Abstract
In large part, cancer results from the accumulation of multiple mutations in a single cell lineage that are sequentially acquired and subject to an evolutionary process where selection drives the expansion of more fit subclones. Due to the technical challenge of distinguishing and isolating distinct cancer subclones, many aspects of this clonal evolution are poorly understood, including the diversity of different subclones in an individual cancer, the nature of the subclones contributing to relapse, and the identity of pre-cancerous mutations. These issues are not just important to our understanding of cancer biology, but are also clinically important given the need to understand the nature of subclones responsible for the refractory and relapsed disease that cause significant morbidity and mortality in patients. Recently, advanced genomic techniques have been used to investigate clonal diversity and evolution in acute leukemia. Studies of pediatric acute lymphoblastic leukemia (ALL) demonstrated that in individual patients there are multiple genetic subclones of leukemia-initiating cells, with a complex clonal architecture. Separate studies also investigating pediatric ALL determined that the clonal basis of relapse was variable and complex with relapse often evolving from a clone ancestral to the predominant de novo leukemia clone. Additional studies in both ALL and acute myeloid leukemia (AML) have identified pre-leukemic mutations in some individual cases. This review will highlight these recent reports investigating the clonal evolution of acute leukemia genomes and discuss the implications for clinical therapy.
Introduction
Technological advances in genomics have fundamentally redefined what is possible in cancer research. Ten years after the first draft human genome, hundreds of cancer genomes have now been sequenced (1). For the first time, scientists and clinicians can generate complete catalogs of sequence and structural mutations in a given cancer genome (1, 2). Indeed, whole-genome sequencing has already proved valuable in the clinical diagnosis of individual leukemia patients (3, 4). At the population level, the systematic analysis of mutations and copy number alterations (CNA) in many cancer genomes has helped define the genes recurrently involved in a given malignancy. These new experimental possibilities enable traction on questions that have been unresolved in the field of cancer biology for decades.
In large part, cancer results from the accumulation of multiple mutations in a single cell lineage (5). Clonal progression is an evolutionary process in which mutations provide genetic diversity within a cell lineage and selection drives the expansion of more fit subclones (Figure 1) (6). In order to observe clonal evolution, cancer subclones must be distinguished, which has been a major technical challenge. Examples of overcoming this challenge have been informative, but few in number; thus, many aspects of clonal evolution are unknown. How diverse and numerous are different subclones of an individual cancer? Which subclones contribute to relapse? What are the minimum mutations necessary to result in the cancer? Which of these mutations are pre-cancerous? What is the order in which these mutations are acquired?
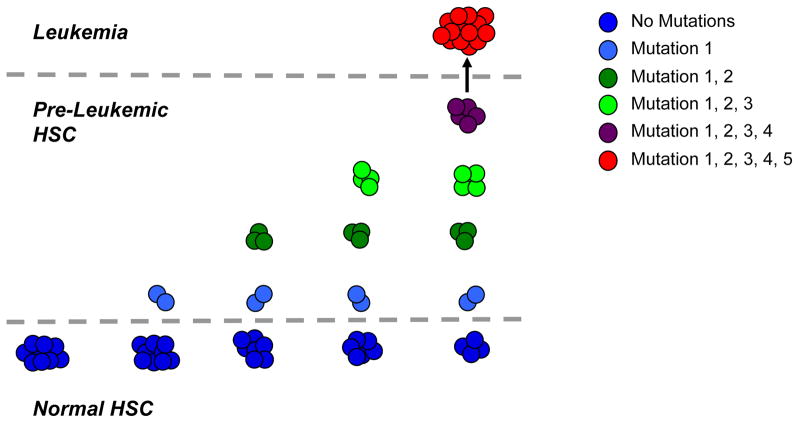
Figure 1. Model for the Clonal Progression of HSC into a Frankly Leukemic Clone.
