Abstract
Landscape ecology examines the relationships between the spatial arrangement of different landforms and the processes that give rise to spatial and temporal patterns in local community structure. These relationships that underlie the patterns of the microbial communities that inhabit the human body, and in particular, those of the nose, mouth and throat, deserve greater attention. Important questions include what defines the size of a population (i.e., ‘patch’) in a given body site; what defines the boundaries of distinct patches within a single body site, and where and over what spatial scales within a body site are gradients detected. This review looks at the landscape ecology in the upper respiratory tract and mouth, and seeks greater clarity about the physiological factors, whether immunological, chemical or physical, that govern microbial community composition and function, and the ecological traits that underlie health and disease.
Keywords: landscape ecology, spatial ecology, biogeography, microbiota, gradient, nares, dental plaque, supragingival, subgingival
Introduction
In an effort to discern the role of fundamental ecological processes in community assembly, early ecological models assumed organisms are distributed across a spatially homogenous environment. Yet, nearly every ecosystem, including the human microbial ecosystem, exhibits distinct patterns of community structure and assembly across gross anatomic sites suggesting rich habitat differentiation (Costello et al., 2009). One explanation for these distinct patterns is underlying spatial heterogeneity in topographical anatomy (i.e., the ‘landform’), local chemistry, or both, and in the case of host-associated environments, physiology, as well as tissue type and associated structures, desquamation rates, immune processes, temperature, moisture, and other local conditions. Thus, in treating the human body as a microbial landscape – we must consider the underlying spatial heterogeneity in landforms and environmental features as selective factors that give rise to spatial patterns in microbial community structure and function; these communities in turn, generate additional spatial heterogeneity.
The theory of landscape ecology, which emerged in the 1960s, sought to explain the spatial patterns and processes operating on a landscape rather than assuming space to be homogenous (Wiens et al., 1993). When surveying a landscape, the observed types of spatial (and temporal) patterns depend critically on the scale of observation (e.g., a micron, a meter, a kilometer; and a second, a day, a year). In turn, the scale of observation informs a variety of parameters – such as patch size, patch density, patch quality, inter-patch distances, and stability. By quantifying these parameters experimentally ecosystem stability and expected resilience in the face of a variety of disturbances can be modeled using the mathematical underpinnings of landscape ecology. Yet, with few exceptions (Bouslimani et al., 2015; Mark Welch et al., 2016; Swidsinski et al., 2007) most molecular studies of the host-associated microbiota continue to discuss biogeography not as a function of geography per se, but as variation across gross anatomic sites and treat these sites as categorical, discrete entities. For this reason in part, our understanding of the spatial and temporal scale(s) important in the ecology of the microbes and viruses that inhabit the human body is limited.
In this review, we begin by providing a primer on landscape ecology before discussing some of the processes that give rise to patches, while highlighting the importance of spatial scale in commensal microbial communities. Next, we discuss the landscape heterogeneity of the upper respiratory tract and mouth, an excellent anatomic region for studying spatial ecology due to its easy accessibility. Then, we consider the possibility that baseline immune function represents a disturbance regime that is perturbed during acute or chronic infections, and associated with detectable pulse or press perturbations in community structure. We conclude by presenting a few ways in which landscape theory might be applied to the microbiota of the oral and nasal cavities in order to increase our understanding of how pattern and process contribute to the structure and function of host associated communities.
Primer on landscape ecology
Landscape ecology examines the processes that give rise to spatial patterning in communities across a landscape, ‘the landforms of a region in the aggregate’ (Turner, 1989). The chief proposition of landscape ecology is that the features and spatial arrangement of the landscape dynamically interact with ecosystem function, each shaping the other, such that it is difficult to understand a community without considering the context of its associated landscape (Wiens, 1995). To facilitate a discussion of this proposition, in this section, we review the basic history, terms and definitions of landscape ecology while identifying the types of spatial patterns that might be observed in the ecology of the microbiota.
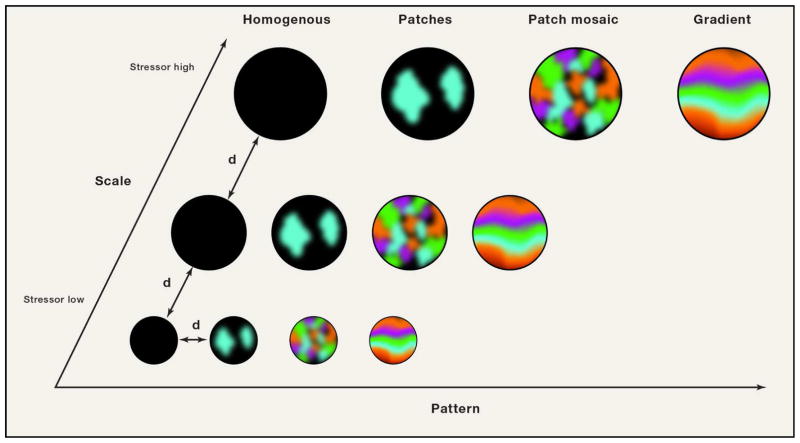
In population biology, a ‘patch’ was historically defined as a spatially clustered population (i.e., a group of interacting organisms of the same species) that could be distinguished from its surroundings (Hutchinson, 1953). The spatial boundaries of a given patch (e.g., city limits) are given by the boundaries of suitable habitat, and each patch is embedded in an inhospitable or neutral matrix (Fig. 1). With the development of the theory of island biogeography and meta-population theory, ecologists began considering the interaction of patches (i.e., populations) with each other through migration, dispersal, colonization and extinction, processes that mediate inter-patch dynamics. In so doing, meta-population biologists solved a fundamental problem in patch biology: that spatially isolated populations have a stochastic, non-zero probability of going extinct and should do so by chance over different time scales (Levins, 1969).
Figure 1. Relationship between spatial scales, spatial patterns, and dispersal distance.
As the scale of observation increases from a small spatial area to a larger area, patterns can be detected across the landscape. A homogenous pattern reflects a set of unoccupied patches or, alternatively, a set of patches entirely occupied by a single species. Fragmented patchiness on the other hand reflects the occurrence of spatially segregated patches or patch types at various sites across the landscape, while a patch mosaic consists of a set of patch types in which patch types do not vary in a discernable pattern with respect to each other at increasing spatial scales of observation. A gradient is observed when a pattern can be detected when comparing patch types or mosaics at increasing spatial scales. The variable “d” indicates the dispersal distance, which can be defined as the distance between patch types (large circles) or the distance microbes traverse when dispersing across the landscape or across sites within a patch type. Adapted from Wiens, J.A. (1995). Habitat fragmentation: island v landscape perspectives on bird conservation. Ibis 137, S97–S104.
Community ecology in turn extends the domain of meta-population studies from populations of one species to the ‘community’, an assemblage of interacting organisms of two or more species. In spatial ecology, a ‘patch type’ is analogous to a patch but represents a spatially-clustered community rather than a population (Fig. 1). Patch types are often defined by the dominant organism or land usage regime (Pickett and Cadenasso, 1995), such as an oak woodland, pine woodland or coastal sage scrub. As such, the concept of a ‘community state type’ (CST) might be considered as a basic description of a patch type in human microbial ecology, since most researchers use it to denote a community dominated by one organism (e.g., a vaginal community dominated by Lactobacillus crispatus) (Ravel et al., 2011). In the human body, dominance by one species is most apparent in the vagina. It is unclear in general whether most surveys of the microbiota characterize single patch types or whether they pool multiple patch types since the spatial extent of CSTs is rarely defined. Further, there is some debate in the field of microbial ecology as to how community state types should be defined (Callahan et al., 2016a).
