Abstract
The ribosomal proteins play important roles in the growth and development of organisms. This study aimed to explore the function of NlRPL5 (GenBank KX379234), a ribosomal protein L5 gene, in the brown planthopper Nilaparvata lugens. The open reading frame of NlRPL5 was cloned from N. lugens based on a previous transcriptome analysis. The results revealed that the open reading frame of NlRPL5 is of 900 bp, encoding 299 amino acid residues. The reverse transcription quantitative PCR results suggested that the expression of NlRPL5 gene was stronger in gravid females, but was relatively low in nymphs, males, and newly emerged females. The expression level of NlRPL5 in the ovary was about twofolds of that in the head, thorax, or fat body. RNAi of dsNlRPL5 resulted in a significant reduction of mRNA levels, ∼50% decrease in comparison with the dsGFP control at day 6. Treatment of dsNlRPL5 significantly restricted the ovarian development, and decreased the number of eggs laid on the rice (Oryza sativa) plants. This study provided a new clue for further study on the function and regulation mechanism of NlRPL5 in N. lugens.
Keywords: Nilaparvata lugens, NlRPL5 gene, RNA interference, ovarian development
Brown planthopper, Nilaparvata lugens Stål, is one of the most destructive inset pests in the subtropical areas of Asia. N. lugens feeds on rice (Oryza sativa) crop and can cause hopperburn on rice plant, whenever its density is high enough. N. lugens can also transmit the grassy stunt disease, which will further reduce rice yield (Nault and Ammar 1989, Zhu et al. 2004, Bao et al. 2009). Various types of resistant rice varieties have been used to control this insect pest as an effective and environment-friendly strategy for protecting rice (Wang et al. 2015). However, several N. lugens strains have recovered their virulence to these resistant rice varieties due to a continuous cultivation of planthopper-resistant varieties and a high level application of nitrogen fertilizer (Tanaka 1999, Horgan et al. 2016, Rashid et al. 2016). Therefore, it is important to develop the strategy for controlling N. lugens, including identifying some new genes as targets.
In order to screen suitable genes as targets, we analyzed the gene expression profile of N. lugens, and found that the “ribosome” term in KEGG pathway showed significant difference between the Rh colony reared on resistant rice variety Rathu Heenati (RHT) and the Tn colony reared on susceptible variety TN1 (Unpublished data Hao, et al). In this pathway, a ribosomal protein gene L5 (NlRPL5) was upregulated about twofolds in the Rh colony, compared with that in the Tn colony, indicating that NlRPL5 may play a role in the interaction between N. lugens and resistant rice. As several ribosomal proteins including RPL5 have been implicated in stress amelioration besides house-keeping in several species (Grewal et al. 2007, Li et al. 2010, Moin et al. 2016), NlRPL5 gene was selected for further investigation.
Ribosomes are cellular machines essential for protein synthesis. The biogenesis of ribosomes is a highly complex and energy consuming process that initiates in the nucleolus (Goudarzi and Lindström 2016). In recent research, ribosomal proteins were found also involved in functions beyond ribosome, such as transcriptional regulation, cell development, and cell differentiation process (Ferguson et al. 2015, Orelle et al. 2015, Takada and Kurisaki 2015). Ribosomal protein L5 (RPL5) is a part of the 60S ribosomal subunit and is localized in both cytoplasm and nucleus of eukaryotic cells. Acting as a nucleocytoplasmic shuttle protein, RPL5 plays an important role in 5S rRNA intracellular transport during assembly of the large ribosomal subunit (Rosorius et al. 2000). RPL5, together with RPL11, can monitor ribosome biogenesis and control cell proliferation and growth (Liao et al. 2013). Teng et al (2013) found that the loss of RPL5 strongly suppressed cell cycle progression, and the effects on cell cycle progression stemmed from reduced ribosome content and translational capacity, which suppressed the accumulation of cyclins at the translational level. Although ribosomal proteins have been extensively studied, especially in vertebrates and plants, the functions beyond ribosome of NlRPL5 remain largely unclear in N. lugens.
In this study, the complete open reading frame (ORF) of NlRPL5 was cloned, and the physicochemical properties of the protein sequence were predicted. The reverse transcription quantitative PCR (RT-qRCR) analysis was conducted to detect the relative mRNA expression levels of NlRPL5 in different stages and different populations of N. lugens. RNA interference (RNAi) was performed to explore the effect of NlRPL5 knockdown on N. lugens. We aimed to explore the function of NlRPL5 in N. Lugens.
Materials and Methods
Insects and Plants
Rice varieties include susceptible variety TN1 and resistant varieties ASD7 (with a brown planthopper resistance gene, bph2) and RHT (with a brown planthopper resistance gene, Bph3). Test insects were collected from the following N. lugens colonies: Tn colony reared on TN1 continuously for >80 generations; As colony reared on ASD7 continuously for >70 generations; Rh colony reared on RHT continuously for >70 generations; All test insects and plants were maintained in an controlled conditions room at 26 ± 2 °C, and 75–80% relative humidity, with a 16:8 (L:D) h photoperiod.
Isolation of Total RNA and Synthesis of First Strand cDNA
Total RNA was extracted using Takara MiniBEST Universal RNA Extraction Kit cot9767 (Takara, Tokyo, Japan). The RNA quality was determined with a spectrophotometer Nanodrop 2000 (Thermo, Wilmington, DE, USA) and then the integrity of RNA was checked using the 1% agarose gel electrophoresis. About 1 µg RNA was used to synthesize the first-strand cDNA, using the reverse transcription kit PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Tokyo, Japan).
