ketol. The nature of the enzyme activities needed for allene oxide formation and cyclization is unknown.
The first step is likely oxidation of 18:3n-3 to 13S-HPOTrE. As far as is known, the fungal DOXs of the cyclooxygenase gene family do not oxidize 18:3n-3 at C-13, but there are two possible LOX candidates (Fig. 2). These LOXs have only been studied by recombinant expression and expressed sequence tags have not yet been detected in F. oxysporum.2 Fo-MnLOX is likely secreted and forms 11-HPOTrE as a major metabolite (22). The latter could not be detected. The 13S-LOX with catalytic iron is therefore the obvious candidate (26).
We have little information on the other fungal enzymes leading to biosynthesis of (+)-JA-Ile. AOS, AOC, 12-OPDA reductase (OPR3), and JAR1 proteins of Arabidopsis thaliana are well-characterized (39–42). AOS can be aligned with putative CYP of F. oxysporum at the C-terminal end with 27% identity, and OPR3 with NADPH-dependent reductases with 46% identity.3 We could not detect homologs of AOC or JAR1 in the genome of F. oxysporum.4 Both plant enzymes lack catalytic metals. The active site of JAR1 relies on an adaptable three-dimensional scaffold (8) and the crystal structure of AOC indicates an active site inside a barrel cavity (10). It seems likely that fungal AOC and acyl amino acid synthase could be examples of parallel evolution of active sites.
α-Ketols are formed by nonenzymatic hydrolysis of allene oxides taking place with predominant inversion of configuration (43). The α-ketol of the present work was mainly 13R, suggesting its formation from 12,13S-EOT. The latter is likely formed from 13S-HPOTrE by a 13S-AOS. AOSs belong to two families of heme proteins, CYP of plants and fungi and catalases of corals and cyanobacteria (36, 44, 45). Fungal AOSs have so far only been detected in DOX-CYP fusion proteins, e.g., 8R-, 8S-, 9R-, and 9S-DOX-AOS and not in fungal catalases (24, 46–48). In contrast to plant CYP74, which contains a characteristic insertion sequence of seven to nine amino acids in the Cys pocket, the fungal AOSs of DOX-AOS enzymes do not share common characteristic sequences (9, 47, 49). It will be a challenging task to identify the putative 13S-AOS. The efficient synthesis of jasmonates in fungi suggests a tight coupling between the AOS and cyclase activities and even the possibility that they may reside in the same protein (compare Ref. 50).
The 12-OPDA is formed in plants by AOS and AOC in plastids. The location of the fungal AOS and AOC is unknown. Eukaryotic CYP enzymes are typically membrane-bound and found in the endoplasmic reticulum. β-Oxidation in fungi only occurs in peroxisomes (51). In analogy with plants, 12-OPDA could be transferred to peroxisomes where the ring double bond could be reduced and the side chain shortened by β-oxidation.
We expected N2 powder of mycelia to oxidize 18:3n-3 to 13S-HPOTrE, but this was not detected with certainty. This may explain why the fungal biosynthesis of JA has been enigmatic. Biosynthesis of (+)-JA-Ile in shaking cultures in PDB was associated with prominent expression of 10R-DOX-EAS and bikaverin, which colored the mycelium dark red. Bikaverin biosynthesis in Fusarium and JA biosynthesis by L. theobromae start when glucose and/or nitrogen have been partly consumed and can be optimized in different ways (17, 18). It is not unlikely that the biosynthesis of jasmonates by Fot also can be optimized.
How can the enzymes in the fungal cascade to (+)-JA-Ile be identified? The classical method to purify enzymes from cell-free preparations does not appear to be feasible due to undetectable enzyme activities. This is in striking similarity to the undetectable prostaglandin biosynthesis by subcellular fractions of the coral, Plexaura homomalla (52). This enigma was solved by recombinant expression of the coral cyclooxygenase (52). The 13S-LOX of F. oxysporum has also been expressed (26) and we detected biosynthesis of 13S-HOTrE by the mycelium. The enzymes further down the biosynthetic pathway to (+)-JA-Ile are unknown. It is possible that comparison of mRNA expression under growth conditions with little and augmented JA biosynthesis might generate hypotheses for subsequent recombinant enzyme expression and analysis.