Landscape ecology examines ecosystems across a wide variety of spatial scales surveying the dynamics of spatial patterns known as ‘patch mosaics’ and ‘gradients’. A ‘patch mosaic’ is a collection of patch types within a spatial territory where patch types exhibit a definite but seemingly random spatial distribution with respect to each other (Fig. 1). In the mouth, the recently described ‘cauliflower’ arrangement (Fig. 1) of bacteria is one of the most beautiful visuals of a patch type, consisting of spatially segregated patches of organisms including Lautropia, Veillonella, Haemophilus/Aggregatibacter, Capnocytophaga, and Streptococcus spp., among others (Mark Welch et al., 2016). In one representative image, Lautropia formed clusters that rarely included other members of the community while Streptococcus was interspersed amongst clusters of Haemophilus/Aggregatibacter, and Veillonella occupied peripheral regions where gaps could be found. Large gaps between many patches can be seen and when larger spatial areas were viewed, different patch types including ‘corncob’, ‘cauliflower’ and ‘hedgehog’ structures were found to repeat across space. The repetition of these patch types across space is indicative of a patch mosaic.
Examining the spatial arrangement of patch types and patch mosaics, in turn, leads to some of the fundamental questions in landscape ecology, such as, how many distinct patches can be found in a mosaic and why; how large is each patch and why; do patch or patch types have sharp or indistinct boundaries; how connected are the distinct patches; and to what extent do these quantitative features predict the occurrence of spatial arrangements between and across sites? The answers to these questions would enable the prediction of ecosystem stability and resilience (Wiens et al., 1993) allowing the field to move beyond qualitative narratives of spatial patterns towards quantitative descriptions of spatial dynamics and predictions about function.
By examining patch mosaics across the entire landscape, the size, orientation and arrangement of patches and patch mosaics can be defined (Turner, 1989). These features must be understood to identify ‘gradients’, ordered arrangements of patch mosaics (Fig. 1). Patch types in a gradient tend to have indistinct boundaries (Wiens et al., 1993) as a result of gradation in underlying environmental factors such as pH, temperature, and moisture. The recognition and understanding spatial gradients in microbial community structure across the human body is still in an early phase. Meanwhile, debate in soil microbiology concerns the question as to whether microbes follow biogeographical patterns that are fundamentally different than those of macroscopic organisms (Fierer et al., 2011{Tripathi, 2017 #150).
Processes that give rise to patchiness
What processes give rise to spatial heterogeneity? This question was first answered by Alan Turing who sought to understand how spatial patterns emerge from a uniform surface (Turing, 1952). Turing studied the unfolding of pattern during morphogenesis by modeling reaction diffusion dynamics. In that seminal work, spatial patterns developed after irregularities were amplified due to system instability in chemical reaction dynamics. These irregularities may have been stochastic, or they may have been emergent features of the system, as is the case with differential gene expression during morphogenesis. In ecosystems, such “Turing irregularities” can give rise to spatial pattern in community structure – in this section, we review some of the factors that may give rise to spatial patterns in ecosystems including disturbance, abiotic factors, and biotic factors (Hutchinson, 1953). Ecological processes – dispersal, selection, diversification, and drift, as well as priority effects – that give rise to heterogeneity have been reviewed elsewhere (Costello et al., 2012; Fukami, 2015; Martiny et al., 2006).
One of the biggest sources of spatial heterogeneity is disturbance. ‘Disturbance’ is defined as an irregularity that perturbs the ecosystem as well as community or population structure. An ecological disturbance is thought to induce spatial heterogeneity by making space available for new colonists and by inducing a temporal irregularity that disrupts the natural course of succession (Levin and Paine, 1974). Disturbances can be one-off events, or they can occur either regularly or stochastically with a specific periodicity and intensity, in which case the periodic oscillations define the disturbance regime (Relman, 2012). One single disturbance event impacting a region may have a different impact on different sites since the “spread of disturbance across a landscape is influenced by spatial heterogeneity” (Turner, 1989). For example, tooth-brushing removes biofilms fairly well from the cheek and tongue-facing surfaces of teeth, but it tends to be less effective at removing biofilms from the biting surfaces of molars and pre-molars, permitting higher biomass accumulation at those sites. In other words, the intensity of brushing as a disturbance is higher at one site than it is at the other because of spatial heterogeneity in the landscape.
Besides disturbances, a second cause of patchiness is the underlying partitioning of environmental resources or stressors – for example, resident anaerobes in the mouth that prefer a lower redox potential will likely be found where oxygen is limiting in the subgingival crevice, especially in severe cases of periodontitis, or on the dorsal tongue. Other examples include stressors that structure gradients as previously discussed. Biotic interactions – competitive, social or reproductive – provide a third cause of patchiness. For example, bacteriocins, anti-competitor proteins employed by the microbiota that typically target conspecifics (Zheng et al., 2015), diffuse away from those cells that produce them, creating a concentration gradient and a zone of competitive exclusion, which is observed as the repulsion of the producer and sensitive strains. Similarly, social behaviors typified in bacteria by quorum sensing may generate patchiness by inducing dispersal of surface-associated cells or attachment of planktonic cells, two processes that would naturally lead to different spatial patterns at different time scales. Finally, certain reproductive strategies also give rise to patchiness – an example can be seen in the Cathedrals of the California redwood Sequoia sempervirens in which a circle of clones surrounds the mother tree when that individual reproduces clonally.
Spatial scale in the landscape ecology of the microbiota
One critical question in microbiome research is how to couple our analytical techniques with the spatial or temporal scale(s) required to identify patterns and underlying mechanisms important in the ecology of the microbes and viruses that inhabit the human body. The grain of observation profoundly influences our ability to observe patterns. If the grain is too coarse, such as what is gained by using nasal lavage or oral rinses to sample the nasal or oral cavities, then the community that is observed is a statistical sample of a heterogenous landscape. If the grain of observation is too fine, a single patch or patch type might be misinterpreted as representative of the entire landscape.
This raises the question, what spatial scales are relevant to the genesis of spatial patterns and processes in the communities of the human body? Our ability to detect spatial patterns depends on the spatial scale of organisms in a given ecosystem. For plants, patches are typically observed at small spatial scales (1 m), patch mosaics are observed at larger spatial scales (50 km) and gradients are observed at the most expansive spatial scales (200 km) (Joan G. Ehrenfeld, 1997). Microbial ecology, on the other hand, must focus on variation between entities separated in space on a scale that is appropriate to their body size. Viruses range in size from 20–450 nanometers while bacteria vary in size from 0.3 microns for Mycoplasma (diameter) to the average cell, Escherichia coli, which is 1.1 to 1.5 microns wide and 2 to 6 microns long. The spatial extent of a population of micron-sized organisms will depend on the scale of the processes that give rise to their spatial arrangement across space, as already discussed.
Imaging studies provide initial insights into the scales important in the spatial organization of the microbiota. Microbial colonization of the nasal turbinates may be sparse and patchy with single bacterial cells seeding the surface of epithelia at seemingly random locations (Swidsinski et al., 2007). On the other hand, inflamed adenoids are punctuated by focal patches and highly confluent polymicrobial biofilms that sometimes disrupt epithelial surface integrity. A similar survey revealed that bacterial biofilms collected from a single tooth surface in the absence of disease range up to hundreds of microns in radius (Mark Welch et al., 2016). Patch sizes on epithelial surfaces are known to be limited compared to those on non-shedding surfaces of teeth. Similarly, desquamation rates which differ between tissues may restrict patch size even on epithelial surfaces. Taken together, these observations suggest that additional in vivo or ex vivo work (e.g., on whole teeth rather than tooth swabs) is needed to characterize the size(s) of individual biofilms in each of the major body site habitats.