The Full-Length ORF cDNA Amplification of NlRPL5
A sequence predicted as RPL5 gene (particial), obtained from our previous N. lugens transcriptome, was used to do BLAST search in the database of the whole genome of N. lugens (Xue et al. 2014). As a result, a hit scaffold (scaffold3211_7) was selected and submitted to Augustus (http://bioinf.uni-greifswald.de/augustus/submission.php) for the ORF prediction, and Acyrthosiphon pisum was selected as reference specie. The whole ORF and the untranslated regions were predicted firstly, and then primers of NlRPL5-flF and NlRPL5-flR were designed at both ends of the ORF to test the validation of the prediction. All primers were designed using Primer Premier 5.0 software and synthesized by Sunny Biotechnology Co. Ltd. (Shanghai, China). PCR amplification was performed in 50 µl reaction volumes using the following protocol: 95 °C for 4 min, followed by 35 cycles of 95 °C for 30 s, 59 °C for 30 s, 72 °C for 100 s, and a final extension at 72 °C for 10 min. The PCR product was confirmed by electrophoresis in the 1% agarose gel and stained by ethidium bromide, and then purified by the DNA Gel Extraction Kit (Axygen, Union City,CA,USA), cloned into the pMD18-T vector (Takara, Tokyo, Japan) and sent to Sunny Company (Shanghai, China) for sequencing.
Sequence and Structure Analysis of the ORF of NlRPL5
The cloned cDNA sequence of NlRPL5 was analyzed by using DNAMAN 5.0 software. The identity of Amino acid sequences was performed on NCBI by BLAST (http://www.ncbi.nlm.nih.gov/), using the ORF Finder (http://www.ncbi.nlm.nih.gov/orffinder/) to get the sequence information of ORF. Molecular mass of the predicted proteins was calculated using the Compute pI/Mw program available online (http://www.expasy.ch/tools/pi_tool.html).
RT-qPCR Analysis
Specific primer pairs (Table 1) for RT-qPCR analysis were designed based on the cloned sequence of NlRPL5. RT-qPCR was performed in 20 µl reactions that included 10 µl SYBR Premix EX Taq (Takara, Tokyo, Japan), 0.4 µl of each of the forward and reverse primers, 0.4 µl ROX Reference Dye (Takara, Tokyo, Japan), 6.8 µl ddH2O, and 2 µl of cDNA template. PCR cycling conditions were as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 55 °C for 30 s. A melting curve analysis was performed by heating the PCR product from 65 °C to 95 °C to ensure the specificity of the amplified product (Supp Fig. S1 [online only]). The 2-ΔΔCT method (Livak and Schmittgen 2001) was used to calculate the relative quantity of NlRPL5 mRNA, based on a pilot experiment result that the efficiencies of the PCR reactions are almost the same (Actin: E = 100.3%, R2 = 0.999; NlRPL5: E = 100.6%, R2 = 0.998; Supp Fig. S2 [online only]). The mRNA level was normalized using Actin gene as an endogenous control as in previous expression profile analysis (Chen et al. 2010).
Table 1.
Primers used in gene cloning, real-time PCR and dsRNA synthesis
| Use of primers | Primer name | Primer sequence |
|---|---|---|
| cDNA cloning | NlRPL5 -fl-F | CCGTGTTCATATGGTAGGGC |
| NlRPL5 -fl-R | ACAGACATTTACGATTCAGCG | |
| Real-time PCR | QNlRPL5-F | CAAGAGGCGTAGGGAAGGTAAA |
| QNlRPL5-R | CGATACGAGAGTAAGCGATCTGG | |
| QActin-F | TGCGTGACATCAAGGAGAAGC | |
| QActin-R | CCATACCCAAGAAGGAAGGCT | |
| dsRNA synthesis | dsNlRPL5-F |
|
| dsNlRPL5-R |
|
|
| dsGFP-F |
|
|
| dsGFP-R |
|
RNA Interference
For RNAi, dsRNAs were synthesized using the MEGAscript T7 Transcription Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Using the primers listed in Table 1, the cDNA for the dsNlRPL5 synthesis was amplified, and then cloned into the pMD18-T vector (TaKaRa, Tokyo, Japan), transferred into JM109 for sequencing. The ORF of NlRPL5 was amplified using two primers (Table 1) with a T7 promoter sequence. The clone with T7 promoter was screened out, propagated, and extracted for plasmids to prepare dsNlRPL5 templates. The primers of dsGFP were designed according to the sequence of a green fluorescent protein GFP (GenBank AF234298). The dsNlRPL5 and dsGFP fragments did not cover the fragments for RT-qPCR so that it will not affect the detection of NlRPL5 mRNA after RNAi. PCR was performed under the following conditions: 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 90 s; and a final step at 72 °C for 10 min.
