Abstract
The single class I myosin (MYOA) of Aspergillus nidulans is essential for hyphal growth. It is generally assumed that the functions of all myosins depend on their actin-activated MgATPase activity. Here we show that MYOA mutants with no more than 1% of the actin-activated MgATPase activity of wild-type MYOA in vitro and no detectable in vitro motility activity can support fungal cell growth, albeit with a delay in germination time and a reduction in hyphal elongation. From these and other data, we conclude that the essential role(s) of myosin I in A. nidulans is probably structural, requiring little, if any, actin-activated MgATPase or motor activity, which have long been considered the defining characteristics of the myosin family.
Myosins comprise a large superfamily of mechanochemical enzymes defined by their capability “either of translocating actin filaments or of translocating vesicles or other cargo on fixed actin filaments” (1) coupled to actin-activated hydrolysis of ATP by the myosin. All myosins contain either one or two (identical) heavy chains and one or more light chains. By phylogenetic analysis of their complete amino acid sequences, ≈150 myosin heavy chains have been divided into at least 18 classes (2–4). Myosins occur in all eukaryotes, and a single cell can contain multiple myosin classes and multiple isoforms of a single class. Why there are so many different forms of myosin and what the functions of each myosin are remain major questions in cell biology. The two largest myosin classes, with respect to both the number of isoforms and the number of species in which they occur, are the filamentous muscle and nonmuscle class II and class I myosins, the first unconventional myosins to be described (1–4).
Class I myosins have a single heavy chain that, like the heavy chains of all myosins, can be divided into head (motor), neck (IQ), and tail domains (1). The motor domain contains an ATPase site and an ATP-sensitive F-actin-binding site. The neck domain binds one or more copies of a single light chain (often, but not always, calmodulin). The tail of all class I myosins, which is short compared with the tails of other myosins, contains a basic subdomain, TH1, that binds to phospholipids and membranes. Tails of “classic” class I myosins contain two additional subdomains: a proline-rich region, TH2, which contains a second, ATP-insensitive actin-binding site and a src homology 3 (SH3) domain, TH3 (5–7). Although class I myosins are monomeric under all conditions in vitro, by virtue of the actin-binding site in the head and the membrane and actin-binding sites in the tail, classic class I myosins can crosslink actin filaments to membranes or other actin filaments and, at least in vitro, use the force generated by their actin-activated hydrolysis of ATP to move one relative to the other.
Aspergillus nidulans has a single myosin I, MYOA, that localizes to actin-rich cortical patches, especially at hyphal tips and sites of septum formation (8, 9). MYOA is essential. Conidia of MYOA null cells swell and undergo nuclear division but do not initiate hyphal growth and ultimately die (8). Importantly, MYOA is one of a small subset of myosins (class I myosins of A. nidulans, Acanthamoeba castellanii, Dictyostelium discoideum, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and the eight class VI myosins that have been sequenced) that have either a serine or threonine residue in the motor domain at a position [the TEDS site (10), Ser-371 in MYOA] in an actin-binding surface loop (11) where all other myosins contain either an aspartate or glutamate residue (10, 12, 13). The actin-dependent MgATPase and in vitro motility activities of A. castellanii (14) and D. discoideum (15) class I myosins are regulated by phosphorylation of the TEDS-site serine or threonine by p21-activated kinases (16, 17). Replacement of the TEDS-site serine of A. castellanii myosin IC by glutamate or alanine (18) mimics, in vitro, the phosphorylated (active) and unphosphorylated (inactive) states, respectively.
It was surprising, therefore, to find that S371E and S371A mutants of MYOA are equally functional when expressed in A. nidulans lacking wild-type MYOA (19). Germination and hyphal branching are delayed, and growth rates are slower for both S371E and S371A mutant cells grown in liquid culture, but morphology and growth of both are normal on solid media (19). These results suggest either that, in contrast to A. castellanii myosin IC, these point mutations have little effect on the actin-dependent MgATPase activity of MYOA, or that the essential function(s) of MYOA may not require actin-dependent MgATPase activity. The initial purpose of the work described in this paper was to answer this question by characterizing the ATPase and in vitro motility activities of wild-type MYOA and the S371E and S371A mutants expressed in SF9 cells. The studies were then extended to characterize the in vitro and in vivo properties of other MYOA mutants that have even less actin-dependent MgATPase activity than the S371A mutant.
Materials and Methods
Construction of Genes.