In order to refine our scale of observation it may be useful to collect samples along annotated georeferenced transects, obtaining sample site coordinates and topographical data with imaging as is done in macro-ecology, rather than simply describing anatomic regions. In the nasal cavity, for instance, geographic variation in nasal flow velocity is thought to create focal hotspots of high sheer stress at specific points throughout the mucosa (Doorly et al., 2008). A hypothesis that might be tested is that these hot spots influence microbial colonization of epithelia in the nasal cavity. If so, geographic location may be as important as the type of epithelial surface on which a community is found.
One way to identify the appropriate scale is to determine the amount of “detail that can be ignored without producing results that contradict specific sets of observations” (Levin, 1992). In this light, interpersonal variation can sometimes be viewed as a confounding variable, as it is too expansive a spatial scale across which to make inferences about the spatial organization of nasal or oral microbial communities. The extent to which interpersonal variation overshadows inter-site variation has sparked debate about whether communities inhabiting the anterior nares and the nasal mucosa differ (Yan et al., 2013) (Ramakrishnan et al., 2017)(Kaspar et al., 2016; Wos-Oxley et al., 2016). The degree to which interpersonal variation obscures finer-scale spatial or temporal patterns is evident in studies that present separate ordination plots for each subject (Hauser et al., 2016; Kaspar et al., 2016; Ramakrishnan et al., 2017) or that present a different ordination for each site coloring samples by subjects (Sato Y1, 2015; Sato et al., 2015; Wos-Oxley et al., 2016). In recognizing that interpersonal variation is a confounding variable, in these analyses, rather than a comparator, it becomes possible to manage its effects using numerical ecology. As an example, separate ordinations could be performed, as above, but rather than presenting individual ordinations for each subject, a multiple-tables analysis could be performed to examine the consistency of spatial patterns in community structure across subjects. Another approach where geographic coordinates have been obtained, through imaging or modeling, would be to perform a trend surface analysis which would enable identification of large scale spatial structures.
Landscape ecology of the human nasal cavity
The spatial heterogeneity of the nasal cavity likely shapes the distribution of organisms across its surfaces. Microbes inhabiting the anterior nares (Fig 2) confront a wide variety of host features that are not present elsewhere in the nasal canal including stiff, coarse hairs known as vibrissae that litter a keratinized stratified squamous epithelium, specialized tissue with known microbial associations. At ambient temperature, this region tends to be cooler than other nasal sites and sites throughout are subject to upwelling from sweat and sebaceous glands, which percolate through the region. These micro-environmental differences may give rise to spatial patterns in community structure. For example, sebum secreted from the sebaceous glands appears to be an important determinant of the preferential colonization of Propionibacterium spp. (Mukherjee et al., 2016).
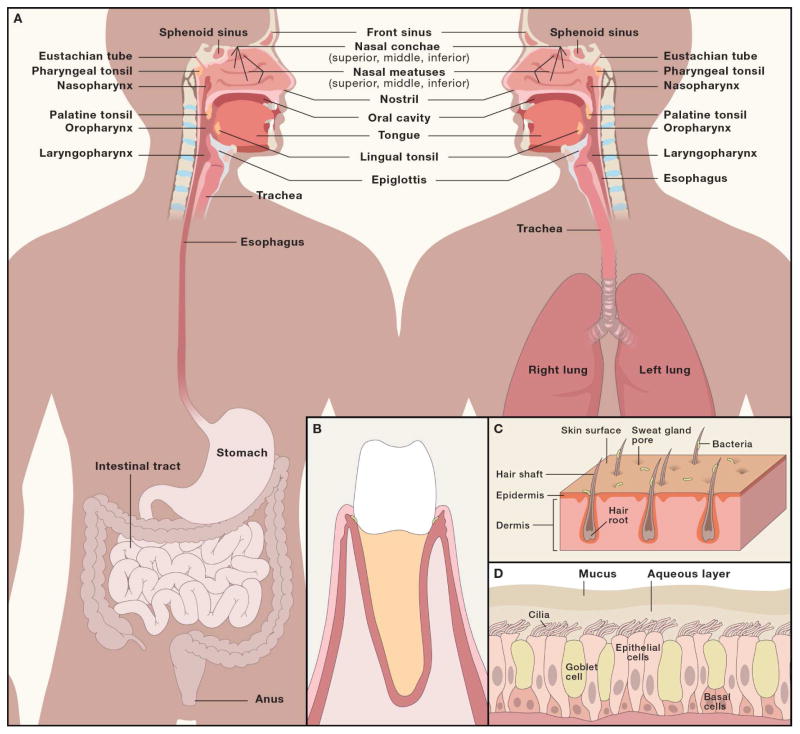
Figure 2. Examples of spatial heterogeneity in the oral and nasal cavity landforms likely to influence spatial patterning of microbial communities.
A) Topography of the anterior nares. Sweat glands, sebaceous glands and nasal hair puncture the epithelial surface of the anterior nares leading to upwelling from the dermal layers at focal points throughout the tissue. The roots of individual hair follicles represent a unique habitat that is not found elsewhere in the nasal cavity. B) Spatial organization of the nasal mucosal surfaces. Inspired air is warmed as it moves across the inferior, middle and superior turbinates. The maxillary sinuses drain into the middle meatus, and the ethmoidal sinuses drains into the sphenoethmoidal recess, which presumably creates local patches reflective of the sinus source pool. C) Topography of the nasal mucosa. Cilia interact with the aqueous phase of nasal mucus, moving the mucus blanket across the mucosal surface, which consists of antimicrobials and mucins not present in the anterior nares. D) Tooth microenvironments. The junctional epithelium is the most permeable tissue of the gingival epithelium allowing immune cells and gingival crevicular fluid to leak out of the subgingival crevice into the oral cavity. It is non-keratinized stratified squamous epithelial tissue that surrounds each tooth.
The nasal mucosa, which lies just 2 cm beyond the anterior nares, differs from the nares in several ways that may considerably influence microbial community structure. Yan et al. found that communities at three nasal sites, anterior naris, middle meatus, and sphenoethmoidal recess (Fig 2) differed according to epithelium type (Yan et al., 2013). A variety of anatomic and physiological features may explain this finding. While microbes in the nares are subject to ambient temperature, temperatures in the turbinates increase by ~4.5°C, reaching ~33°C by the time air reaches the nasopharynx (Keck et al., 2000). This temperature gradient, which induces the formation of a moisture gradient, may differentially influence gene expression in both pathogens and commensals at different locations along the nasal passages. In addition, some bacteria colonize the crypts of the pseudostratified columnar ciliated epithelium of the mucosa (Swidsinski et al., 2007), a niche that is inaccessible to inhabitants of the nares. Likewise, drainage from sinuses occurs at specific locations throughout the mucosa, e.g., maxillary sinus into the middle meatus, and ethmoidal sinuses into the sphenoethmoidal recess, which presumably creates local patches reflective of the sinus source pool. These features are also lacking in the nares.
Overlaying the cilia is a shifting blanket of mucus, which is comprised of two distinct layers, an aqueous fluid with which cilia interact (Fig. 2) and an overlaying mucus layer comprised of more than 100 proteins, including mucins which agglomerate in “rafts”, antimicrobial proteins like lactoferrin, lysozyme and peroxidase, as well as secretory IgA (sIgA), albumin, fibrinogen, and lipid binding proteins (Casado et al., 2005). Myriad mechanisms underlying interactions between microbes and mucins have been identified – the ability or inability of an organism to interact with mucus likely influences whether it persists on a mucosal surface (Zanin et al., 2016). The blanket of mucus is transported through the nasal cavity before being drained into the nasopharynx (Fig 2) where microbes may gain access to the soft and hard palates of the oral cavity.