Fifth-instar nymphs were used for the injection of RNAi, and each group of dsNlRPL5 or dsGFP includes 120 individuals divided into three replications. In total, 0.15 µg dsNlRPL5 or dsGFP of 1.5 µg µl−1 was injected into each individual under a microscope. After injection, the injected insects were maintained on TN1 rice plant in a controlled conditions room at 26 ± 2 °C for 2, 4, 6, 8 days and then sampled for RT-qPCR using the extracted RNA as template. The RNAi treated insects held on TN1 rice plant for 6 days in a parallel experiment were used for isolating the ovary. Five ovaries in both dsNlRPL5 treated group and dsGFP control group were dissected and taken photo under a stereoscope equipped with a digital image acquisition system (Nikon SMZ1500, Tokyo, Japan). Eggs laid on TN1 rice plants were counted at day 2, 4, 6, and 8.
Data Analysis
The data of mRNA expression level were analyzed by ANOVA (LSD) for different time points of a given population or for different populations. However, data of mRNA expression level and number of eggs from different treatment groups in RNAi experiments were evaluated using t-test.
Results
Sequence Analysis of NlRPL5
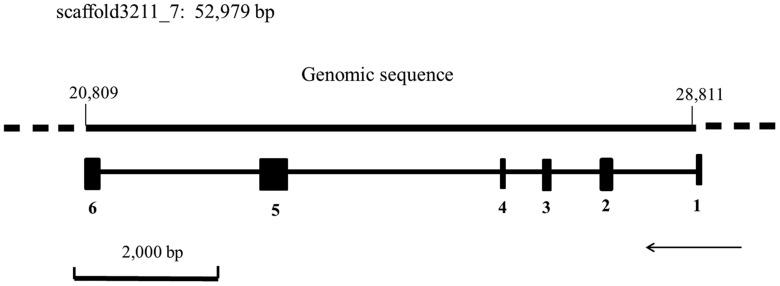
A cDNA sequence of 959 bp predicted as RPL5 was cloned from N. lugens, and named NlRPL5 (GenBank KX379234). The NlRPL5 gene contains six exons and five introns, with an ORF of 900 bp ((Figs. 1 and2). ProtParam (http://web.expasy.org/cgi-bin/protparam/) analysis showed that the sequence encodes a 299-amino acid protein with a calculated molecular weight of 33.9 kDa and its theoretical pI is 9.84. The chemical formula of NlRPL5 protein is C1526H2442N436O427S8. The total number of negatively charged residues (Asp + Glu) is 33, and the total number of positively charged residues (Arg + Lys) is 60. Homology analysis revealed that the NlRPL5 protein is highly conserved in insects, and the deduced amino acid sequence shares a high similarity with the RPL5 of other species, for example, 99% identity with Laodelphax striatella (ADQ73874.1) and 82% identity with Riptortus pedestris (BAN20290.1; Supp Fig. S3 [online only]).
Fig. 1.
Gene structure of NlRPL5 from Nilaparvata lugens. Arrows indicate the direction of the gene transcription; Numbers 1–6 represent exons.
Fig. 2.
cDNA and deduced amino acid sequence of NlLRPL5 gene in N. lugens. The start codon and stop codon are in boldface.
Expression of NlRPL5 in N. lugens at Different Developmental Stages
RT-qPCR revealed that the mRNA expression level of NlRPL5 in gravid female of N. lugens was the highest among all developmental stages analyzed. When the relative expression of NlRPL5 in first–second instar nymph was set as reference (1.0), then the expression of NlRPL5 in gravid female was 1.95, about onefold more than that of the reference. The mRNA expressions of third–fourth instar and fifth instar nymphs were near to that of first–second instar ones. Meanwhile, the expressions of male adult and newly emerged female were relatively low, only about half of that in first–second instar nymphs (Fig. 3).
Fig. 3.
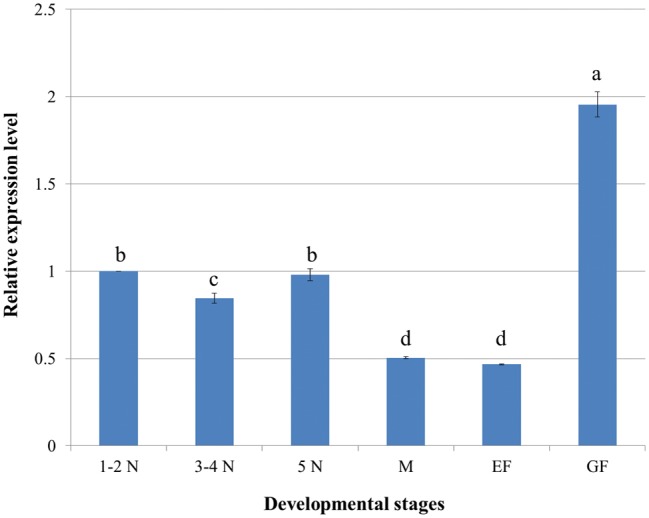
Expression pattern of NlRPL5 in different developmental stages of N. lugens. N. lugens samples were collected from Tn colony. The mRNA level of NlRPL5 was detected using RT-qPCR. The mRNA level was normalized relative to the Actin transcription levels and the reference was the mRNA level of first–second instar nymph. 1–2N: first–second instar nymph; 3–4N: third–fourth instar nymph; 5N: fifth instar nymph; M: Male adult; EF: Newly emerged female adult; GF: Gravid female adult. Data were presented as mean ± SD. Symbols of a, b, c and d indicate significant differences between each two groups (ANOVA, LSD, P < 0.05).
Expression of NlRPL5 in Different Parts and Tissues of N. lugens
RT-qPCR revealed that the mRNA expression level of NlRPL5 in the head, thorax and fat body showed no significant differences between each two groups. Meanwhile, the mRNA expression level of NlRPL5 in the ovary (2.07) was relatively higher, about twofold of that in the head (1.0), suggesting that NlRPL5 may play an important role in ovarian development (Fig. 4).