Genomic and functional studies have demonstrated that multiple mutations are necessary to transform normal cells into a leukemic clone (here depicted as 5 mutations). In the case of acute leukemia, these mutations occur in hematopoietic cells in the bone marrow, most of which are short-lived. As hematopoietic stem cells (HSC) are the only self-renewing cells among bone marrow progenitors, a model has been proposed that mutations must sequentially accumulate within distinct clones of HSC over time (x-axis), eventually resulting in the generation of a frankly leukemic clone (y-axis). According to this model, the HSC compartment at the time of diagnosis is heterogeneous with both genetically normal HSC and numerous intermediate pre-leukemic HSC clones, possessing some, but not all of the mutations found in the leukemic clone.
These questions are not just important to our understanding of cancer biology, but are also important clinically. Indeed, identifying the subclonal origin of relapsed disease may shift the target of therapy to include both the dominant diagnostic clone and a potentially minor relapse clone. Moreover, rare pre-cancerous clones may contribute to relapse through the acquisition of new mutations, suggesting that these cells may also need to be targeted for cure. One consequence of the emerging capacity for deep characterization of cancer genomes is the ability to discriminate subclones by using acquired mutations as genetic markers. With this technological advance, further insight into clonal evolution is now possible. This review will highlight recent reports investigating the clonal evolution of acute leukemia genomes and discuss the implications for clinical therapies.
Acute Myeloid Leukemia
Acute myeloid leukemia (AML) is an aggressive malignancy of the bone marrow with a five-year overall survival between 30% and 40% (7, 8). AML is predominantly a disease of adults, and patients over age 65 have especially poor outcomes (7, 8). Prior to the advent of genome-scale sequencing, a great deal had been learned about somatically acquired genetic abnormalities and mutations in AML. Recurrent cytogenetic abnormalities have been identified in AML including translocations resulting in oncogenic fusion proteins and chromosome copy number variants, many of which are prognostic for patients (9, 10). However, in the majority of patients, AML blasts have a normal karyotype (NK-AML). Investigation of these cases has led to the identification of a number of recurrent molecular mutations, many of which are prognostic among NK-AML patients (11, 12).
Investigation of the nature of these mutations suggested that they could be classified into two complementation groups: (1) mutations that impair normal differentiation, and (2) mutations that increase proliferation and/or impair cell death (13). Analysis of multiple AML cases determined that an individual leukemia often has one ‘differentiation’ mutation and one ‘growth’ mutation, but will rarely have multiple mutations within a single complementation group (13). These results suggested a two-hit model for the development of AML.
This two-hit model has undergone significant revision due to recent AML genome and exome sequencing studies. The first complete cancer genome to be reported was from a patient with AML (14). This work advanced computational methods for the detection of somatic mutations by short-read genome re-sequencing and heralded the current era of explosive growth in cancer genome sequencing. In three separate reports, this group detailed results from two AML patients, which were primarily focused on analysis of single nucleotide polymorphisms (SNPs) and small insertions or deletions (Indels) in the protein coding sequences or exons (14–16). In these cases, 10–12 mutations were identified, and in each, 2 were known to be recurrent in AML. The remaining 8–10 mutated genes were sequenced in a large cohort of AML specimens, and only two, in IDH1 and DNMT3A, were found to be recurrent (14–16). On average, it appears that AML genomes contain approximately 10 mutations that disrupt protein sequences, a minority of which are proving to be recurrent, and therefore likely to be “driver” mutations involved in leukemogenesis. It remains to be seen if the remainder are either “driver” mutations specific to individual patients or are “passenger” mutations retained in the leukemic clone, but not involved in disease pathogenesis.
Given that most whole genome sequencing analysis has focused on protein coding exons, targeted exome capture has been used to focus sequencing analyses and costs on this 1% of the genome (17, 18). Targeted exome sequencing of AML M5 patients identified five recurrent mutations including DNMT3A (19). Several other recurrent mutations have recently been identified through investigation of karyotypic abnormalities in myeloid malignancies including TET2 (20) and EZH2 (21). Interestingly, many of the newly identified mutations occur in genes involved in the regulation of the epigenome, and are often mutually exclusive as evidenced by the fact that mutations in TET2 and IDH1/2 are not detected in the same patient (22). These results suggest that a third complementation group of mutations in epigenome regulators should be added to the two-hit model described above.