The construction of MYOA heavy chain mutants S371A and S371/E (18) and cloning of the A. nidulans calmodulin gene (20) were previously reported. The S199T, E428V, and E445K mutants were prepared by using a PCR-based mutagenesis method (21). The primers used to make the mutations in myoA were: S199T (5′-GACGTTGCGCAACAATACCTCACGGTTTGG-3′), E428Vup-Pi (5′-GATTACGAAACCGTAGATATC-3′), and E445Kup-Pi (5′-CTTCTTATTGACATAGTTGAT-3′). The flanking primer used for S199T was myo7(5′-CTACAATCCAGCGAAGA-3′), and for E428V and E445K myo24 (5′-ATCAGTGGTTGACTTCGTCG-3′). PCR reactions were performed in 50 mM KCl/10 mM Tris⋅HCl, pH 7.4/2.5 mM MgCl2/0.25 mM dNTPs with 1 M of each primer at 95°C for 5 min to denature the plasmid, followed by 25 cycles of 95°, 55°, and 72°C for 1 min each. The final PCR products were gel-purified and cloned into pmyo3 in a three-way ligation. For the S199T mutation, the PCR fragment was first digested with FspI and SpeI and ligated with a HindIII/FspI upstream genomic fragment and pmyo3 cut with HindIII/SpeI to generate pmyo3-S199T. For the E428V and E445K mutations, the PCR fragment was first digested with SpeI and ligated with a second KpnI cut PCR-derived fragment downstream of the mutation into SpeI/KpnI cut pmyo3 to generate pmyo3-E428V and pmyo3-E445K. The presence of the specific mutations and the absence of other mutations were confirmed by DNA sequencing. These plasmids were used for transformation of A. nidulans strain GR5.
Strains, Media, and Growth Conditions.
The GR5 strain (genotype: pyrG89; wA3; pyroA4) was the parental strain used throughout this study. S199T, E428V, and E445K “motor mutant” strains (wA3; pyroA4) were obtained by transformation of GR5 with the appropriate plasmid containing point mutations in myoA and the pyr-4 gene, thus complementing the pyrG89 mutation that is the selection marker. Yeast extract glucose (YAG) medium (8) was used for the growth of the motor mutant strains. This medium was supplemented with 5 mM uridine and 10 mM uracil (YAGUU) for GR5, because the pyrG89 mutant strain requires uridine and uracil for optimal growth.
Southern and Western Analysis.
Cultures of the mutants and the GR5 control strain were grown in liquid YAG or YAGUU medium, respectively. For Southern analysis, genomic DNA was isolated as described previously (22). The genomic DNAs were digested with BamHI and probed sequentially with a SpeI-BamHI fragment of myoA and the EcoRI fragment of pyr-4 radiolabeled by using the random prime method. Hybridization conditions were those described previously (23). For Western analysis, cells were harvested, pressed dry between paper towels, frozen in liquid nitrogen, and lyophilized overnight. The lyophilized cell mass was pulverized in a 1.5-ml microcentrifuge tube in the presence of glass beads and suspended in 0.5 ml of boiling SDS sample buffer containing 8 M urea. The samples were boiled for 5 min and centrifuged for 15 min at 15,000 × g to remove the insoluble cell debris, and 20 g of total soluble proteins was separated by 6% SDS/PAGE gel. The proteins were transferred to a nitrocellulose membrane by electroblotting. MYOA proteins were detected by using an affinity-purified polyclonal MYOA antibody raised against the COOH-terminal portion of MYOA expressed in Escherichia coli by using the pRSET expression system (8). Peroxidase-conjugated goat anti-rabbit antibody was used as the secondary antibody. Chemiluminescent detection of the proteins by enhanced chemiluminescence (Amersham Pharmacia) was used to verify the presence of the various mutant proteins.
Growth Studies.
Radial growth studies were performed by plating the strains at ≈10 spores/plate on solid YAG plates and measuring the diameter of the colonies at various times. Approximately 50 independent colonies were measured for each time point. Hyphal growth studies were performed by plating 106 spores/ml onto sterile coverslips in YAG medium. At various times, a coverslip was removed and analyzed. Recorded electronic images were measured for germination, hyphal length, and branching morphology. Approximately 50 independent germlings were measured for each time point.
Microscopy.
Spores (106/ml) were germinated on sterile coverslips in liquid YAG medium at 37°C. At various times, a coverslip was removed for differential interference contrast microscopy by using a Nikon microscope fitted with a Hamamatsu (Middlesex, NJ) model C2400 camera by using a SCSI adapter (Adaptec, Santa Clara, CA) and twain driver. Pictures were transferred into Adobe photoshop 3.0 (Adobe Systems, Mountain View, CA) for further analysis.
Expression and Purification of Proteins.
The genes and mutations were constructed by using standard methods for expression in Sf9 cells. A FLAG-tag (DYKDDDDK) was fused to the N terminus of the heavy chain constructs to facilitate purification of the expressed myosins. The wild-type, S371E, and S371A heavy chain DNAs (18) and A. nidulans calmodulin (the MYOA light chain) DNA were cloned into transfer vector pFastBac (Life Technologies, Rockville, MD). S199T, E428V, and E445K heavy chain DNAs were cloned into transfer vector pBlueBac4.5 (Invitrogen). Recombinant viruses were purified and amplified according to the manufacturer's protocols and kept at 4°C. Sf9 cells were cultured on plates by using Grace's medium supplemented with 10% fetal calf serum (Life Technologies), transfected by mixing 2–4 μg of plasmid DNA with 0.5 μg of linearized baculovirus DNA, according to the manufacturer's protocol, and the myosins and myosin I heavy chain kinase purified by affinity chromatography on a FLAG-antibody column as described (18, 24). The purity of the expressed proteins was evaluated by SDS/PAGE, and the purified proteins were kept in liquid nitrogen in the presence of 50% glycerol until use. Wild-type and mutant MYOAs were dephosphorylated with λ protein phosphatase (New England Biolabs) and phosphorylated by myosin I heavy chain kinase as described (24).