Access to strains of L. theobromae with a high capacity to form (+)-JA is restricted by environmental and commercial considerations. In contrast, Fot is generally available for future studies from the ARS culture collection. Our study raises many questions, which could be investigated in mycelia of Fot, e.g., the effect of gene deletion of 13S-LOX, the subcellular location of the biosynthesis of 12-OPDA, the reduction of the ring double bond, and conditions for optimal biosynthesis of (+)-JA-Ile in liquid cultures.
CONCLUSIONS
We have identified 12-OPDA as an intermediate, which was formed from 18:3n-3 and 13S-HPOTrE, in the biosynthesis of jasmonates in F. oxysporum. An α-ketol was also detected, which provides evidence for an allene oxide serving as the immediate precursor of 12-OPDA. It seems likely that further conversions of 12-OPDA take place as established for higher plants, i.e., by reduction of the ring double bond, β-oxidation, and conjugation with Ile.
Supplementary Material
Footnotes
Abbreviations:
- AOC
- allene oxide cyclase
- AOS
- allene oxide synthase
- α-ketol
- 12-oxo-13-hydroxy-9Z,15Z-octadecadienoic acid
- α-ketol-Ile
- N-[12-oxo-13-hydroxy-9Z,15Z-octadecadienoyl]-S-isoleucine
- CDB
- Czapec-Dox broth
- CP
- chiral phase
- CYP
- cytochrome P450
- DOX
- dioxygenase
- EAS
- epoxy alcohol synthase
- 12
- 13S-EOT, 12(13S)epoxy-9Z,11,15Z-octadecatrienoic acid
- Fo-MnLOX
- lipoxygenase containing catalytic manganese
- Fot
- F. oxysporum f. sp. tulipae
- FoxLOX
- lipoxygenase containing catalytic iron
- HOTrE
- hydroxyoctadecatrienoic acid
- HPODE
- hydroperoxy-octadecadienoic acid
- HPOTrE
- hydroperoxyoctadecatrienoic acid
- JA
- jasmonic acid
- (+)-JA
- (+)-7-iso-jasmonic acid
- (−)-JA
- (−)-jasmonic acid
- (+)-JA-Ile
- (+)-7-iso-jasmonoyl-(S)-isoleucine
- KOTrE
- keto-octadecatrienoic acid
- LOX
- lipoxygenase
- MO
- O-methyloxime
- 18:3n-3-Ile
- N-[9Z,12Z,15Z-octadecatrienoyl]-S-isoleucine
- 12-OPDA
- 12-oxo-10,15Z-phytodienoic acid
- PDA
- potato dextrose agar
- PDB
- potato dextrose broth
- RP
- reversed phase
This work was supported by Vetenskapsrådet Grant K2013-67X-06523-31-3, Knut and Alice Wallenberg Foundation Grant KAW 2004.0123, and Uppsala University.
The online version of this article (available at http://www.jlr.org) contains a supplement.
The dbEST of NCBI (https://www.ncbi.nlm.nih.gov).
BLAST analysis at NCBI showed that AOS (GenBank AAF00225) and 12-OPDA reductase (OPR3; GenBank OAP09565) of A. thaliana could be aligned with putative CYP (27% identity; 24% query cover) and with a putative NADPH-dependent dehydrogenase (45% identity; 97% query cover) of F. oxyporum Fo47.
BLAST analysis at NCBI showed no significant homology between proteins of F. oxysporum and AOC (GenBank CAC83764) and JAR1 (GenBank OAP073631) of A. thaliana.