During the first year of life, the density of the nasal microbiota increases with age while alpha diversity decreases (Mika et al., 2015). Nasopharyngeal communities of the infant tend to be dominated by taxa associated with the skin, including Staphylococcus and Corynebacterium spp. (Teo et al., 2015). These taxa are later succeeded in the nasopharynx by Moraxella or Alloiococcus spp., which when dominant in a community tend to be fairly stable (Biesbroek et al., 2014). When either Haemophilus or Streptococcus spp. colonize the nasal cavity and ascend to dominance in a community, they tend to be displaced quickly, at least in infancy. Early patterns of community succession in the nasal cavity, in turn, determine the likelihood a given patch type will transition to another since different invading colonists seem to have different probabilities of displacing dominant strains. For example, in adults, the ability of Staphylococcus aureus to invade patch types dominated by Streptococcus pneumoniae is limited (Bogaert et al., 2004; Chien et al., 2013; Cremers et al., 2014) though other colonists can and do displace S. pneumoniae as the dominant member.
The mechanisms governing CST transitions in nasal communities are now being defined. One mechanism is the elaboration of proteins or small molecules by one organism to antagonize or inhibit the growth of another (Abreu et al., 2012; Zipperer et al., 2016). Similar antagonism can be achieved as a side-effect of metabolism as demonstrated by an elegant study showing the commensal Corynebacterium accolens impairs S. pneumoniae colonization by metabolizing host lipids to oleic acid, a potentially mutualistic action that may limit the density of this ‘pathobiont’ on our epithelial surfaces (Bomar et al., 2016). Other mechanisms including immune modulation by the microbiota. For example, in the presence of S. pneumoniae, Haemophilus influenzae appears to upregulate the expression of chemokines that lead to the complement-mediated phagocytic removal of S. pneumoniae from the neighborhood (Lysenko et al., 2005).
Landscape ecology of the human oral cavity
The human oral cavity also provides a unique opportunity to study the spatial ecology of microbial communities due to its accessibility and wealth of unique microbial habitats. The clearest topographical feature that distinguishes microbial communities is whether the superficial tissue layer of a given site is shedding (oral mucosa) or non-shedding (dental enamel) (Human Microbiome Project Consortiu, 2012). In this section we discuss the features of the oral landscape which may give rise to spatial patterning in oral microbial communities.
Keratinization of the oral mucosa creates spatial heterogeneity. The stratified squamous epithelia of the oral mucosa can be subdivided into several functional types - the masticatory, lining and specialized mucosa – each distinguished by functional histologic features. The superficial layer of the masticatory mucosa that lines the hard palate, the dorsal tongue surface and keratinized gingiva proximal to supragingival tooth surfaces consists of a cornified envelope of orthokeratinized (i.e., the superficial cell layer lacks nuclei) or parakeratinized (i.e., the superficial cell layer is pyknotic) cells. By contrast, the lining mucosa of flexible tissues like the soft palate, ventral tongue surface, floor of the mouth, and labial mucosa lacks keratinization. An example of the specialized mucosa is found in the region of the papilla on the dorsal surface of the tongue, which give it a bumpy appearance. These features are not present on the ventral or lateral surfaces of the tongue.
Importantly, spatial heterogeneity in keratinization and the spatial arrangement of papilla at sites across the dorsal tongue have been shown by microscopy to be associated with spatial patterning in microbial colonization of that surface (Aufdemorte and Cameron, 1981). More recent work has demonstrated that community structure differs between the dorsal tongue and the lateral or ventral tongue surfaces (Aas et al., 2005; Mager et al., 2003). These patterns likely arise as a consequence of the surface topography of sites, which determines the intimacy with which different sites come into contact with each other.
In addition to these differences in topography, surfaces vary with respect to their proximity to the nearest salivary gland, a major source of environmental disturbance in the mouth. The minor salivary glands form a dense and expansive network that punctuates the labial, palatal and buccal mucosa, releasing viscous, highly proteinaceous secretions with poor buffering capacity (Dawes and Wood, 1973). These secretions bathe the surfaces from which they emanate as well as opposing surfaces creating heterogeneity that likely explains, in concert with other factors, the observation that communities found on cheek-facing aspects of individual teeth differ from those on tongue-facing aspects (Sato et al., 2015; Simon-Soro et al., 2013).
The three major salivary glands differ in their secretory rates and composition (Schneyer and Levin, 1955), giving rise to gradients in salivary film velocity, oral clearance and intra-plaque pH across the teeth (Dawes et al., 1989; Kleinberg and Jenkins, 1964; Wolff and Kleinberg, 1998). Moreover, the salivary glands also give rise to spatial variation in patterns of wetness and dryness across different geographic regions of the mucosa (Fig. 2) suggesting that microbial communities inhabiting soft tissues may vary along a moisture or pH gradient (Wolff and Kleinberg, 1998) although to our knowledge this has not been tested.
Despite the existence of multiple known compartments in the mouth most surveys of oral communities provide limited insight into the spatial patterning of supragingival communities across expansive spatial scales. This is because most extant studies including our own (Bik et al., 2006) have either reported findings of biofilms pooled from multiple tooth surfaces; or used saliva as a sample of supragingival surfaces; or used rinsing samples instead. The grain of resolution afforded by such techniques does not permit interrogation of the fine-scale spatial variation of communities across sites. Of the studies that have analyzed independent samples of each tooth surface most treat the unit of spatial variation – the physical location of a tooth in the mouth – as a categorical variable such as tooth number, tooth class or tooth aspect (Haffajee et al., 2009; Mager et al., 2003). We have recently shown in a pilot experiment that microbial communities inhabiting the exposed tooth surfaces of healthy humans vary not only based on tooth aspect and tooth class, but as a function of the physical distance separating sites in a manner that is consistent with a spatial gradient (Callahan et al., 2016a).
The oral cavity is a dynamic ecosystem that varies over time in ways that influence spatial patterns of microbial community assembly. The eruption of our dentition can be compared to the uplift of mountains, as both processes describe the dynamics of landform development and the emergence of new habitat into an existing ecosystem. Infants enter the world toothless and remain that way for ~6 months when teeth begin erupting. The deciduous teeth erupt over the first two years of life and are gradually shed and replaced by the permanent dentition between the ages of 6–12. Importantly, different tooth classes (e.g., molars, incisors) erupt in a stereotypic sequence at different developmental ages, making some of our teeth older than others within an individual, yet comparable in morphology and tooth-age between individuals. Once teeth break through the gumline, gingival crevicular fluid, complement, phagocytes, and other components from the bloodstream begin, in minute measure, leaking into the mouth, providing novel growth substrates for some organisms at the same time as adding additional mechanisms of immune control. In infants, community assembly in the oral cavity reflects this extended process of geomorphogenesis; within a day of birth, Streptococcus salivarius and Streptococcus mitis colonize the oral mucosa while Streptococcus sanguinis, which preferentially colonizes dental enamel, is not seen until after the teeth erupt.
Dispersal across anatomic sites
Different gross anatomic sites are connected to each other. Some of these sites may serve as sources of colonists for other sites which serve as ‘sinks’. The nasal and oral cavities for example both drain to the pharynx, which ultimately connects through the trachea to the lungs or through the esophagus to the stomach which is connected to the gut (Fig. 2). In this section, we highlight several studies of dispersal between body sites and identify the obstacles researchers face in characterizing these dynamics.