ATPase Assays.
Steady-state ATPase activities were determined at 30°C by measuring the radioactivity of Pi released from [γ32P]ATP, as described (25). The reaction mixtures for the assay of MgATPase activity contained 20 mM imidazole, pH 7.5, 4 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 3 mM [γ32P]ATP (120 cpm/nmol) with or without F-actin, as indicated. The reaction mixtures for the assay of NH4/EDTA-ATPase activity contained 25 mM Tris, pH 7.5, 400 mM NH4Cl, 35 mM EDTA, 1 mM DTT, and 3 mM [γ32P]ATP (120 cpm/nmol). The reactions were started by the addition of myosin at 30°C. Rabbit skeletal muscle actin was prepared from rabbit skeletal muscle acetone powder according to Spudich and Watt (26). Myosin concentrations were determined by the Bradford method by using BSA as the standard (27). Actin concentrations were determined spectrophotometrically by using an extinction coefficient of 0.62 cm2/ml at 290 nm.
Actin-Binding Assay.
The binding of wild-type and mutant myoA and Acanthamoeba myosin IC to F-actin was assayed in solutions containing 10 mM Tris, pH 7.5, 3.5 mM MgCl2, 1 mM EGTA, 0.2 mg/ml BSA, 12.5 mM NaCl, and 25% glycerol (the NaCl and glycerol were required to keep MYOA soluble) with or without 2.5 mM ATP, as indicated. Wild-type, S199T, E428V, and E445K MYOA (each 360 nM) and Acanthamoeba myosin IC (790 nM) were incubated alone and with F-actin (10 μM) with and without ATP (2 mM) for 5 min at room temperature. The samples were then centrifuged for 20 min at 30,000 rpm in a Beckman TL100 centrifuge at 4°C.
In Vitro Motility Assay.
The modified protocol of Sellers et al. (28) was used at 3°C in buffer containing 3 mM MgCl2 and 2 mM ATP. Myosin was phosphorylated while it was bound to the slide by adding Acanthamoeba myosin I heavy chain kinase in kinase buffer (24) and incubating for 4 min.
Results
Biochemical Characterization.
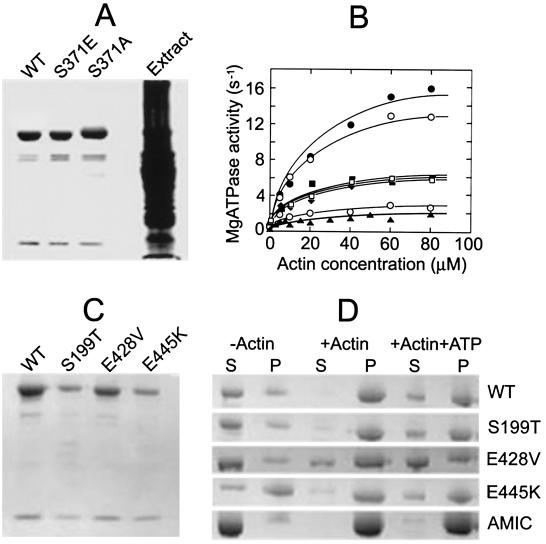
Purified wild-type MYOA and the S371E and S371A mutants are shown in Fig. 1A. Wild-type and mutant myosins had similar nonphysiological NH4ATPase activities (Table 1 Upper). Like class I amoeba myosins, the actin-dependent MgATPase activity of wild-type MYOA is regulated by phosphorylation. Wild-type MYOA was active as expressed, inhibited by treatment with phosphatase and reactivated when phosphorylated (1 mol/mol) by A. castellanii myosin I heavy chain kinase (Fig. 1B). Although the site of phosphorylation was not definitively established, that the activity of the S371E mutant was unaffected by phosphatase and kinase treatments, and that a synthetic peptide with the sequence of the putative phosphorylation site of MYOA (RGGRGSVYEV) is an excellent substrate for myosin I heavy chain kinase in vitro (data not shown), establish Ser-371 as the phosphorylated residue in wild-type MYOA.
Figure 1.