Fig. 4.
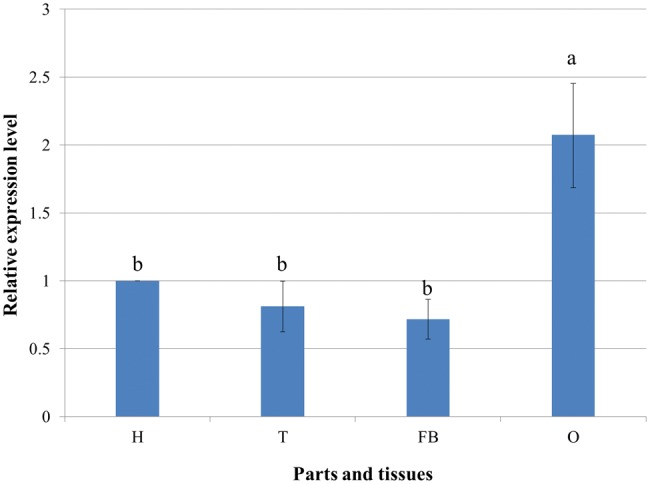
Expression pattern of NlRPL5 in different parts and tissues of N. lugens. The mRNA level of NlRPL5 was detected using RT-qPCR. The mRNA level was normalized relative to the Actin transcription levels and the reference was the mRNA level in head of N. lugens. H: Head; T: Thorax; FB: Fat body; O: Ovary. Data were presented as mean ± SD. Symbols of a and b indicate significant differences between each two groups (ANOVA, LSD, P < 0.05).
Expression of NlRPL5 in N. lugens on Resistant and Susceptible Rice Varieties
As to gravid female N. lugens, the expression levels of NlRPL5 on resistant rice varieties (ASD7 and RHT) were higher than that on susceptible rice variety TN1. When the relative expression of NlRPL5 in N. lugens on susceptible rice varieties TN1 was set as reference (1.0), then the expressions of NlRPL5 in N. lugens on resistant rice varieties were 1.66 (ASD7) and 1.85 (RHT), respectively (Fig. 5). The result suggests that NlRPL5 might be an important responsive gene in N. lugens feeding on resistant rice, and the upregulation of NlRPL5 might contribute to the adaptation of N. lugens to resistant rice.
Fig. 5.
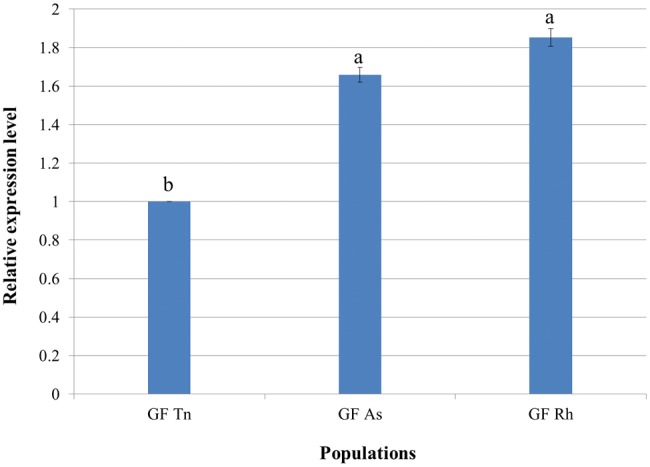
Expression of NlRPL5 in gravid female of different N. lugens populations. The mRNA level of NlRPL5 was detected using RT-qPCR. The mRNA level was normalized relative to Actin and the reference was the mRNA level in head of N. lugens. H: Head; T: Thorax; FB: Fat body; O: Ovary. Data were presented as mean ± SD. Symbols of a, b, and c indicate significant differences between each two groups (ANOVA, LSD, P < 0.05).
Effects of RNAi on mRNA Expression of NlRPL5 and Ovarian Development
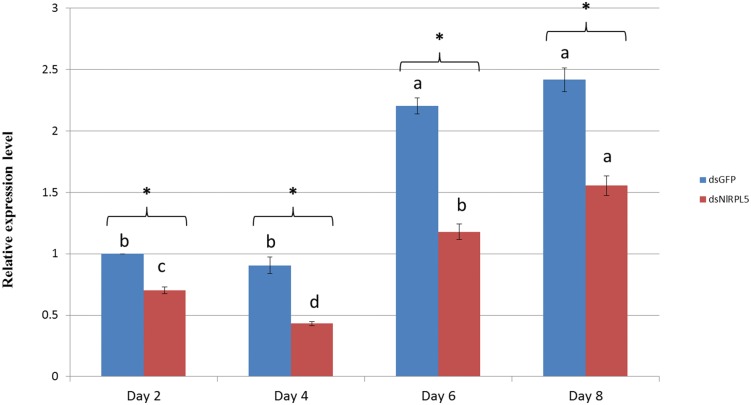
After RNAi was initiated by injecting the insects with dsRNA solution, samples were collected every two days. RT-qPCR results revealed that the mRNA expression of NlRPL5 showed no significant difference between day 2 and day 4, but then clearly increased from day 6 in both dsGFP control group and dsNlRPL5 treated group. It also showed that RNAi with dsNlRPL5 significantly inhibited the expression of NlRPL5, resulting in significantly lower mRNA expression of NlRPL5 in dsNlRPL5 treated groups from day 2 to day 6. At day 4 or 6, a decrease ∼50% was achieved in dsNlRPL5 injected group in contrast to that of dsGFP control group (Fig. 6).