Acute Lymphoblastic Leukemia
Acute lymphoblastic leukemia (ALL) is the second major aggressive malignancy of the bone marrow and is predominantly a disease of children. Precursor B-ALL comprises the majority of cases, and treatment with combination chemotherapy results in a five-year event-free survival of 80% for children and 40% for adults (23). Like AML, multiple recurrent cytogenetic abnormalities have been identified in B-ALL, including t(12;21) resulting in a TEL-AML1 fusion, t(9;22) resulting in a BCR-ABL fusion, several translocations involving the MLL gene, and global chromosome copy number changes. As with AML, these cytogenetic abnormalities specify prognosis and response to chemotherapy regimens (23).
Beyond cytogenetics, understanding of B-ALL pathogenesis has been greatly advanced through the recent identification of multiple recurrent genetic mutations in B-ALL. This advance was initiated through global analyses of somatic copy number alterations (CNA) by SNP microarray in 242 cases of pediatric ALL, which identified an average of 6.5 CNA per patient (24). These results are in contrast to similar analyses of AML, where the mean number of CNA per patient was 2.3 (25). Investigation of the loci with recurrent CNA determined that approximately 40% of B-ALL cases contained mutations in B-lineage specification genes, most frequently PAX5, EBF1, and IKZF1 (24). A follow-up study identified mutation of IKZF1 as a near-obligate event, occurring in 84% of cases, in BCR-ABL B-ALL, and as a mutation acquired during the progression from chronic phase chronic myeloid leukemia (CML) to lymphoid blast crisis (26). Whole genome sequencing is yet to be reported in ALL, however, a candidate gene re-sequencing study of matched diagnosis and relapse samples identified CREBBP as a gene mutated in 18% of B-ALL cases that relapsed, but was essentially unmutated in cases that did not relapse (27). Together, the recent identification of genetic abnormalities in AML and ALL at the molecular and single gene level not only provide a greater understanding of the pathogenesis of these diseases, but also provide the genetic markers necessary to begin to advance our understanding of clonal evolution in acute leukemia.
The Genetic Diversity of Leukemia Subclones
One of the most important questions in understanding clonal progression in leukemia is to determine the nature and number of different subclones within an individual cancer. In order to approach this question, distinct genetic subclones must be distinguished, which is now possible with the recent identification of CNA abnormalities in ALL. Two recent publications utilized CNA and/or chromosomal structural abnormalities as genetic markers to discriminate subclones in pediatric ALL, both demonstrating that in individual patients there are multiple genetic subclones of leukemia-initiating cells, with a complex clonal architecture (Figure 2).
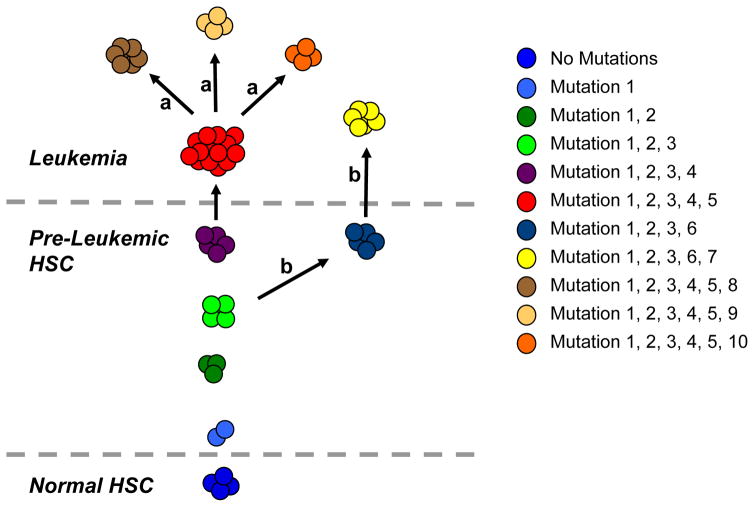
Figure 2. Model for Clonal Heterogeneity in Acute Leukemia.
Recent evidence has demonstrated that in some cases of acute leukemia, multiple genetic subclones of leukemia-initiating cells exist within a complex clonal architecture. This clonal heterogeneity usually consists of distinct subpopulations with a dominant leukemic clone (red). There are several possibilities for the generation of the additional leukemic clones. (a) The dominant clone can undergo additional mutational events to generate distinct daughter subclones (brown, tan, and orange). (b) Alternatively, a common pre-leukemic clone can undergo divergent evolution to generate a distinct, but related, leukemic clone (yellow). Ultimately, the total population of leukemic cells consists of these multiple subclones with a complex genetic relationship, and both common and divergent mutations.