Biochemical characterization of wild-type and mutant A. nidulans myosin I (MYOA). (A) SDS/PAGE of purified wild-type (WT), S371E, and S371A MYOA and crude extract of Sf9 cells expressing wild-type MYOA. (B) MgATPase activity of wild-type and mutant MYOA as a function of actin concentration. Dephosphorylated (⊙), as isolated (○), and phosphorylated (●) wild-type; S371A MYOA (▴); S371E MYOA as isolated (□) and after incubation with kinase (■) or phosphatase (⧫, partially obscured by □ and ■). This result is typical of two separate experiments with different preparations of myosins. (C) SDS/PAGE of purified WT, S199T, E428V, and E445K MYOA. (D) Binding of wild-type and mutant MYOA to F-actin in the presence and absence of MgATP. WT, S199T, E428V, and E445K MYOA (each 360 nM) and Acanthamoeba myosin IC (AMIC) (790 nM) were incubated alone and with F-actin (10 μM) with and without ATP (2 mM) for 5 min at room temperature in 10 mM Tris, pH 7.5, containing 1 mM EGTA, 1 mM DTT, 3 mM MgCl2, 12.5 mM NaCl, and 25% glycerol (NaCl and glycerol were added to increase MYOA solubility). The samples were then centrifuged for 20 min at 30,000 rpm in a Beckman TL100 centrifuge at 4°C and the supernatants and pellets analyzed by SDS/PAGE.
Table 1.
ATPase and in vitro motility activities of wild-type and mutant A. nidulans myosin I (MYOA)
| MYOA | Basal | MgATPase, s−1 + Actin*
|
NH4ATPase, s−1 | Motility, μm s−1 | |
|---|---|---|---|---|---|
| 80 μM | Vmax | ||||
| Wild type, dephos. | 0.41 | 2.31 (15)† | 3.80 | ND‡ | |
| Wild type, phos. | 0.44 | 15.55 (100) | 17.24 | 11.18 | 0.102 ± 0.044 |
| S371E | 0.43 | 6.06 (39) | 7.24 | 11.78 | 0.050 ± 0.005 |
| S371A | 0.25 | 1.76 (11) | 2.95 | 8.86 | ND |
| Wild type, phos. | 0.32 | 14.25 (100) | 11.5 | 0.114 ± 0.029 | |
| S199T, phos. | 0.15 | 0.27 (1.9) | ND | ND | |
| E428V, phos. | 0.07 | 0.15 (1.1) | ND | ND | |
| E445K, phos. | 0.08 | 0.19 (1.3) | ND | ND | |
Wild-type and mutant MYOA, prepared as described in Fig. 1, were assayed for ATPase and in vitro motility activities as previously described (17). (Upper) One of two very similar sets of data for two different preparations of each myosin. (Lower) The enzyme data are the average of results of four different preparations of each myosin; the motility data are one of two sets of similar results for different preparations of each myosin.
Activity plus actin minus basal activity.
Percent of phosphorylated wild-type activity.
ND, none detected: in the motility assay, less than 0.01 μm s−1; in the ATPase assays, less than 0.01 s−1. 1314/120
MYOA MgATPase activity displays a simple hyperbolic dependence on actin concentration (Fig. 1B). This differs from the triphasic actin dependence observed for A. castellanii class I myosins, which is thought to result from the second actin-binding site in the tail of the amoeba myosins (29). The actin-dependent MgATPase activities of the S371E and S371A mutants were about 39 and 11%, respectively, as high as wild-type MYOA activity (Fig. 1B, Table 1 Upper), and the S371A mutant was slightly less active than dephosphorylated wild type (Fig. 1B, Table 1 Upper). Double reciprocal plots of these data (not shown) showed the differences in activities to be caused entirely by differences in Vmax, Table 1) with the same KATPase for F-actin of 20 μM for phosphorylated and dephosphorylated wild type and the two mutants. The in vitro motility activities of wild-type MYOA and the S371E mutant were approximately proportional to their actin-dependent MgATPase activities (Table 1 Upper), but neither dephosphorylated wild-type nor S371A MYOA had detectable activity in this assay (the actin filaments were bound in an apparently immobile state).
Although substantially lower than wild-type activity, we thought the actin-dependent MgATPase activity of the S371A mutant (10–15% of phosphorylated wild type and 30–40% of S371E) might be sufficient to support the essential function(s) of MYOA. Therefore, we wished to express a MYOA mutant expected to have essentially no actin-dependent MgATPase activity. Chicken smooth muscle myosin II mutant S245T was reported to have no detectable actin-dependent MgATPase activity (and only 30% of wild-type nonphysiological KATPase activity) (30), and D. discoideum myosin II mutants E459V and E476K were reported to have no detectable ATPase activity under any conditions of assay and no detectable in vitro motility activity (31). All three of the myosin II mutants were reported to bind to F-actin in the absence of MgATP but, as expected, not in the presence of MgATP (30, 31). For these reasons, we constructed and expressed the corresponding mutants of MYOA—S199T, E428V, and E445K, respectively—and determined their catalytic and motility activities in vitro and their effects on fungal cell growth when expressed in vivo.
None of the three purified expressed mutants (Fig. 1C) had detectable NH4ATPase activity (Table 1 Lower); the S199T mutant had only 2% and the E428V and E445K mutants only 1% of wild-type actin-dependent MgATPase activity (Table 1 Lower), activities too low to determine their Vmax or KATPase values. None of these MYOA mutants had detectable in vitro motility activity (Table 1 Lower); actin filaments bound to the myosins but were immobile. Although anything less than 10% of wild-type motility activity would probably not have been detectable, it is likely that the motility activities are proportional to the actin-dependent MgATPase activities, i.e., no more than 1–2% of the motility activity of wild-type MYOA.