Fig. 6.
Effects of RNAi with NlRPL5 on mRNA expression. The mRNA level of NlRPL5 was detected using RT-qPCR. The mRNA level was normalized relative to the Actin transcription levels and the reference was the mRNA level of dsGFP injected N. lugens on day 2. dsGFP: RNAi treatment of dsGFP (1.5 µg µl−1, total 0.15 µg). dsNLRPL5: RNAi treatment of dsNLRPL5 (1.5 µg µl−1, total 0.15 µg). Data were presented as mean ± SD. Symbols of a, b, c indicate significant differences between each two groups (ANOVA, LSD, P < 0.05). The statistical significance of the gene expression between two different processing samples was evaluated using t-test. Asterisks represent significant differences between controls and treatments at P < 0.05.
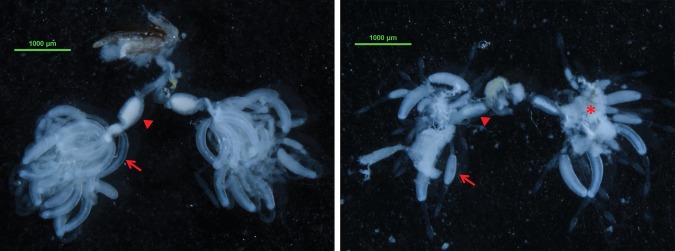
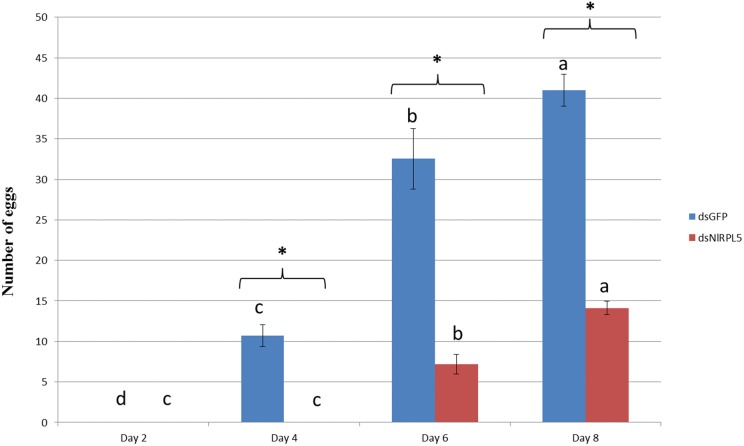
N. lugens was dissected at day 6 to elevate the ovarian development situation. The results showed that the ovaries in the dsGFP control group were well developed, but those in dsNlRPL5 treated group were clearly inhibited with less and smaller eggs (Fig. 7). The number of eggs laid per female of dsNlRPL5 treated group was 14.1 at day 8, significantly less that of dsGFP control 41.0 (Fig. 8). The results suggest that NlRPL5 is important to the ovarian development and female fecundity.
Fig. 7.
Effects of NlRPL5 RNAi on ovarian development. Left, dsGFP treated group; Right, dsNlRPL5 control group. Arrows indicate eggs in ovarian tubules. Arrow heads indicate lateral oviduct. Asterisks indicate fat body tissue.
Fig. 8.
Eggs laid on TN1 rice plants. Data were presented as mean ± SD. Symbols of a, b, c indicate significant differences between each two groups (ANOVA, LSD, P < 0.05). The statistical significance of the egg number between two different processing samples was evaluated using t-test. Asterisks represent significant differences between controls and treatments at P < 0.05.
Discussion
Ribosomal RNAs and ribosomal proteins assemble into ribosomes, which play an important role for the maintenance of normal life activities and biological regulation of organism growth and development (Ferreira-Cerca and Hurt 2009). The regulation of ribosome biogenesis involves the coordination of multiple steps including synthesis and processing of ribosomal RNA (rRNA), synthesis of ribosomal proteins and their import into the nucleus, assembly of ribosome subunits, transport of the mature 40S and 60S subunits to the cytoplasm, and assembly of 80S ribosome in the cytoplasm (Tschochner and Hurt 2003). The ribosomal subunits subsequently bind to and translate mRNAs into polypeptides. In a vigorously growing cell, ribosome biogenesis is a major consumer of cellular energy and resource. Thus, as growth conditions change, cells must rapidly rebalance ribosome production with the availability of resources (Kressle et al. 2010, Tschochner and Hurt 2003). In this study, we found that the expression of NlRPL5 in gravid female was higher than that of other developmental stages, and it was also higher in ovary than other parts of N. lugens. These results indicated that the higher expression of NlRPL5 in the ovary of gravid female may be due to the fast cell proliferation and a large demand for protein synthesis. In this process, NlRPL5 plays role probably in a normal way, being assembled into ribosome to translate enough proteins. Under RNA interference, the ovarian development in dsNlRPL5 group was seriously inhibited, indicating that knockdown of NlRPL5 may interfere the balance between ribosomal RNA and ribosomal protein, and subsequently affect the assembling of ribosome and the translation of protein.