In the first paper, multicolor fluorescence in situ hybridization (FISH) was used to track multiple genetic abnormalities identified in bulk ALL cells, yielding quantitative single cell resolution of the relative frequency of genetically distinct leukemia subclones (28). The authors investigated 30 cases of pediatric ALL with the TEL-AML1 translocation, a pre-natal and putative founding mutation (29), and additional deletions of the remaining TEL allele, CDKN2A, and/or PAX5. By applying multicolor FISH for these genetic abnormalities, multiple subclones were identified in each case with diverse patterns and frequencies, which are likely to be an underestimate given the further subclonal heterogeneity that surely exists from single nucleotide variants not sampled in this study. In these cases, minor subclones represented at least 14% of leukemic cells from any patient. Furthermore, 80% of cases contained non-linear branched hierarchies of subclones with no stereotypic order of recurrent CNA. Moreover, subclones with more CNA were not necessarily more prevalent than those with fewer abnormalities. Importantly, some relationships between subclones could only be explained by multiple independent CNA affecting a single gene, suggesting that oncogenic mutations may occur more frequently than is often assumed. Lastly, a similar pattern of subclonal diversity was observed in leukemia cells engrafted into immunodeficient mice, establishing the genetic heterogeneity of leukemia-initiating/propagating ALL cells from a single patient. One caveat to be noted in assessing the data from this report, is that FISH is a low-resolution tool to indicate clonal relationships, and it is possible that cells with similar FISH patterns have distinct mutations at the DNA sequence level. Indeed, the authors concluded that in some cases, multiple independent events occurred at the same locus in different subclones.
In the second study, CNA analysis by SNP array in conjunction with the xenotransplantation assay were used to dissect genetically distinct leukemia subclones in BCR-ABL-positive ALL (30). Primary and secondary xenografts were established for multiple cases, and engrafted cells along with the primary patient sample were profiled for CNA. Interestingly, 50% of cases engrafted leukemia clonally related to the dominant leukemic clone based on shared CNA and antigen receptor (AgR) rearrangements, but contained some divergent CNA. Moreover, limiting dilution analyses further identified subclones lacking a competitive advantage, formally demonstrating the genetic heterogeneity of leukemia-initiating cells. Although CNA discovered in xenografts were not identified by PCR or other methods in the initial patient sample, the authors argued that these mutations were likely present as a rare subclone (as opposed to emerging during the course of the xenograft experiment) because novel CNA were often found in multiple xenografts from the same patient sample. Finally, in two cases, analysis of shared and distinct CNA yielded evolutionary relationships among leukemia-initiating subclones that were non-linear and branched, similar to those observed with TEL-AML1 ALL. Together, these two ground-breaking studies illustrate the complex evolutionary history of ALL and definitively demonstrate the genetic diversity of leukemia-initating subclones from individual patients.
The Clonal Origin of Relapse
The increased knowledge of individual gene mutations in acute leukemia has made it possible to investigate the clonal origin of relapse. The first study to address this issue used genome-wide analyses of CNA in 61 paired diagnostic and relapse pediatric ALL samples to describe the evolutionary relationship between the predominant diagnostic and relapse leukemic clones (31). The authors determined that 6% of relapse clones were genetically distinct, 8% carried identical CNA, and 34% evolved from the diagnostic clone to acquire new CNA. Importantly, 52% of relapse clones shared some, but not all CNA identified in the diagnostic clone, suggesting that relapse most often evolves from a clone ancestral to the predominant de novo leukemia clone (31). Moreover, most relapse-specific CNA were identified in the diagnostic specimen at a subclonal level using PCR, suggesting both that the leukemia at diagnosis contains genetically diverse subclones and that therapy drives the selection of the eventual dominant relapse clone (31). Several other recent reports utilized a similar genome-wide investigation of CNA in paired diagnostic and relapse samples, and also found that there are multiple clonal evolutionary paths to relapse, although the majority of cases shared some CNA with the dominant diagnostic clone (32–34). In one of these reports investigating patients with TEL-AML1 ALL, the discordant relapse clone could be detected at low levels in the diagnostic sample by FISH, again indicating that there is subclonal diversity at diagnosis that is subject to selective pressures imposed by therapy (32).