We also compared the ability of wild-type, S199T, E428V, and E445K MYOA to bind to F-actin in the absence and presence of MgATP by using expressed A. castellanii myosin IC as a control (Fig. 1D). Although wild-type MYOA was more insoluble than A. castellanii myosin IC in the absence of F-actin (even though 12.5 mM NaCl and 25% glycerol were added to the reaction mixtures to increase solubility), it clearly bound to F-actin in the absence of MgATP and also, but more weakly than A. castellanii myosin IC, in the presence of MgATP. The S199T mutant MYOA bound to F-actin similarly to wild-type MYOA, both in the presence and absence of MgATP. The E428V mutant had less affinity for F-actin than wild-type MYOA but did show some binding, probably even in the presence of MgATP. Because of the greater insolubility of E445K in the absence of actin, binding assays of this mutant were impossible to evaluate. Similar results were obtained when the myosin solutions were centrifuged under identical conditions without F-actin and the supernatant solutions used for the binding assays.
Phenotypic Characterization.
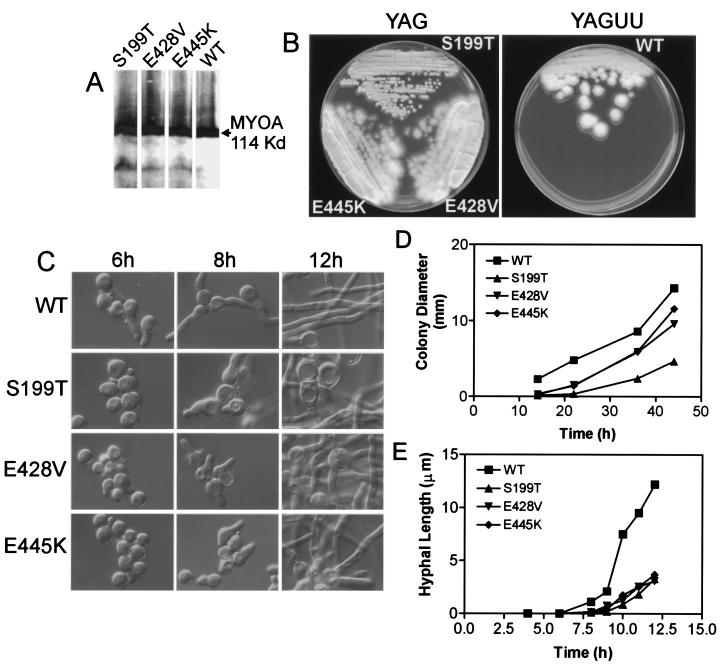
By using the same approach of integrating the mutant MYOA genes at the homologous site in the A. nidulans chromosome as was previously used for the S371E and S371A TEDS-site mutants (ref. 19; see Materials and Methods), we investigated the phenotypes of cells expressing MYOA mutants S199T, E428V, and E445K. Wild-type, S199T, E428V, and E445K MYOA were expressed at similar levels (Fig. 2A). [MYOA appears as a doublet on the gels because the phosphorylated form migrates more slowly than the unphosphorylated form (8)]. Colonies expressing the mutant myosins appeared to grow more slowly on solid medium than wild-type cells with the S199T mutant displaying the greatest growth deficit (Fig. 2B).
Figure 2.
myoA motor mutants are viable but display subtle growth defects. (A) Expression of S199T, E428V, and E445K-MYOA protein in A. nidulans strains. Protein extraction and analysis were performed as previously described (34). MYOA proteins were detected by using an affinity-purified polyclonal MYOA antibody (8). (B) Growth of wild-type and myoA motor mutant A. nidulans strains streaked on agar plates and incubated for 2 days at 37°C. (C) Differential interference contrast microscopy reveals that the initiation of hyphal growth on liquid medium is delayed in the myoA motor mutant strains. S199T cells also exhibit aberrant swollen hyphae and extensive vacuolization, whereas E428V and E445K hyphae display these effects to a lesser degree. (D) Quantification of radial growth of wild-type, S1992, E428V, and E445K strains on agar plates at 37°C. All of the mutants show a 2-h lag in hyphal emergence, whereas S199T also exhibits a decreased rate of radial growth as compared with the wild-type control strain. Approximately 50 independent colonies were measured for each time point. (E) myoA motor mutants exhibit delayed germination and decreased rates of hyphal elongation as compared with a wild-type control strain when grown in liquid culture at 37°C. Values plotted are the means measured for ≈50 independent germlings.
When characterized in greater detail, the growth rates of the mutants on solid medium, as measured by colony diameter (Fig. 2 B and D), were similar to wild type except for S199T, although all of the mutants displayed a lag before hyphal elongation began (Fig. 2D). Other than delayed hyphal emergence and shorter hyphal length, the mutant cells were morphologically normal on solid medium and completed normal conidial development.