In the state of nutritional deficiencies, organisms will inhibit the synthesis of ribosomal RNA, which may break the balance between ribosomal proteins and ribosomal RNA, causing the ribosome stress (Grummt et al. 1976, Grewal et al. 2007, Warner and McIntosh 2009, Kim et al. 2014, Liu et al. 2014). Impaired ribosome assembly generates a feedback signal to cell cycle regulators, resulting in cell cycle arrest or apoptosis (Kressle and Bassler 2010). In this study, NlRPL5 transcription was upregulated in N. lugens fed on resistant rice varieties, which seems unreasonable given that the N. lugens females usually show lower fecundity on the resistant rice than that on the susceptible one. However, when consider that ribosomal proteins also play roles beyond the ribosome, for example, slowing down the cell cycle to optimize allocation of resources and energy, it is reasonable to suggest that the upregulation of NlRPL5 may contribute to the adaptation of N. lugens on resistant rice. When N. lugens feeds on resistant rice, the condition of food and nutrition is different from that on susceptible rice (Wang et al. 2012). Generally, resistant rice varieties have lower levels of free amino acids, higher levels of phenolic compounds, and lower levels of reducing sugars (Das 1976. Grayer et al. 1994, Thayumanavan 1990, Jung and Im 2005, Chen et al. 2011). Although the exact mechanism of rice resistance to N. lugens has not been completely demonstrated yet, at least several mainstream views below should be concerned. First, the nutrition, especially the variation in amino acid composition and abundance, may affect N. lugens fitness and development as well (Chino et al. 1987, Sogawa and Pathak 1970, Chen et al. 2011). On nitrogen-deficient or resistant rice plants, N. lugens excreted less honeydew, and most of the amino acids in honeydew were significantly decreased, compared with those fed on susceptible TN1 plants (Peng et al. 2016). On the contrary, application of a nitrogen fertilizer can increase the free amino acids available in the phloem sap, which may affect resistance of rice plants to N. lugens (Jairin et al. 2017). Second, the inhibiting factors in the phloem sap will also cause nutritional deficiency to N. lugens. Some secondary metabolites, such as flavanoids, were found different in resistant and susceptible rice varieties, and can inhibit the feeding behavior of N. lugens at concentrations high enough (Grayer et al. 1994, Stevenson et al. 1996). N. lugens feeding on the resistant variety RHT generally ingested lower quantity of phloem sap, which indicated that the N. lugens was in nutritional deficiency states (Ghaffar et al. 2011, Peñalver et al. 2011, Xiao et al. 2016). Third, the endosymbionts are suggested to have evolved to complement the amino acids needs of their host, so that N. lugens can survive on a nutritionally imbalanced food source (Chen et al. 2011, Xue et al. 2014). On the other hand, if the endosymbionts were inhibited by some components in resistant rice, N. lugens may be in amino acids starvation. Therefore, when N. lugens feeds on resistant rice varieties, it will probably be faced to nutrition deficiency directly or indirectly.
Under the conditions of nutrition deficiency or stress, such as feeding on the resistant rice varieties, N. lugens may be encountered with a ribosome stress, and up-regulates the expression of NlRPL5 gene, so that it can meet the energy and resource demand for survival. In this process, NlRPL5 should play its role in a different way, by regulating adaptation-related genes expression rather than translating more protein as it does in the ovarian development. Similarly, in Drosophila, it was found that starvation for dietary amino acids leads to dynamic changes in transcript levels of ribosome pathway (Li et al. 2010). Ribosomal genes also enhanced in the resistance of the whitefly Bemisia tabaci to phenylproapnoids/flavonoids pathway (Alon et al. 2012). Lior et al. (2014) found when the cell was faced to the ribosome stress, the expression of RPL5 also upregulated. Although, at present, there is very few information available about RPL5 participates in regulating the adaptation of insect to its resistant host, there still are some clues for us to suggest that NIRPL5 may play roles in: (1) monitoring the ribosome biogenesis and inhibiting the ovarian development to reduce the energy and resource consumption, probably by miRNAs just as the ribosomal proteins L5 and L11 do, co-operatively inactivate the transcription factors Myc via RNA-induced silencing complex (Liao et al. 2013); (2) playing a moonlighting role in the unfolded protein response (UPR) signaling to maintain cellular protein homeostasis and reutilize amino acids (Anshu and Dey 2015); (3) mobilizing the resources and energy, for example, as it was reported that free ribosomal proteins play roles in promoting lipolysis, enhancing β oxidation of mitochondrial fatty acids, and activating ATP production (Liu et al. 2014). Take together, the results of this work suggest that nutritional status of the resistant rice potentially modulate the molecular biology and biology of the N. lugens, in turn, NIRPL5 participates in the regulation of N. lugens adaptation to resistant rice. This should have profound ramifications in terms of insect biology and pest control on both the basic and applied levels.
Future work need to be undertaken to demonstrate that NlRPL5 actually contributes to the adaptation of N. lugens to resistant rice varieties, and to reveal how NlRPL5 plays its role in this process. Therefore, NlRPL5 protein and its antibody need to be prepared, so that effects on NIRPL5 at the protein level can be investigated in order to establish a functional relationship between NIRPL5 and ovarian development. It is also necessary to identify the possible nutrition/resistant factors in rice which can induce NlRPL5 upregulation. For this purpose, feeding experiments need to be conducted by changing the amount of certain amino acid in the artificial diet, or adding potential metabolites/flavonoids into the diet. Currently, it is difficult to generate genetically modified insect of N. lugens to explore whether the overexpression of NlRPL5 contributes to the ability to overcome resistant rice varieties, alternatively, overexpressing NIRPL5 in the culture cell should be more practicable. With cell culture systems, further investigations, such as subcellular location of NlRPL5 protein, cell division, UPR signaling, activity of mitochondria, and ATP production, are able to be conducted, and thus the regulation mechanism of NlRPL5 will be better understood.