Researchers have examined whether the middle ear and/or the adenoids serve as a reservoir for the bacterial agents of otitis media with effusion (OME) not only because the nasal canal is connected to the middle ear via the eustachian tube (ET) (Fig. 2) but also because the nasopharynx is often colonized by organisms implicated in OME, including S. pneumoniae, H. influenzae, Moraxella catarrhalis, and Alloiococcus otitidis. In one study, community composition of the middle ear closely resembled that of the external auditory canal (EAC) both in abundance and in community similarity, leading the authors to consider the EAC to be a likely source for OME (Chan et al., 2017). A competing theory with supporting evidence from microscopy is that the middle ear is seeded by the adenoids (Torretta et al., 2013). Patch types comprised of S. aureus, M. catarrhalis and S. pneumoniae – were found on the adenoid adjacent to the ostia of the ET more frequently than in the region of the NP dome. Interestingly, for unknown reasons, microbial clusters near the ET were more often polymicrobial than were the clusters on the NP dome. And interestingly, isolates derived from the ET region were more likely to form biofilms in vitro than were the isolates from the NP region. Taken together, these data led the authors to conclude that the adenoids are a more likely source for OME. By contrast, Chan et al. tested the adenoid theory using 16S rRNA data, concluding the adenoids to be an unlikely source of colonists to the middle ear since middle ear effusions (MEF) were dissimilar in community structure to adenoids (Chan et al., 2016). Dissimilarity appeared to be driven by the differential abundance of Alloiococcus found in high and low abundance in the MEF and adenoids, respectively. Implicit in this conclusion is the hypothesis that the size of different Alloiococcus populations in this ecosystem drive dispersal dynamics between these sites.
The sinuses experience flooding during respiratory colds as well as during physiological reflexes like coughing or sneezing. As a result of transient spikes in intranasal pressure, nose-blowing pushes as much as 1 ml of nasal mucus into the ostiomeatal complex as well as the ethmoid and sphenoid sinuses (Gwaltney et al., 2000). The periodic flooding of the sinuses with mucins, a nutrient source, as well as microbes trapped in the mucus suggests this habitat is functionally similar to a floodplain. The nasal mucus may transport colonists to the paranasal sites; and the nutrient influx may cause blooms in the sinus microbiota, a community found even in healthy humans (Abreu et al., 2012; Aurora et al., 2013). To determine the source pool for the sinus microbiota, one group assessed the similarity of communities in the anterior nares (AN), nasopharynx (NP) and ethmoid sinus (ES) before and at 2 and 6 weeks after sinus surgery (Hauser et al., 2016). Communities of the ES 6 weeks after surgery were most similar to those of the AN and ES pre-surgery, leading the authors to conclude that the AN may serve as the source of post-disturbance colonists. Communities of the NP on the other hand were ecologically dissimilar to those of the ES, leading the authors to conclude that the NP is an unlikely source of sinus colonists. Of interest, communities inhabiting the NP were as dissimilar to the ES as the ES was to itself before and 2-weeks after the disturbance, raising the question as to whether the observed level of dissimilarity that excluded the NP must also exclude the ES as a likely source of its own repopulation.
In attempting to understand the relationship between the bacteria found in the upper and lower respiratory tracts researchers discovered a biomass gradient that distinguishes between the two anatomical sites (Charlson et al., 2011). The adapted island model was subsequently proposed to explain decreasing community richness at lung sites as a function of increasing distance to the supraglottis, the proposed source (Dickson et al., 2015). Later work led the authors to conclude that the oral cavity may seed both the lungs and the stomach since these sites are more similar to each other than they are to nasal communities (Bassis et al., 2015). Prior work from our group has also shown a large overlap in the composition of the microbiota of the stomach, mouth and esophagus (Bik et al., 2006). An operational taxonomic unit (OTU) level analysis of Human Microbiome Project (HMP) 16S rRNA amplicon data revealed high levels of similarity between communities of the distal colon and oral cavity, but not of the colon and nasal or skin communities (Ding and Schloss, 2014), leading these workers to postulate that the oral cavity may seed the gastrointestinal tract, an attractive but as yet unproven proposition.
The most convincing examples of bacterial dispersal across the human landscape come from culture-based studies demonstrating that the S. aureus strains found in the anterior nares of an individual are the strains found in S. aureus bacteremia of the same individual (von Eiff et al., 2001). Yet, since S. aureus inhabits multiple regions in the nasal cavity, including the anterior nares, the middle meatus and other turbinates (Yan et al., 2013), it is hard to say that the nares serves as the source for S. aureus bacteremia, as has often been claimed, and the processes giving rise to dispersal from the true source(s) remain insufficiently characterized. Similarly, carriage of S. aureus in the anterior nares predisposes individuals to soft tissue and skin infections, such as cutaneous abscesses, at sites far removed from the nose – what gives rise to this phenomenon and whether the anterior nares per se is the source remains unclear (Johnson et al., 2015).
Valuable insights into the obstacles researchers face when examining dispersal across anatomic sites have come into focus from these early studies. First, community-based similarities (or dissimilarities) and population abundances do not provide direct evidence that a site is (or is not) the source of colonists for another site. Analyzing exact sequences and finding the same ribosomal sequence variations (Callahan et al., 2016b) at each site would provide more compelling evidence but may not yet meet the gold standard of culture-based tests and full genome sequencing, due to amplicon bias, potential sequencing errors, and the limited resolution of the highly-conserved 16S rRNA gene.
Future work using strain-resolved metagenomics (Donati et al., 2016) may improve our understanding of how often and how far microbes travel across the human body. Likewise, culture-based and imaging work should supplement 16S rRNA-based surveys to achieve the same effect. Second, our understanding of dispersal across the human body is limited by our incomplete characterization of the spatial patterns and scales important in the ecology of these sites. This is an important point since dispersal-colonization dynamics are influenced by the orientation of patch types relative to each other and to the environment, the distance between patches, the number and quality of patches, and the dispersal capability of the organism(s) in question (Gadgil, 1971).
The immune system as a source of landscape transformation
Interactions between commensals and the immune system appear to be important in shaping the topography of the human landscape. The magnitude of the immune response appears to vary depending on whether a surface is colonized by a commensal or a pathogen. In mice, nasal colonization by the commensal Lactobacillus murinus induced Th1 immune responses of the nasal cavity to a measurable but lesser degree than Streptococcus pyogenes colonization (Costalonga et al., 2009). Furthermore, germ-free mice have reduced epithelial and mucosal thickness, more collagen, fewer goblet cells, and smaller nasal-associated lymphoid tissue compared to pathogen free mice (Jain et al., 2016).
A baseline level of cell-mediated immune function dramatically shapes the landscape of the oral cavity. Diminshed alveolar bone in the oral cavities of germ-free mice and rats was thought to be paradoxical (Baer and Fitzgerald, 1966) given that bone loss was presumed to follow chronic inflammation triggered by an aberrant subgingival community, as in generalized periodontitis. In an effort to address this paradox, other work identified hallmarks of inflammation – mast cells and basophils – in the junctional epithelia of germ-free animals (Wolf et al., 1973). And recent molecular studies showed that the junctional epithelia constitutively express the pro-inflammatory cytokines IL-1β and TNF-α at comparable levels in germ-free and conventional animals (Tsukamoto et al., 2012). Collectively, these studies imply that the gingiva is subject to low-grade inflammation in the absence of microbial exposure. Moreover, mice reared under conventional conditions were found to upregulate the chemokines, KC/CXCL1 and MIP-2 as compared to germ-free animals (Tsukamoto et al., 2012), suggesting that the commensal microbiota fine-tunes immune function as has been shown in the nose.