On liquid medium, the S199T, E428V, and E445K mutants displayed a variety of relatively minor defects compared with wild-type control. First, all three of the mutants had a 2-h lag in hyphal emergence (Fig. 2C), and hyphal growth was slower for all three mutants (Fig. 2E), with an increased frequency of abnormally shaped hyphae. Furthermore, cells expressing the MYOA mutants had vacuoles that were more prominent in the swollen spore and hyphae (Fig. 2C). Interestingly, all of the abnormalities were most evident in cells expressing S199T, which had the highest (although still very low) actin-dependent MgATPase activity of the three MYOA mutants (Table 1).
Discussion
This is only the second time that Glu and Ala have been substituted for a TEDS-site Ser; although qualitatively similar, the results were less pronounced than those obtained when Glu and Ala were substituted for the TEDS-site Ser of Acanthamoeba myosin IC, which had 100 and 5%, respectively, of the actin-dependent MgATPase activity of phosphorylated wild type (18). In related experiments, substitution of Ala for Asp at the TEDS site of human nonmuscle myosin IIA heavy meromyosin reduced activity to 10% of wild type (32). Although the 1–2% of wild-type activity that we found for the S199T, E428V, and E445K mutants was not observed for the myosin II mutants on which the MYOA mutants were based, this level of activity might have been undetectable because the myosin II wild types have only about 3–10% of the actin-dependent MgATPase activity of wild-type MYOA.
The important result of our experiments is that MYOA mutants that, in vitro, have only 1% of the actin-dependent MgATPase of wild type and no detectable motility activity are able to support almost normal growth rates on both solid and liquid media. Cell morphology was normal on solid medium and there were only minor morphological defects on liquid medium. Therefore, these results suggest that enzymatic activity may not be required for the essential function(s) of myosin I in A. nidulans. The minor defects in cells expressing the enzymatically inactive MYOA mutants, which were more pronounced, although still minor, in cells grown on liquid medium than in cells grown on solid medium, were presumably the results of impairment of a nonessential MYOA function(s).
If very little and possibly no enzymatic activity is required for the essential function of MYOA, what is required, and what might the essential function be? Insight into this question might be gained from the general similarities, and specific differences, between our results for A. nidulans MYOA and recent studies of the two class I myosins, Myo3p and Myo5p (33, 34), of budding yeast, S. cerevisiae, and the single class I myosin, Myo1p (13), of fission yeast, S. pombe. Like MYOA, which localizes to actin patches at hyphal tips and sites of septum formation (8, 9), the yeast class I myosins partially colocalize with actin patches at sites of polarized growth at the tips of cells and sites of cell division (13, 34, 35). Double deletion of functionally redundant Myo3p and Myo5p is nearly lethal for some strains of S. cerevisiae, resulting in severe defects in growth and actin cytoskeleton organization (33) and lethality in other strains (36). Deletion of Myo1p causes similar actin cytoskeletal defects in S. pombe (13). Although deletion of MYOA is lethal for A. nidulans (8), thus giving no clue to MYOA's function, partial rescue by MYOA mutants (37) suggests roles for MYOA in cell polarity, septal wall formation, hyphal branching patterns, and hyphal size and shape. Taken together, these observations indicate an important role for fungal class I myosins in the organization and proper functioning of the actin cytoskeleton, specifically in relation to polarized cell growth and cell division.
Notably, all of the fungal class I myosins, and no other myosins yet described, contain a C-terminal acidic A-domain, similar to the C-terminal A-domain of animal WASp/Scar proteins that interacts with and activates the Arp2/3 complex that regulates actin assembly (38). The A-domains of S. cerevisiae Myo3p/Myo5p (39, 40) and S. pombe Myo1p (13) have been shown to bind to Arp2/3 and facilitate its role in actin polymerization. Moreover, the TH3(SH3) tail domains of Myo3p/Myo5p interact with both Bee1p (a WASP homologue) and Vrp1p (a homologue of WIP, WASp-interacting protein) (35, 39–41); thus Bee1p and the A-domain of Myo3p/Myo5p are functionally redundant in their abilities to stimulate actin assembly (39). Although there is yet no experimental evidence for functional interactions between A. nidulans MYOA and an Arp2/3 complex, that MYOA contains a C-terminal A-domain as well as a TH3(SH3) domain makes such interactions seem likely.