REFERENCES
- 1.Ma L. J., Geiser D. M., Proctor R. H., Rooney A. P., O’Donnell K., Trail F., Gardiner D. M., Manners J. M., and Kazan K.. 2013. Fusarium pathogenomics. Annu. Rev. Microbiol. 67: 399–416. [DOI] [PubMed] [Google Scholar]
- 2.Michielse C. B., and Rep M.. 2009. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10: 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ploetz R. C. 2015. Fusarium wilt of banana. Phytopathology. 105: 1512–1521. [DOI] [PubMed] [Google Scholar]
- 4.Demole E., Lederer E., and Mercier D.. 1962. Isolement et determination de la structure du jasmonate de methyle, constituant odorant caracteristique de l’essence de jasmin. Helv. Chim. Acta. 45: 675–685. [Google Scholar]
- 5.Nishida R., and Acree T. E.. 1984. Isolation and characterization of methyl epijasmonate from lemon (Citrus limon Burm.). J. Agric. Food Chem. 32: 1001–1003. [Google Scholar]
- 6.Miersch O., Preiss A., Sembdner G., and Schreiber K.. 1987. (+)-7-Isojasmonic acid and related compounds from Botryodiplodia theobromae. Phytochemistry. 26: 1037–1039. [Google Scholar]
- 7.Miersch O., Brückner B., Schmidt J., and Sembdner G.. 1992. Cyclopentane fatty acids from Gibberella fujikuroi. Phytochemistry. 31: 3835–3837. [Google Scholar]
- 8.Westfall C. S., Zubieta C., Herrmann J., Kapp U., Nanao M. H., and Jez J. M.. 2012. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science. 336: 1708–1711. [DOI] [PubMed] [Google Scholar]
- 9.Lee D. S., Nioche P., Hamberg M., and Raman C. S.. 2008. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature. 455: 363–368. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann E., and Pollmann S.. 2008. Molecular mechanism of enzymatic allene oxide cyclization in plants. Plant Physiol. Biochem. 46: 302–308. [DOI] [PubMed] [Google Scholar]
- 11.Wasternack C., and Kombrink E.. 2010. Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 5: 63–77. [DOI] [PubMed] [Google Scholar]
- 12.Wasternack C., and Strnad M.. 2016. Jasmonate signaling in plant stress responses and development - active and inactive compounds. N. Biotechnol. 33: 604–613. [DOI] [PubMed] [Google Scholar]
- 13.Schaller A., and Stintzi A.. 2009. Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochemistry. 70: 1532–1538. [DOI] [PubMed] [Google Scholar]
- 14.Gimenez-Ibanez S., Chini A., and Solano R.. 2016. How microbes twist jasmonate signaling around their little fingers. Plants (Basel). 5: E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole S. J., Yoon A. J., Faull K. F., and Diener A. C.. 2014. Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Mol. Plant Pathol. 15: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skrzypek E., Miyamoto K., Saniewski M., and Ueda J.. 2005. Identification of jasmonic acid and its methyl ester as gum-inducing factors in tulips. J. Plant Res. 118: 27–30. [DOI] [PubMed] [Google Scholar]
- 17.Dhandhukia P. C., and Thakkar V. R.. 2008. Response surface methodology to optimize the nutritional parameters for enhanced production of jasmonic acid by Lasiodiplodia theobromae. J. Appl. Microbiol. 105: 636–643. [DOI] [PubMed] [Google Scholar]
- 18.Eng F., Haroth S., Feussner K., Meldau D., Rekhter D., Ischebeck T., Brodhun F., and Feussner I.. 2016. Optimized jasmonic acid production by Lasiodiplodia theobromae reveals formation of valuable plant secondary metabolites. PLoS One. 11: e0167627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng F., Gutiérrez-Rojas M., and Favela-Torres E.. 1998. Culture conditions for jasmonic acid and biomass production by Botryodiplodia theobromae in submerged fermentation. Process Biochem. 33: 715–720. [Google Scholar]
- 20.Miersch O., Bohlmann H., and Wasternack C.. 1999. Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry. 50: 517–523. [Google Scholar]