Pathogens as disturbance: mechanisms of interplay between host and microbe
Acute infections may be viewed as a ‘pulse disturbance’ when a pathogen directly or indirectly modifies community composition or structure. Viral infections, for example, have been shown to modulate the structure of the nasopharyngeal (NP) microbiota. In healthy children, a single upper respiratory infection (URI) can reduce the phylogenetic diversity of NP communities (Santee et al., 2016). Moreover, children who experience a large number of URIs tend to have lower NP community richness and diversity than children who experience fewer URIs, suggesting a high frequency of disturbance can have persistent effects. Different patterns can be expected to arise as a consequence of different infections; human rhinovirus (HRV) depresses community richness less than does respiratory syncytial virus (RSV) (Rosas-Salazar et al., 2016).
Interestingly, the commensal microbiota affects the likelihood that either a URI or a lower respiratory infection (LRI) will occur. Children with NP communities dominated by Moraxella, Haemophilus or Streptococcus were more likely to experience LRIs when infected with either HRV or RSV as compared to those with other community types (Teo et al., 2015). Moraxella, Haemophilus and Streptococcus, moreover, were independent predictors of acute respiratory symptoms, including fever, suggesting that the invading viral pathogen is not the only organism that modulates immune function and inflammation during acute respiratory infections. S. pneumoniae appears to take advantage of inflammation in viral-induced asymptomatic URI (Glennie et al., 2016). In the presence of virus, S. pneumoniae increased mucosal factor H (FH) but not SLP1 or β-defensin-2 or lactoferrin; high levels of FH in turn induced inflammation allowing S. pneumoniae population size to bloom to a much higher density as compared to individuals with low FH levels. Mechanistically, FH appeared to facilitate the adherence and subsequent internalization of S. pneumoniae in nasopharyngeal epithelial cells where population growth is not restricted by complement-mediated opsonophagocytosis. S. pneumoniae often colonizes the upper respiratory tract in healthy individuals, though this organism does of course cause disease, a transition that may be strongly influenced by acute viral infection. Collectively, these data suggest that acute viral infections modulate microbial community structure and function.
Chronic inflammation may be viewed as a ‘press disturbance’ if it causes long-standing and persistent changes in the composition or structure of microbial communities. One such example is chronic rhinosinusitis (CRS), characterized by prolonged inflammation of the sinuses and a shift from a Th1 to a Th2 response (Aurora et al., 2013). Microbial communities in the paranasal sinuses of CRS patients are less rich and less diverse than commensal communities in healthy individuals (Wagner Mackenzie et al., 2017). Linear discriminant analysis identified the genus Corynebacterium as a potential biomarker that was over-represented in CRS. Abreu et al. has beautifully shown that mice challenged with Corynebacterium tuberculostearicum after antibiotic-mediated depletion of the commensal community developed goblet cell hyperplasia and mucin hypersecretion, two hallmarks of CRS (Abreu et al., 2012). In that work, the commensal community protected against this immunopathology since depletion of the microbiota was required to see the emergence of sinonasal pathology. In stark contrast, another group found that nasal lavage samples of the microbiota collected from patients with CRS, but not healthy controls, stimulated the induction of Il-5 in peripheral leukocytes isolated from healthy controls as well as from the same host (Aurora et al., 2013). This work suggests that chronic inflammatory conditions, such as CRS, represent an altered ecological landscape, one that is both enforced by aberrant immune cells and responses, and reinforced by a dysfunctional microbiota.
In the oral cavity, the prolonged absence of salivary flow induces a press disturbance. The movement of saliva through the mouth underlies the variable exchange rates between whole saliva and plaques on different dental surfaces as well as oral clearance from larger compartments (Dawes, 1989). Not only do such heterogeneities make certain dental surfaces more or less susceptible to demineralization but they also provide a primary basis (e.g., pH) for structuring the biogeography of the oral microbiota. In healthy individuals, dental caries usually takes years to develop but caries can manifest on the timescale of months in individuals with chronic low salivary flow (i.e., hyposalivation) (Sreebny and Valdini, 1988). Moreover, individuals with hyposalivation have more decayed, filled and missing teeth compared to controls even in patient populations that practice ultra-fastidious oral hygiene (Abraham et al., 1998). And, the pattern of caries attack in these individuals shifts from the biting surfaces of teeth in the posterior towards the smooth and root surfaces of teeth in the anterior compartment, sites that are infrequently attacked in otherwise healthy individuals (Dreizen et al., 1977).
A rich history surveying this phenomenon extends back to the 1950s. There is general consensus that hyposalivation selects for caries-associated bacteria such as Lactobacillus spp., Candida albicans and Streptococcus mutans (Almstahl et al., 2003), indicating that the loss of salivary flow represents a sustained ecological disturbance that alters ecosystem function. Given that the pattern of dental caries shifts in a site-specific manner in these individuals relative to healthy controls a natural question is whether or not the loss of salivary flow exerts site-specific effects on oral microbial communities. All extant studies examining the impact of hyposalivation on the microbiota have relied on pooled plaque samples from multiple tooth surfaces or rinsing samples or sampling of just a handful of sites to survey supragingival community structure, thereby obscuring the extent to which shifts in community composition and structure occur, if any, at different biogeographic sites across the dentition.
Perspectives and future directions
An important unanswered question in the field of microbiome research is, what spatial (and temporal) scales are relevant to the bacteria that inhabit the human body? To define scales for the microbiota that are analogous to those defined for macro-ecology sufficiently powered observational studies need to be undertaken with the goal of ascertaining the average size of a single microbial population or community, the spatial extent of patch mosaics, and the scales along which gradients occur on the human body. Community function will be as important to measure as community structure. Once the spatial scales for a given habitat have been determined, it will be possible to describe the types, sizes, and extent of spatial patterns observed in the microbiota. Coupled with perturbation experiments, the underlying processes – stochastic, biotic, disturbance – driving spatial patterns can be elucidated. The nose, mouth and throat are particularly amenable to such lines of inquiry because these habitats are more easily accessible to sample collection as compared to other body sites.
The application of landscape ecology to the field of microbiome research also requires a shift from describing sample sites as categorical variables (e.g., anterior nares, middle meatus) towards thinking of them as georeferenced ones. Obtaining geographic coordinates, as well as a model of the topography of anatomic site, through imaging, would allow investigators to estimate critical ecological parameters, including dispersal distance, defined here as the geometric distance between two patch types. Moreover, having geographic coordinates would enable researchers to test variation in community features as a function of the physical distance separating sites or separating a site with respect to some environmental stressor. Another important and unknown parameter in the ecology of the microbiota is the frequency at which different patches (and patch types) go extinct, as well as the frequency with which they repopulate within and between different gross anatomic sites. Knowledge of the dispersal, colonization and extinction parameters would enable modeling of community dynamics, for example, in the wake of antibiotic disturbance. With these and other studies of landscape ecology in the nose, mouth and throat, a more comprehensive understanding will be acquired of the environmental parameters in health, setting the stage for more mechanistically informed and predictive interventions in disease.
Figure 3. Understanding dispersal across gross anatomic sites.
A) The nasal sinuses drain to the turbinates and similarly receive periodic influxes of mucus. B)The Eustachian tube may be a route for the dispersal of organisms between the nasal cavity, nasopharynx and the middle ear and vice versa. C) The nasal and oral cavities both drain to the pharynx and may serve as sources of colonists to the trachea and subsequently to the lungs. Alternatively, each may seed the stomach and ultimately the intestinal tract via the esophagus.
Theory, methods and principles of landscape ecology enhance our understanding of the spatial scales, patterns and processes that underlie host-microbiota interactions. As examined in this review by Proctor and Relman, the human nose, mouth, and throat are attractive study sites for elucidating microbial biogeography, host physiology and immune function.