The only known interactions between fungal class I myosins and the Arp2/3 complex involve the TH3(SH3) domain and the A-domain. However, although these domains are important for Myo3p/Myo5p-induced actin polymerization and the proper localization of Myo5p in S. cerevisiae (35, 41), just the head and TH1 tail-domain are sufficient for proper localization of Myo1p in S. pombe and complete rescue of all Myo1p-deletion defects (13). Moreover, although rescue of the S. pombe synthetic lethal double deletion of Myo1p and Wsp1p (the fission yeast homologue of WASP) requires the TH2 and TH3(SH3) tail domains, in addition to the head and TH1 tail domain, the A-domain is not required (13). Similarly, MYOA mutants missing either the TH3 domain or A-domain can substitute for wild-type MYOA in A. nidulans [the double deletion of the TH3(SH3)-domain and A-domain has not been tested], but both the TH1 domain and a C-terminal proline-rich segment of TH2 are required (37); no requirement for the TH-2 domain of S. cerevisiae Myo3p/Myo5p has yet been demonstrated. Although our data provide evidence that actin-dependent MgATPase activity is probably not required for the essential function(s) of myosin I in A. nidulans, the requirement for TEDS-site serine phosphorylation and the ability of TEDS-site mutant S357D, and the inability of mutant S357A, to substitute for wild-type Myo3p indicate that catalytic activity may be required for the essential function of Myo3p in S. cerevisiae (36, 39). However, as neither wild-type nor mutant Myo3p has been purified and characterized biochemically, their enzymatic activities are not known. The possibility that enzymatic activity may be required for the essential function(s) of Myo1p in S. pombe has not yet been investigated.
Differences in the myosin I domains that are required in the three fungal systems may result from species-specific functionally redundant proteins or differences in the essential functions of the myosins. Thus, the essential function of MYOA in A. nidulans, and perhaps also in S. pombe, may be the proper localization of the actin cytoskeleton rather than its role in Arp2/3-initiated actin polymerization. Proper localization may involve the membrane-binding tail domain (TH1), the ATP-insensitive actin-binding TH2 tail domain (although our data indicate only relatively weak ATP-insensitive actin-binding for MYOA in vitro), and the head domain that, although not tested in A. nidulans, is required for proper localization of Myo1p in S. pombe (13).
If very little or no catalytic activity is required for the essential function(s) of myosin I in A. nidulans, why does MYOA have such high actin-dependent MgATPase activity, higher than almost any myosin other than the class I myosins of A. castellanii (1, 18, 24)? When one considers that its in vitro motility activity is almost the lowest of any myosin (1), it seems possible that a major function of MYOA may be to generate cortical tension (analogous to isometric muscle contraction) and not motility (analogous to isotonic muscle contraction). Indeed, maintenance of cortical tension has been shown to be a major function of class I myosins in D. discoideum (42, 43) and was suggested (42) to be the basis for the role of class I myosins in pseudopod formation, macropinocytosis, and phagocytosis in amoebae (44). This interpretation is consistent with the observation that pinocytosis in A. nidulans is impaired by the TEDS-site mutants of MYOA (19) and that, as for D. discoideum, cells expressing catalytically inactive myosin I show substantially fewer defects when grown on solid medium than in liquid culture where pinocytosis is more pronounced.
Acknowledgments
We thank Dr. James Sellers for advice during the course of this research and Drs. Sellers, Robert Adelstein, and John Hammer for suggestions that improved the manuscript.
Abbreviations
- YAG
yeast extract glucose
- YAGUU
yeast extract with uridine and uracil
References
- 1.Sellers J R. Myosins. Oxford, U.K.: Oxford Univ. Press; 1999. [Google Scholar]
- 2.Hodge T, Cope M J T V. J Cell Sci. 2000;113:3353–3354. doi: 10.1242/jcs.113.19.3353. [DOI] [PubMed] [Google Scholar]
- 3.Korn E D. Proc Natl Acad Sci USA. 2000;97:12559–12564. doi: 10.1073/pnas.230441597. . (First Published October 31, 2000; 10.1073/pnas.230441597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita R A, Sellers J R, Anderson J B. J Muscle Res Cell Motil. 2000;21:491–505. doi: 10.1023/a:1026589626422. [DOI] [PubMed] [Google Scholar]
- 5.Coluccio L M. Am J Physiol. 1997;273:C347–C359. doi: 10.1152/ajpcell.1997.273.2.C347. [DOI] [PubMed] [Google Scholar]
- 6.Barylko B, Binns D D, Albanesi J P. Biochim Biophys Acta. 2000;1946:23–35. doi: 10.1016/s0167-4889(00)00006-9. [DOI] [PubMed] [Google Scholar]