In this study, we cloned the cDNA of NlRPL5 from N. lugens and analyzed it functions by using RT-qPCR and RNAi. The results suggest that the NlRPL5 might play roles in regulating the development of N. lugens, and participating in the adaptation to the host. From this point of view, the NlRPL5 gene has some potential to be developed as a target for controlling N. lugens. In the future, RNAi of NlRPL5 may be undertaken by dsRNA-transgenic plant-mediated RNA interference (Zha et al. 2011), so that a more effective and lasting RNAi to N. lugens can be achieved.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grant numbers 31672026, 31171860, 31501632]; the 973 Program for Key Basic Research of the Ministry of Science and Technology, China [grant numbers 2012CB114100, 2011CB111602]; and the Research on Public Welfare Technology Application Projects of Zhejiang Province, China [grant number 2012C22041].
References Cited
- Alon M., Elbaz M., BenZvi M. M., Feldmesser E., Vainstein A., Morin S. 2012. Insights into the transcriptomics of polyphagy: Bemisia tabaci adaptability to phenylpropanoids involves coordinated expression of defense and metabolic genes. J. Insect Biochem. Mol. Biol. 42: 251–263. [DOI] [PubMed] [Google Scholar]
- Anshu F. A., Dey M. 2015. Ribosomal protein L5 plays a moonlighting role in HAC1 mRNA. FASEB J. 29: 1–2.25561464 [Google Scholar]
- Bao H. B., Hua L. S., Hua G. J., Zhe W. X., Liang L. X., Wen L. Z. 2009. Sublethal effects of four insecticides on the reproduction and wing formation of brown planthopper, Nilaparvata lugens. J. Pest Manage. Sci. 65: 170–174. [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang D., Yao Q., Zhang J., Dong X., Tian H., Chen J., Zhang W. 2010. Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 19: 777–786. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Bernal C. C., Tan J., Horgan F. G., Fitzgerald M. A. 2011. Planthopper “adaptation” to resistant rice varieties: changes in amino acid composition over time. J. Insect Physiol. 57: 1375–1384. [DOI] [PubMed] [Google Scholar]
- Chino M., Hayashi H., Fujumorita T. 1987. Chemical composition of rice phloem sap and its fluctuation. J. Plant Nutr. 10: 1651–1661. [Google Scholar]
- Das Y. T. 1976. Some factors of resistance to Chilo-suppressalis in rice varieties. J. Entomol. Exp. Appl. 20: 131–134. [Google Scholar]
- Ferguson A., Yi W. L., Roger A., Daniel T. S., Manuel J. F., Benjamin B. J., Jose A. L., Randall D. A., Matthew P. M., Theresa V. C., et al. 2015. Functional dynamics within the human ribosome regulate the rate of active protein synthesis. J. Mol. Cell. 60: 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffar M.A.B., Prichard J., Ford-Lloyd B. 2011. Brown planthopper (N. lugens Stal) feeding behaviour on rice germplasm as an indicator of resistance. J. PLoS ONE. 6: e22137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi K. M., Lindström M. S. 2016. Role of ribosomal protein mutations in tumor development. Int. J. Oncol. 48: 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayer R. G., Kimmins F. M., Stevenson P. C., Harborne J. B., Wijayagunesekera M. 1994. Phenolics in rice phloem sap as sucking deterrents to the brown planthopper, (Nilaparvata lugens). J. Acta Horticult. 381: 691–694. [Google Scholar]
- Grewal S. S., Evans J. R., Edgar B. A. 2007. Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J. Cell Biol. 179: 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Smith V. A., Grummt F. 1976. Amino acid starvation affects the initiation frequency of nucleolar RNA polymerase. J. Cell. 7: 439–445. [DOI] [PubMed] [Google Scholar]
- Horgan F. G., Srinivasan T. S., Naik B. S., Ramal A. F., Bernal C. C., Almazan M.L.P. 2016. Effects of nitrogen on egg-laying inhibition and ovicidal response in planthopper-resistant rice varieties. Crop Protect. 89: 223e230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairin J., Teangdeerith, Leelagud S.P.K., Phengrat A., Vanavichit, Toojinda T. 2007. Physical mapping of Bph3, a brown planthopper resistance locus in rice. 01: 166–177. [Google Scholar]
- Jung J. K., Im D. J. 2005. Feeding inhibition of the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae) on a resistant rice variety. J. Asia-Pacific Entomol. 8: 301–308. [Google Scholar]
- Kim T. H., Leslie P., Zhang Y. 2014. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. J. Oncotarget. 5: 860–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressle D., Hurt E., Bassler J. 2010. Driving ribosome assembly. J. Biochim. Biophys. Acta. 1803: 673–683. [DOI] [PubMed] [Google Scholar]
- Li L., Edgar B. A., Grewal S. S. 2010. Nutritional control of gene expression in Drosophila larvae via TOR, Myc and a novel cisregulatory element. J. BMC Cell Biol. 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Zhou M. X., Gatignol A., Lu H. 2014. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. J. Oncogene. 33:4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior G., Sinisa V., Moshe O. 2014. p53 and ribosome biogenesis stress: the essentials. J. FEBS Lett. 588: 2571–2579. [DOI] [PubMed] [Google Scholar]