Acknowledgments
The authors wish to thank Gary Armitage and Susan Holmes for instrumental guidance as well as an anonymous reviewer for comments that significantly improved the manuscript. This work was supported by the National Institutes of Health [DP1OD000964 and R01DE023113 to D.A.R.], and by the Thomas C. and Joan M. Merigan Endowment at Stanford University [D.A.R.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham CM, al-Hashimi I, Haghighat N. Evaluation of the levels of oral Candida in patients with Sjogren’s syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1998;86:65–68. doi: 10.1016/s1079-2104(98)90151-2. [DOI] [PubMed] [Google Scholar]
- Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, Lynch SV. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almstahl IA, Wikstrom M, Stenberg I, Jakobsson A, Fagerberg-Mohlin B. Oral microbiota associated with hyposalivation of different origins. Oral Microbiol Immunol. 2003;18:1–8. doi: 10.1034/j.1399-302x.2003.180101.x. [DOI] [PubMed] [Google Scholar]
- Aufdemorte TB, Cameron IL. The relation of keratinization to bacterial colonization on the baboon tongue as demonstrated by scanning electron microscopy. J Dent Res. 1981;60:1008–1014. doi: 10.1177/00220345810600060201. [DOI] [PubMed] [Google Scholar]
- Aurora R, Chatterjee D, Hentzleman J, Prasad G, Sindwani R, Sanford T. Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg. 2013;139:1328–1338. doi: 10.1001/jamaoto.2013.5465. [DOI] [PubMed] [Google Scholar]
- Baer N, Fitzgerald RJ. Periodontal disease in the 18-month-old germfree rat. J Dent Res. 1966;45:406. doi: 10.1177/00220345660450023401. [DOI] [PubMed] [Google Scholar]
- Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rumke HC, Verbrugh HA, Hermans PW. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio. 2016;7:e01725–01715. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouslimani A, Porto C, Rath CM, Wang M, Guo Y, Gonzalez A, Berg-Lyon D, Ackermann G, Moeller Christensen GJ, Nakatsuji T, et al. Molecular cartography of the human skin surface in 3D. Proc Natl Acad Sci U S A. 2015;112:E2120–2129. doi: 10.1073/pnas.1424409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B, Proctor D, Relman D, Fukuyama J, Holmes S. Reproducible research workflow in R for the analysis of personalized human microbiome data. Pac Symp Biocomput. 2016a;21:183–194. [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016b;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casado B, Pannell LK, Iadarola P, Baraniuk JN. Identification of human nasal mucous proteins using proteomics. Proteomics. 2005;5:2949–2959. doi: 10.1002/pmic.200401172. [DOI] [PubMed] [Google Scholar]
- Chan CL, Wabnitz D, Bardy JJ, Bassiouni A, Wormald PJ, Vreugde S, Psaltis AJ. The microbiome of otitis media with effusion. Laryngoscope. 2016;126:2844–2851. doi: 10.1002/lary.26128. [DOI] [PubMed] [Google Scholar]
- Chan CL, Wabnitz D, Bassiouni A, Wormald PJ, Vreugde S, Psaltis AJ. Identification of the bacterial reservoirs for the middle ear using phylogenic analysis. JAMA Otolaryngol Head Neck Surg. 2017;143:155–161. doi: 10.1001/jamaoto.2016.3105. [DOI] [PubMed] [Google Scholar]
- Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YW, Vidal JE, Grijalva CG, Bozio C, Edwards KM, Williams JV, Griffin MR, Verastegui H, Hartinger SM, Gil AI, et al. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J. 2013;32:72–77. doi: 10.1097/INF.0b013e318270d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalonga M, Cleary PP, Fischer LA, Zhao Z. Intranasal bacteria induce Th1 but not Treg or Th2. Mucosal Immunol. 2009;2:85–95. doi: 10.1038/mi.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers AJ, Zomer AL, Gritzfeld JF, Ferwerda G, van Hijum SA, Ferreira DM, Shak JR, Klugman KP, Boekhorst J, Timmerman HM, et al. The adult nasopharyngeal microbiome as a determinant of pneumococcal acquisition. Microbiome. 2014;2:44. doi: 10.1186/2049-2618-2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C. An analysis of factors influencing diffusion from dental plaque into a moving film of saliva and the implications for caries. J Dent Res. 1989;68:1483–1488. doi: 10.1177/00220345890680110301. [DOI] [PubMed] [Google Scholar]
- Dawes C, Watanabe S, Biglow-Lecomte P, Dibdin GH. Estimation of the velocity of the salivary film at some different locations in the mouth. J Dent Res. 1989;68:1479–1482. doi: 10.1177/00220345890680110201. [DOI] [PubMed] [Google Scholar]
- Dawes C, Wood CM. The composition of human lip mucous gland secretions. Arch Oral Biol. 1973;18:343–350. doi: 10.1016/0003-9969(73)90157-x. [DOI] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, Curtis JL. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12:821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati C, Zolfo M, Albanese D, Tin Truong D, Asnicar F, Iebba V, Cavalieri D, Jousson O, De Filippo C, Huttenhower C, et al. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nat Microbiol. 2016;1:16070. doi: 10.1038/nmicrobiol.2016.70. [DOI] [PubMed] [Google Scholar]
- Doorly DJ, Taylor DJ, Gambaruto AM, Schroter RC, Tolley N. Nasal architecture: form and flow. Philos Trans A Math Phys Eng Sci. 2008;366:3225–3246. doi: 10.1098/rsta.2008.0083. [DOI] [PubMed] [Google Scholar]
- Dreizen S, Brown LR, Daly TE, Drane JB. Prevention of xerostomia-related dental caries in irradiated cancer patients. J Dent Res. 1977;56:99–104. doi: 10.1177/00220345770560022101. [DOI] [PubMed] [Google Scholar]
- Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM, Silman MR, Knight R. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology. 2011;92:797–804. doi: 10.1890/10-1170.1. [DOI] [PubMed] [Google Scholar]
- Fukami T. Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Ann Rev Ecol Evol Syst. 2015;46:1–23. [Google Scholar]
- Gadgil M. Dispersal: population consequences and evolution. Ecology. 1971;52:253–261. [Google Scholar]
- Glennie S, Gritzfeld JF, Pennington SH, Garner-Jones M, Coombes N, Hopkins MJ, Vadesilho CF, Miyaji EN, Wang D, Wright AD, et al. Modulation of nasopharyngeal innate defenses by viral coinfection predisposes individuals to experimental pneumococcal carriage. Mucosal Immunol. 2016;9:56–67. doi: 10.1038/mi.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwaltney JM, Jr, Hendley JO, Phillips CD, Bass CR, Mygind N, Winther B. Nose blowing propels nasal fluid into the paranasal sinuses. Clin Infect Dis. 2000;30:387–391. doi: 10.1086/313661. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Teles RP, Patel MR, Song X, Yaskell T, Socransky SS. Factors affecting human supragingival biofilm composition. II. Tooth position. J Periodontal Res. 2009;44:520–528. doi: 10.1111/j.1600-0765.2008.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser LJ, Ir D, Kingdom TT, Robertson CE, Frank DN, Ramakrishnan VR. Investigation of bacterial repopulation after sinus surgery and perioperative antibiotics. Int Forum Allergy Rhinol. 2016;6:34–40. doi: 10.1002/alr.21630. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson GE. The Concept of Pattern in Ecology. Proc Acad Nat Sci Phila. 1953;105:1–12. [Google Scholar]
- Jain R, Waldvogel-Thurlow S, Darveau R, Douglas R. Differences in the paranasal sinuses between germ-free and pathogen-free mice. Int Forum Allergy Rhinol. 2016;6:631–637. doi: 10.1002/alr.21712. [DOI] [PubMed] [Google Scholar]