- 7.Osherov N, May G S. Cell Motil Cytoskeleton. 2000;47:163–173. doi: 10.1002/1097-0169(200011)47:3<163::AID-CM1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.McGoldrick C A, Gruver C, May G S. J Cell Biol. 1995;128:577–587. doi: 10.1083/jcb.128.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita R, Osherov N, May G S. Cell Motil Cytoskeleton. 2000;45:163–172. doi: 10.1002/(SICI)1097-0169(200002)45:2<163::AID-CM7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.Bement W H, Mooseker M S. Cell Motil Cytoskeleton. 1995;31:87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- 11.Rayment I, Rypniewski W R, Schmidt-Bäse K, Smith R, Tomchick D R, Benning M M, Winkelmann D A, Wesenberg G, Holden H M. Science. 1993;261:50–65. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 12.Brzeska H, Korn E D. J Biol Chem. 1996;271:16983–16986. doi: 10.1074/jbc.271.29.16983. [DOI] [PubMed] [Google Scholar]
- 13.Lee W, Bezanilla M, Pollard T D. J Cell Biol. 2000;151:789–799. doi: 10.1083/jcb.151.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch T J, Brzeska H, Miyata H, Korn E D. J Biol Chem. 1989;264:19333–19339. [PubMed] [Google Scholar]
- 15.Lee S F, Côté G P. J Biol Chem. 1995;270:11776–11782. doi: 10.1074/jbc.270.20.11776. [DOI] [PubMed] [Google Scholar]
- 16.Brzeska H, Szczepanowska J, Hoey J, Korn E D. J Biol Chem. 1996;271:27056–27062. doi: 10.1074/jbc.271.43.27056. [DOI] [PubMed] [Google Scholar]
- 17.Lee S F, Egelhof T T, Mahasneh A, Côté G P. J Biol Chem. 1986;271:27044–27048. doi: 10.1074/jbc.271.43.27044. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z-Y, Wang F, Sellers J R, Korn E D, Hammer J A., III Proc Natl Acad Sci USA. 1998;95:15200–15205. doi: 10.1073/pnas.95.26.15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita R, May G S. J Biol Chem. 1998;273:14644–14648. doi: 10.1074/jbc.273.23.14644. [DOI] [PubMed] [Google Scholar]
- 20.Means R L, Lu K P, May G S, Means A R. J Biol Chem. 1990;265:13767–13775. [PubMed] [Google Scholar]
- 21.Ho S N, Hunt H D, Pullen J K, Pease A R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 22.Osmani S A, May G S, Morris N R. J Cell Biol. 1987;104:1495–1504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May G S, Gambino J, Weatherbee J A, Morris N R. J Cell Biol. 1985;101:712–719. doi: 10.1083/jcb.101.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Brzeska H, Korn E D. J Biol Chem. 2000;275:24886–24892. doi: 10.1074/jbc.M004287200. [DOI] [PubMed] [Google Scholar]
- 25.Pollard T D, Korn E D. J Biol Chem. 1973;248:4682–4690. [PubMed] [Google Scholar]
- 26.Spudich J A, Watt S. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 27.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Sellers J R, Cuda G, Wang F, Homsher F. Methods Cell Biol. 1993;39:3923–3949. doi: 10.1016/s0091-679x(08)60159-4. [DOI] [PubMed] [Google Scholar]
- 29.Albanesi J P, Fujisaki H, Korn E D. J Biol Chem. 1985;260:11174–11179. [PubMed] [Google Scholar]
- 30.Li X, Rhodes T E, Ikebe R, Kambara T, White H D, Ikebe M. J Biol Chem. 1998;273:27404–27411. doi: 10.1074/jbc.273.42.27404. [DOI] [PubMed] [Google Scholar]
- 31.Ruppel K M, Spudich J A. Mol Biol Cell. 1996;7:1123–1136. doi: 10.1091/mbc.7.7.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Harvey E V, Conti M A, Wei D, Sellers J R. Biochemistry. 2000;39:5555–5560. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- 33.Goodson H V, Spudich J A. Cell Motil Cytoskeleton. 1985;30:73–84. doi: 10.1002/cm.970300109. [DOI] [PubMed] [Google Scholar]
- 34.Goodson H V, Anderson B L, Warrick H M, Pon L A, Spudich J A. J Cell Biol. 1996;133:1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson B L, Boldogh I, Evangelista M, Boone C, Greene L A, Pon L A. J Cell Biol. 1998;141:1357–1370. doi: 10.1083/jcb.141.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C, Lytvyn V, Thomas D Y, Leberer E. J Biol Chem. 1997;272:30623–30626. doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- 37.Osherov N, Yamashita R, Chung Y-S, May G S. J Biol Chem. 1998;273:27017–27025. doi: 10.1074/jbc.273.41.27017. [DOI] [PubMed] [Google Scholar]
- 38.Wear M A, Schafer D A, Cooper J A. Curr Biol. 2000;10:R891–R895. doi: 10.1016/s0960-9822(00)00845-9. [DOI] [PubMed] [Google Scholar]
- 39.Lechler T, Shevchenko A, Li R. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evangelista M, Klebl B M, Tong A H Y, Webb B A, Leeuw T, Leberer E, Whiteway M, Thomas D Y, Boone C. J Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geli M I, Lombardi R, Schmelzl B, Riezman H. EMBO J. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai J, Ting-Berall H P, Hochmuth R M, Sheetz M P, Titus M A. Biophys J. 1999;77:1168–1176. doi: 10.1016/s0006-3495(99)76968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz E C, Neuhaus E M, Kistler C, Henkel A W, Soldati T. J Cell Sci. 2000;113:621–633. doi: 10.1242/jcs.113.4.621. [DOI] [PubMed] [Google Scholar]
- 44.Tuxworth R I, Titus M A. Traffic. 2000;1:11–18. doi: 10.1034/j.1600-0854.2000.010103.x. [DOI] [PubMed] [Google Scholar]