Pre-Leukemia and Initiating Mutations
Cancer genome re-sequencing efforts have determined that most leukemia cases harbor multiple mutations that have sequentially occurred in a single cell lineage to generate a dominant leukemic clone. One important question in leukemia genomics is the identity of leukemia-initiating mutations that result in pre-leukemic clones. In general, our knowledge of initiating mutations in many cancers, including leukemia, is limited (35). The antecedent, pre-leukemic cells that could inform our knowledge of leukemia-initiating mutations are generally clinically undetected and are outcompeted by their malignant descendants, such that isolation and investigation of these rare cells has been difficult. Consequently, “backtracking” studies that enable the direct investigation of pre-leukemia are both rare and important (36).
Although difficult to investigate, the early evolutionary history of a cancer is likely to prove clinically relevant. For example, mutations were identified from sequencing >20,000 protein-coding exons in “index” metastatic lesions from seven patients with pancreatic cancer (37, 38). These mutations, of which an average of 61 were found in each patient, were screened in the primary pancreatic tumor and other metastases, such that mutations could be classified as either “founder” mutations detected in all samples from the same patient, or “progressor” mutations that must have evolved after the inception of metastasic subclones. Lastly, by modeling the rate of mutation and cell division, various intervals of tumor evolution were estimated based on the accumulation of neutral passenger mutations. The time from initiation to generation of the parental cancer clone was estimated to be approximately twelve years, subsequent evolution of the index metastatic clone was estimated to span almost seven years, and subsequent progression of the disease to the patient’s death only 2.7 years. The authors lamented the frequent diagnosis of pancreatic cancer after metastatic progression, but also noted the longer-than-expected pre-metastatic phase during which improved diagnostics may have clinical impact.
Backtracking studies in leukemia have investigated cases of pediatric ALL and identified leukemia-initiating events occurring in utero, as evidenced by the presence of clonotypic chromosomal translocations in both archived neonatal blood samples and monochorionic twins with concordant disease (39). Strikingly, this group identified the separation of a clonal antecedent pre-leukemic ALL cell population from frankly leukemic cells, in a monochorionic twin pair with one ‘leukemic’ and one ‘healthy’ twin (40). In pediatric ALL, a Lin-CD34+CD38−/lowCD19+ population, absent from normal bone marrow, has been detected and determined to have leukemia-initiating activity in xenograft assays (41). In the monochorionic twin pair, rare Lin-CD34+CD38−/lowCD19+ cells were found in the ‘healthy’ twin that shared TEL-AML1 fusion transcripts and clonotypic DJ recombination sequences with the ‘leukemic’ twin. These results identified a clinically silent pre-leukemic population in the ‘healthy’ twin that was clonally related to the sibling’s leukemia. Moreover, modeling the effect of TEL-AML1 by retroviral transduction in normal cord blood suggested that the founding chromosomal translocation was likely sufficient to induce the pre-leukemic population found in the ‘healthy’ twin. A follow-up study in the same twin pair used genome-wide CNA profiling to identify three potential “driver” CNA in the frankly leukemic cells. FISH analysis did not detect these three CNA in the ‘healthy’ twin’s pre-leukemic cells, supporting the hypothesis that the pre-leukemic cells diverged genetically after the initiating chromosomal translocation, with subsequent events leading to the clonal evolution of the affected twin’s leukemia (42).