- 21.Tsukada K., Takahashi K., and Nabeta K.. 2010. Biosynthesis of jasmonic acid in a plant pathogenic fungus, Lasiodiplodia theobromae. Phytochemistry. 71: 2019–2023. [DOI] [PubMed] [Google Scholar]
- 22.Wennman A., Magnuson A., Hamberg M., and Oliw E. H.. 2015. Manganese lipoxygenase of Fusarium oxysporum and the structural basis for biosynthesis of distinct 11-hydroperoxy stereoisomers. J. Lipid Res. 56: 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sooman L., and Oliw E. H.. 2015. Discovery of a novel linoleate dioxygenase of Fusarium oxysporum and linoleate diol synthase of Colletotrichum graminicola. Lipids. 50: 1243–1252. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann I., and Oliw E. H.. 2013. Discovery of a linoleate 9S-dioxygenase and an allene oxide synthase in a fusion protein of Fusarium oxysporum. J. Lipid Res. 54: 3471–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann I., Jerneren F., and Oliw E. H.. 2014. Epoxy alcohol synthase of the rice blast fungus represents a novel subfamily of dioxygenase-cytochrome P450 fusion enzymes. J. Lipid Res. 55: 2113–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodhun F., Cristobal-Sarramian A., Zabel S., Newie J., Hamberg M., and Feussner I.. 2013. An iron 13S-lipoxygenase with an alpha-linolenic acid specific hydroperoxidase activity from Fusarium oxysporum. PLoS One. 8: e64919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Jernerén F., and Oliw E. H.. Purification and site-directed mutagenesis of linoleate 9S-dioxygenase-allene oxide synthase of Fusarium oxysporum confirms the oxygenation mechanism. Arch. Biochem. Biophys. Epub ahead of print. May 11, 2017; doi:10.1016/j.abb.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., and Solano R.. 2009. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- 29.Koch T., Krumm T., Jung V., Engelberth J., and Boland W.. 1999. Differential induction of plant volatile biosynthesis in the lima bean by early and late intermediates of the octadecanoid-signaling pathway. Plant Physiol. 121: 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monte I., Hamberg M., Chini A., Gimenez-Ibanez S., Garcia-Casado G., Porzel A., Pazos F., Boter M., and Solano R.. 2014. Rational design of a ligand-based antagonist of jasmonate perception. Nat. Chem. Biol. 10: 671–676. [DOI] [PubMed] [Google Scholar]
- 31.Powell W. S. 1980. Rapid extraction of oxygenated metabolites of arachidonic acid from biological samples using octadecylsilyl silica. Prostaglandins. 20: 947–957. [DOI] [PubMed] [Google Scholar]
- 32.Balan J., Fuska J., Kuhr I., and Kuhrova V.. 1970. Bikaverin, an antibiotic from Gibberella fujikoi, effective against Leishmania brasiliensis. Folia Microbiol. (Praha). 15: 479–484. [DOI] [PubMed] [Google Scholar]
- 33.Schneider C., Yu Z., Boeglin W. E., Zheng Y., and Brash A. R.. 2007. Enantiomeric separation of hydroxy and hydroperoxy eicosanoids by chiral column chromatography. Methods Enzymol. 433: 145–157. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama N., Takemae A., and Shoun H.. 1996. Cytochrome P450foxy, a catalytically self-sufficient fatty acid hydroxylase of the fungus Fusarium oxysporum. J. Biochem. 119: 435–440. [DOI] [PubMed] [Google Scholar]
- 35.Bao J., Gao X., and Jones A. D.. 2014. Unusual negative charge-directed fragmentation: collision-induced dissociation of cyclopentenone oxylipins in negative ion mode. Rapid Commun. Mass Spectrom. 28: 457–464. [DOI] [PubMed] [Google Scholar]
- 36.Brash A. R. 2009. Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes. Phytochemistry. 70: 1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmerman D. C. 1966. A new product of linoleic acid oxidation by a flaxseed enzyme. Biochem. Biophys. Res. Commun. 23: 398–402. [DOI] [PubMed] [Google Scholar]