- Liu Y., He Y. Z., Aiwen J., Andrey P. T., Zhou L. S., Laura A. T., Patrick L., Kim T., Lei O., Rosalind A. C., et al. 2014. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. U. S. A. 111: 2414–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. J. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Moin M., Bakshi A., Saha A., Dutta M., Madhav S. M., Kirti P. B. 2016. Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. J. Front. Plant Sci. 7: 1284.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault L. R., Ammar E. D. 1989. Leafhopper and planthopper transmission of plant viruses. J. Annu. Rev. Entomol. 34: 503–529. [Google Scholar]
- Orelle C., Carlson E. D., Szal T., Florin T., Jewett M. C., Mankin A. S. 2015. Protein synthesis by ribosomes with tethered subunits. J. Nat. 7563: 119–124. [DOI] [PubMed] [Google Scholar]
- Peñalver C. A., Arida A., Heong K. L., Horgan F. G. 2011. Aspects of brown planthopper adaptation to resistant rice varieties with the Bph3 gene. J. Entomol. Exp. Appl. 141: 245–257. [Google Scholar]
- Peng L., Zhao Y., Wang H., Zhang J., Song H., Shangguan X., Zhu L., He G. 2016. Comparative metabolomics of the interaction between rice and the brown planthopper. J. Metab. 12: 1–15. [Google Scholar]
- Rashid M. M., Jahan M., Islam K. S. 2016. Response of adult brown planthopper Nilaparvata lugens (Stål) to rice nutrient management. Neotrop. Entomol. 45: 588–596. [DOI] [PubMed] [Google Scholar]
- Rosorius O., Fries B., Stauber R. H., Hirschmann N., Bevec D., Hauber J. 2000. Human ribosomal protein L5 contains defined nuclear localization and export signals. J. Biol. Chem. 275: 12061–12068. [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S., Hurt E. 2009. Cell biology: arrest by ribosome. J. Nat. 459: 46–47. [DOI] [PubMed] [Google Scholar]
- Sogawa K., Pathak M. D. 1970. Mechanisms of brown planthopper resistance in Mudgo variety of rice (Hemiptera: Delphacidae). J. Appl. Entomol. Zool. 5: 145–158. [Google Scholar]
- Stevenson P. C., Kimmins F. M., Grayer R. J., Raveendranath S. 1996. Schaftosides from rice phloem as feeding inhibitors and resistance factors to brown planthoppers, Nilaparvata lugens. J. Entomol. Exp. Appl. 80: 246–249. [Google Scholar]
- Takada H., Kurisaki A. 2015. Emerging roles of nucleolar and ribosomal proteins in cancer, development, and aging. J. Cell. Mol. Life Sci. 72: 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. 1999. Quantitative genetic analysis of biotypes of the brown planthopper Nilaparvata lugens: heritability of virulence to resistant rice varieties. J. Entomol. Exp. Appl. 90: 279–287. [Google Scholar]
- Teng T., Carol A. M., Philip H., George T., Stefano F. 2013. Loss of tumor suppressor RPL5/RPL11 does not induce cell cycle arrest but impedes proliferation due to reduced ribosome content and translation capacity. J. Mol. Cell. Biol. 33: 4660–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayumanavan B. 1990. Phenolic compounds reducing sugars and free amino acids in rice leaves of varieties resistant to rice thrips. J. Int. Rice Res. Newslett. 11: 14–15. [Google Scholar]
- Tschochner H., Hurt E. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. J. Trends Cell Biol. 13: 255–263. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang M., Feng F., He R. 2015. Differentially regulated genes in the salivary glands of brown planthopper after feeding in resistant versus susceptible rice varieties. J. Arch. Insect Biochem. Physiol. 89: 69–86. [DOI] [PubMed] [Google Scholar]
- Wang Y. B., Guo H. M., Li H. C., Zhang H., Miao X. X. 2012. Identification of transcription factors potential related to brown planthopper resistance in rice via microarray expression profiling. J. BMC Genomics. 13: 687.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R., McIntosh K. B. 2009. How common are extraribosomal functions of ribosomal proteins? J. Mol. Cell. 34: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Hu J., Ao Y. T., Cheng M. X., Gao G. J., Zhang Q. L., He G. C., He Y. Q. 2016. Development and evaluation of near-isogenic lines for brown planthopper resistance in rice cv.9311. J. Sci. Rep. 6: 38159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J. X., Zhou, Zhang C. X., Yu L. L., Fan H. W., Wang Z., Xu H. J., Xi Y., Zhu Z. R., Zhou W. W., et al. 2014. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. J. Genome Biol. 15: 521.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha W., Peng X., Chen R., Du B., Zhu L., He G. 2011. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS ONE. 6: e20504.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. R., Cheng J. A., Jiang M. X., Zhang X. X. 2004. Complex influence of rice variety, fertilization timing and insecticide on population dynamics of Sogatella furcifera (Horvath), Nilaparvata lugens (Stål) (Homoptera: Delphacidae) and their natural enemies in rice in Hangzhou, China. J. Pest Sci. 77: 65–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.