In the case of AML, genome re-sequencing suggests that up to 10 mutations are serially acquired in a single cell lineage that ultimately generates a dominant leukemic clone. This finding raises the critical question of how so many mutations can accumulate in a single clone, given the generally low spontaneous mutation rate and lack of hypermutator phenotypes in AML, and the fact that the overwhelming majority of bone marrow cells lack long-term self-renewal ability. Based on these considerations, a model has been proposed in which mutations must be serially acquired in self-renewing HSC, unless they confer self-renewal potential on a downstream cell (Figure 1) (43). This model is derived from investigation of bone marrow cells from patients in long-term remission from AML1-ETO-positive AML. In these patients, the fusion transcript was detectable in FACS-purified B cells, monocytes, and stem cells up to 150 months after therapy, suggesting that the AML1-ETO translocation occurred in a multipotent and self-renewing HSC, generating a pre-leukemic stem cell that underwent subsequent clonal progression to develop into AML (44). Additional support for this model derives from studies of CML, in which the BCR-ABL translocation occurs in HSC and is sufficient for chronic phase disease. Subsequent clonal progression to blast crisis involves the evolution of leukemia stem cells at the level of granulocyte-macrophage progenitors (GMP) that exhibit activation of the Wnt/beta-catenin pathway, known to be involved in HSC self-renewal (45, 46). In AML, most of the known mutations do not occur in pathways implicated in self-renewal. When these mutations occur in non-stem cells, they will quickly be lost from the hematopoietic pool due to the natural course of differentiation and cell death. However, a mutation in a stem cell may persist, and the mutated clone may expand, facilitating further clonal progression until a leukemic stem cell with extensive self-renewal ability develops. We hypothesize that these pre-leukemic HSC containing one, two, three and more clonotypic mutations may still exist in leukemia patients, suggesting the possibility that the prospective isolation and genetic investigation of these cells may reveal the clonal evolution that precedes development of frank AML.
Clonal Evolution of Acute Leukemia Genomes
As with pre-leukemic mutations, investigation has been limited in determining the sequence in which leukemia mutations accumulate, and the potential impact on leukemogenesis. An early report investigating cytogenetic abnormalities in HSC from myeloodysplastic syndrome (MDS) determined that 5q-deficiency preceded trisomy of chromosome 8 in 4 out of 4 cases (47). In AML, mutations that are early or late have previously been defined by their “stability”, that is continued presence in one patient’s leukemia from diagnosis to relapse. FLT3-ITD, for example, is relatively unstable as it is often discordant between diagnosis and relapse (48). In patients with myeloproliferative neoplasms (MPN), which can progress to AML, clonal analyses of hematopoietic progenitors in lympho-myeloid or erythroid-granulocyte promoting in vitro culture conditions determined that mutations in TET2 preceded JAK2 V617F mutations in five patients (20). However, this rigid order was not confirmed by a subsequent report that used a similar strategy of genotyping single cell-derived colonies to determine that TET2 mutations could precede (4/8), follow (2/8), or exist in separate subclones (2/8) from JAK2 V617F (49). Where multiple recurrent mutations occur in a single case, early mutations are present in all leukemic cells, whereas late mutations may be present in subclones.
Apart from acute leukemia, one recent report of a patient with T cell lymphoma paired exome sequencing to identify tumor-specific mutations with genotyping of single cell-derived myeloid colonies from FACS-sorted CD34+ cells to determine that mutations in PLZF, CRIM1, and TET2 preceded a mutation in ZNF774 (50). This approach of exome-wide mutation discovery coupled with single cell clonal analyses for these mutations may also yield the order of events in AML. In contrast to T cell lymphoma, many AML cells express CD34, such that normal or pre-leukemic hematopoietic stem and progenitor cells may be rare relative to leukemia cells within the CD34+ population. Therefore, prospective separation of residual HSC from AML cells, which has recently been accomplished by our group and others (51–54), may make it possible to investigate pre-leukemic subclones and the clonal evolution of de novo AML genomes.