- 38.Floková K., Feussner K., Herrfurth C., Miersch O., Mik V., Tarkowská D., Strnad M., Feussner I., Wasternack C., and Novák O.. 2016. A previously undescribed jasmonate compound in flowering Arabidopsis thaliana - the identification of cis-(+)-OPDA-Ile. Phytochemistry. 122: 230–237. [DOI] [PubMed] [Google Scholar]
- 39.Schaller F., Biesgen C., Mussig C., Altmann T., and Weiler E. W.. 2000. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta. 210: 979–984. [DOI] [PubMed] [Google Scholar]
- 40.Schaller F., Zerbe P., Reinbothe S., Reinbothe C., Hofmann E., and Pollmann S.. 2008. The allene oxide cyclase family of Arabidopsis thaliana: localization and cyclization. FEBS J. 275: 2428–2441. [DOI] [PubMed] [Google Scholar]
- 41.Staswick P. E., Tiryaki I., and Rowe M. L.. 2002. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 14: 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laudert D., Pfannschmidt U., Lottspeich F., Hollander-Czytko H., and Weiler E. W.. 1996. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 31: 323–335. [DOI] [PubMed] [Google Scholar]
- 43.Hamberg M. 1987. Mechanism of corn hydroperoxide isomerase: detection of 12(13)-oxide-9(Z),11-octadecadienoic acid. Biochim. Biophys. Acta. 920: 76–84. [Google Scholar]
- 44.Oldham M. L., Brash A. R., and Newcomer M. E.. 2005. The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proc. Natl. Acad. Sci. USA. 102: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao B., Boeglin W. E., Zheng Y., Schneider C., and Brash A. R.. 2009. Evidence for an ionic intermediate in the transformation of fatty acid hydroperoxide by a catalase-related allene oxide synthase from the cyanobacterium Acaryochloris marina. J. Biol. Chem. 284: 22087–22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann I., Jernerén F., and Oliw E. H.. 2013. Expression of fusion proteins of Aspergillus terreus reveals a novel allene oxide synthase. J. Biol. Chem. 288: 11459–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliw E. H., Arago M., Chen Y., and Jerneren F.. 2016. A new class of fatty acid allene oxide formed by the DOX-P450 fusion proteins of human and plant pathogenic fungi, C. immitis and Z. tritici. J. Lipid Res. 57: 1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teder T., Boeglin W. E., Schneider C., and Brash A. R.. 2017. A fungal catalase reacts selectively with the 13S fatty acid hydroperoxide products of the adjacent lipoxygenase gene and exhibits 13S-hydroperoxide-dependent peroxidase activity. Biochim Biophys Acta. 1862: 706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stumpe M., and Feussner I.. 2006. Formation of oxylipins by CYP74 enzymes. Phytochem. Rev. 5: 347–357. [Google Scholar]
- 50.Grechkin A. N., Mukhtarova L. S., Latypova L. R., Gogolev Y., Toporkova Y. Y., and Hamberg M.. 2008. Tomato CYP74C3 is a multifunctional enzyme not only synthesizing allene oxide but also catalyzing its hydrolysis and cyclization. ChemBioChem. 9: 2498–2505. [DOI] [PubMed] [Google Scholar]
- 51.Martín J. F., Ullán R. V., and García-Estrada C.. 2012. Role of peroxisomes in the biosynthesis and secretion of beta-lactams and other secondary metabolites. J. Ind. Microbiol. Biotechnol. 39: 367–382. [DOI] [PubMed] [Google Scholar]
- 52.Valmsen K., Jarving I., Boeglin W. E., Varvas K., Koljak R., Pehk T., Brash A. R., and Samel N.. 2001. The origin of 15R-prostaglandins in the Caribbean coral Plexaura homomalla: molecular cloning and expression of a novel cyclooxygenase. Proc. Natl. Acad. Sci. USA. 98: 7700–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.