- Ehrenfeld Joan G, XH, Parsons Willian FJ, Zhu Weixing. On the nature of environmental gradients: temporal and spatial variability of soils and vegetation in the New Jersey pinelands. J Ecol. 1997;85:785–798. [Google Scholar]
- Johnson RC, Ellis MW, Lanier JB, Schlett CD, Cui T, Merrell DS. Correlation between nasal microbiome composition and remote purulent skin and soft tissue infections. Infect Immun. 2015;83:802–811. doi: 10.1128/IAI.02664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, Wos-Oxley M, Becker K. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol. 2016;18:2130–2142. doi: 10.1111/1462-2920.12891. [DOI] [PubMed] [Google Scholar]
- Keck T, Leiacker R, Heinrich A, Kuhnemann S, Rettinger G. Humidity and temperature profile in the nasal cavity. Rhinology. 2000;38:167–171. [PubMed] [Google Scholar]
- Kleinberg I, Jenkins GN. The pH of dental plaques in the different areas of the mouth before and after meals and their relationship to the pH and rate of flow of resting saliva. Arch Oral Biol. 1964;9:493–516. doi: 10.1016/0003-9969(64)90015-9. [DOI] [PubMed] [Google Scholar]
- Levin SA. The problem of pattern and scale in ecology: The Robert H. MacArthur Award Lecture. Ecology. 1992;73:1943–1967. [Google Scholar]
- Levin SA, Paine RT. Disturbance, patch formation, and community structure. Proc Natl Acad Sci U S A. 1974;71:2744–2747. doi: 10.1073/pnas.71.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levins R. Some demographic and genetic consequences of environmental heterogeneity for biological control. Am Entomol. 1969;15:237–240. [Google Scholar]
- Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 2005;1:e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113:E791–800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Micro. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Mika M, Mack I, Korten I, Qi W, Aebi S, Frey U, Latzin P, Hilty M. Dynamics of the nasal microbiota in infancy: a prospective cohort study. J Allergy Clin Immunol. 2015;135:905–912. e911. doi: 10.1016/j.jaci.2014.12.1909. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Mitra R, Maitra A, Gupta S, Kumaran S, Chakrabortty A, Majumder PP. Sebum and hydration levels in speciic regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci Rep. 2016;6:36062. doi: 10.1038/srep36062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett ST, Cadenasso ML. Landscape ecology: spatial heterogeneity in ecological systems. Science. 1995;269:331–334. doi: 10.1126/science.269.5222.331. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan VR, Gitomer S, Kofonow JM, Robertson CE, Frank DN. Investigation of sinonasal microbiome spatial organization in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2017;7:16–23. doi: 10.1002/alr.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. The human microbiome: ecosystem resilience and health. Nutr Rev. 2012;70(Suppl 1):S2–9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, Shankar J, Yooseph S, Nelson KE, Halpin RA, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and Respiratory Syncytial Virus in infancy. J Infect Dis. 2016;214:1924–1928. doi: 10.1093/infdis/jiw456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santee CA, Nagalingam NA, Faruqi AA, DeMuri GP, Gern JE, Wald ER, Lynch SV. Nasopharyngeal microbiota composition of children is related to the frequency of upper respiratory infection and acute sinusitis. Microbiome. 2016;4:34. doi: 10.1186/s40168-016-0179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yamagishi J, Yamashita R, Shinozaki N, Ye B, Yamada T, Yamamoto M, Nagasaki M, Tsuboi A. Inter-individual differences in the oral bacteriome are greater than intra-day fluctuations in individuals. PLoS One. 2015;10:e0131607. doi: 10.1371/journal.pone.0131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneyer LH, Levin LK. Rate of secretion by individual salivary gland pairs of man under conditions of reduced exogenous stimulation. J Appl Physiol. 1955;7:508–512. doi: 10.1152/jappl.1955.7.5.508. [DOI] [PubMed] [Google Scholar]
- Simon-Soro A, Tomas I, Cabrera-Rubio R, Catalan MD, Nyvad B, Mira A. Microbial geography of the oral cavity. J Dent Res. 2013;92:616–621. doi: 10.1177/0022034513488119. [DOI] [PubMed] [Google Scholar]
- Sreebny LM, Valdini A. Xerostomia. Part I: Relationship to other oral symptoms and salivary gland hypofunction. Oral Surg Oral Med Oral Pathol. 1988;66:451–458. doi: 10.1016/0030-4220(88)90268-x. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Goktas O, Bessler C, Loening-Baucke V, Hale LP, Andree H, Weizenegger M, Holzl M, Scherer H, Lochs H. Spatial organisation of microbiota in quiescent adenoiditis and tonsillitis. J Clin Pathol. 2007;60:253–260. doi: 10.1136/jcp.2006.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torretta S, Drago L, Marchisio P, Gaffuri M, Clemente IA, Pignataro L. Topographic distribution of biofilm-producing bacteria in adenoid subsites of children with chronic or recurrent middle ear infections. Ann Otol Rhinol Laryngol. 2013;122:109–113. doi: 10.1177/000348941312200206. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Usui M, Yamamoto G, Takagi Y, Tachikawa T, Yamamoto M, Nakamura M. Role of the junctional epithelium in periodontal innate defense and homeostasis. J Periodontal Res. 2012;47:750–757. doi: 10.1111/j.1600-0765.2012.01490.x. [DOI] [PubMed] [Google Scholar]
- Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond B Biol Sci. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MG. Landscape ecology: the effect of pattern on process. Annu Rev Ecol Syst 1989. 1989;20:171–197. [Google Scholar]
- von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- Wagner Mackenzie B, Waite DW, Hoggard M, Douglas RG, Taylor MW, Biswas K. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ Microbiol. 2017;19:381–392. doi: 10.1111/1462-2920.13632. [DOI] [PubMed] [Google Scholar]
- Wiens JA. Landscape mosaics and ecological theory. In: Hansson L, Fahrig L, Merriam G, editors. Mosaic Landscapes and Ecological Processes. London: Chapman & Hall; 1995. [Google Scholar]
- Wiens JA, Stenseth NC, Van Horne B, Ims RA. Ecological Mechanisms and Landscape Ecology. Oikos. 1993;66:369–380. [Google Scholar]
- Wolf JE, App GR, Melfi RC. Mast cells and basophils in the mandibular alveolar connective tissue and bone marrow of germfree albino rats. J Dent Res. 1973;52:1092–1096. doi: 10.1177/00220345730520051801. [DOI] [PubMed] [Google Scholar]
- Wolff M, Kleinberg I. Oral mucosal wetness in hypo- and normosalivators. Arch Oral Biol. 1998;43:455–462. doi: 10.1016/s0003-9969(98)00022-3. [DOI] [PubMed] [Google Scholar]
- Wos-Oxley ML, Chaves-Moreno D, Jauregui R, Oxley AP, Kaspar U, Plumeier I, Kahl S, Rudack C, Becker K, Pieper DH. Exploring the bacterial assemblages along the human nasal passage. Environ Microbiol. 2016;18:2259–2271. doi: 10.1111/1462-2920.13378. [DOI] [PubMed] [Google Scholar]
- Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, Relman DA. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013;14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin M, Baviskar P, Webster R, Webby R. The interaction between respiratory pathogens and mucus. Cell Host Microbe. 2016;19:159–168. doi: 10.1016/j.chom.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Ganzle MG, Lin XB, Ruan L, Sun M. Diversity and dynamics of bacteriocins from human microbiome. Environ Microbiol. 2015;17:2133–2143. doi: 10.1111/1462-2920.12662. [DOI] [PubMed] [Google Scholar]
- Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]