Clinical Implications and Conclusion
The studies highlighted here are not only important to our understanding of leukemogenesis and clonal progression, but also have significant clinical implications. As described above, patients with AML or ALL generally have poor clinical outcomes, particularly adults who have less than 40% long-term survival. In most of these patients, the true killer is relapsed, treatment-refractory disease. This raises the important question of what is the cellular basis of relapse and how does it relate to the original pre-treatment disease? A simple hypothesis is that the predominant leukemic clone, and its leukemia stem cells, persist following therapy, eventually expanding to give rise to the refractory, relapsed clone (Figure 3a). An alternative is that a rare treatment-resistant subclone is selected through the course of therapy that is responsible for relapse (Figure 3b). This scenario is particularly relevant to targeted therapies such as tyrosine kinase inhibitors, in which patients might relapse with a resistant subclone, dampening long-term effectiveness of such therapies. In order to significantly improve long-term patient outcomes, it will be critical to understand heterogeneous subclonal responses to novel targeted therapies. A third possibility is that treatment with DNA-damaging therapeutic agents contributes to genetic evolution of the leukemia, resulting in the relapsed clone (Figure 3c). A final possibility is that DNA-damaging therapeutic agents act on pre-leukemic cells to induce additional mutations resulting in a novel relapsed clone (Figure 3d). As detailed above, there is evidence that several of these mechanisms can be observed in pediatric ALL, where it appears that clonal evolution on a path divergent to the dominant de novo leukemic clone is a common path to relapse. Whether current DNA-damaging treatments that are effective at eradicating the dominant leukemic clone select cells for future emergence, or in fact, drive mutagenesis and clonal evolution is of great importance and is currently unknown.
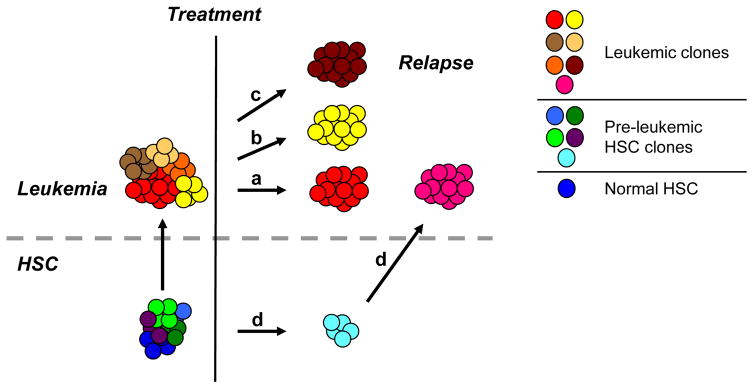
Figure 3. Model for the Clonal Basis of Relapse in Acute Leukemia.
The clonal heterogeneity of pre-leukemic HSC clones and frankly leukemic clones leads to multiple models for the clonal bases of relapse in acute leukemia. (a) A simple option is that the predominant leukemic clone, and its leukemia stem cells, persist following therapy, eventually expanding to give rise to the refractory, relapsed clone (red). (b) An alternative is that a rare treatment-resistant subclone is selected through the course of therapy that is responsible for relapse (yellow). (c) A third possibility is that treatment with DNA-damaging therapeutic agents contributes to genetic evolution of the leukemia, resulting in a genetically novel relapsed clone (dark brown). (d) A final possibility is that DNA-damaging therapeutic agents act on pre-leukemic cells to induce additional mutations resulting in a novel relapsed clone (pink). It is likely that the clonal basis of relapse will differ between individual patients, as suggested by the studies of pediatric ALL described above. It is also possible, that more than one mechanism can be responsible for relapse in the same patient, giving rise to clonal heterogeneity within relapsed leukemia.
While genetic coding mutations clearly play a key role in acute leukemia, growing evidence indicates that epigenetic alterations also contribute to disease pathogenesis. As described above, somatic mutations have been identified in a number of genes involved in the regulation of the epigenome. Accordingly, DNA methylation patterns of bulk blasts can identify distinct previously known and unknown subtypes of AML (55). It remains to be demonstrated that subpopulations of acute leukemia cells exhibit epigenetic heterogeneity, but it seems very likely that epigenetic diversity contributes to subclonal heterogeneity in acute leukemia. Such epigenetic subclones likely differ in their proliferation, self-renewal, differentiation, and response to therapy, adding an additional dimension to the functional heterogeneity of leukemia subclones.
Identification of the subclonal origin of relapsed disease may shift the target of therapy to include both the dominant diagnostic clone and a potentially minor relapse clone. Moreover, rare pre-leukemic clones may contribute to relapse through the acquisition of new mutations, suggesting that these cells may also need to be targeted for cure. Ultimately, a deeper characterization of pre-leukemia will inform our understanding of the path from pre-leukemic cells to relapse, and potentially indicate a need to develop therapies targeting pre-leukemic clones.
Acknowledgments
We would like to acknowledge Ryan Corces-Zimmerman for critical review of the manuscript. M.J. is supported by the Lucille P. Markey Biomedical Research Fellowship and the National Science Foundation Graduate Research Fellowship. R.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Footnotes
CONFLICT OF INTEREST
The authors report no competing interests.
References
- 1.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. Jama. 2011;305:1568–76. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. Jama. 2011;305:1577–84. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–41. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 6.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 7.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 8.Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–62. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 9.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 10.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–33. [PubMed] [Google Scholar]
- 11.Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–48. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 13.Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program. 2004:80–97. doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- 14.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 20.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 21.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 22.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–48. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 24.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 25.Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009;106:12950–5. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 27.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–9. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–61. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 29.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–49. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 30.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–7. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 31.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–80. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuster L, Grausenburger R, Fuka G, Kaindl U, Krapf G, Inthal A, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood. 2011;117:2658–67. doi: 10.1182/blood-2010-03-275347. [DOI] [PubMed] [Google Scholar]
- 33.van Delft FW, Horsley S, Colman S, Anderson K, Bateman C, Kempski H, et al. Clonal origins of relapse in ETV6-RUNX1 acute lymphoblastic leukemia. Blood. 2011;117:6247–54. doi: 10.1182/blood-2010-10-314674. [DOI] [PubMed] [Google Scholar]
- 34.Yang JJ, Bhojwani D, Yang W, Cai X, Stocco G, Crews K, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–83. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 36.Greaves M. Darwin and evolutionary tales in leukemia. The Ham-Wasserman Lecture. Hematology Am Soc Hematol Educ Program. 2009:3–12. doi: 10.1182/asheducation-2009.1.3. [DOI] [PubMed] [Google Scholar]
- 37.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greaves M. Pre-natal origins of childhood leukemia. Rev Clin Exp Hematol. 2003;7:233–45. [PubMed] [Google Scholar]
- 40.Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–9. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 41.Castor A, Nilsson L, Astrand-Grundstrom I, Buitenhuis M, Ramirez C, Anderson K, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11:630–7. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- 42.Bateman CM, Colman SM, Chaplin T, Young BD, Eden TO, Bhakta M, et al. Acquisition of genome-wide copy number alterations in monozygotic twins with acute lymphoblastic leukemia. Blood. 2010;115:3553–8. doi: 10.1182/blood-2009-10-251413. [DOI] [PubMed] [Google Scholar]
- 43.Weissman I. Stem cell research: paths to cancer therapies and regenerative medicine. Jama. 2005;294:1359–66. doi: 10.1001/jama.294.11.1359. [DOI] [PubMed] [Google Scholar]
- 44.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A. 2000;97:7521–6. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abrahamsson AE, Geron I, Gotlib J, Dao KH, Barroga CF, Newton IG, et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci U S A. 2009;106:3925–9. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–67. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 47.Nilsson L, Astrand-Grundstrom I, Anderson K, Arvidsson I, Hokland P, Bryder D, et al. Involvement and functional impairment of the CD34(+)CD38(−)Thy-1(+) hematopoietic stem cell pool in myelodysplastic syndromes with trisomy 8. Blood. 2002;100:259–67. doi: 10.1182/blood-2001-12-0188. [DOI] [PubMed] [Google Scholar]
- 48.Cloos J, Goemans BF, Hess CJ, van Oostveen JW, Waisfisz Q, Corthals S, et al. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia. 2006;20:1217–20. doi: 10.1038/sj.leu.2404246. [DOI] [PubMed] [Google Scholar]
- 49.Schaub FX, Looser R, Li S, Hao-Shen H, Lehmann T, Tichelli A, et al. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 2010;115:2003–7. doi: 10.1182/blood-2009-09-245381. [DOI] [PubMed] [Google Scholar]
- 50.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event during Human Lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Jan M, Chao MP, Cha AC, Alizadeh AA, Gentles AJ, Weissman IL, et al. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci U S A. 2011;108:5009–14. doi: 10.1073/pnas.1100551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kikushige Y, Shima T, Takayanagi S, Urata S, Miyamoto T, Iwasaki H, et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell. 2010;7:708–17. doi: 10.1016/j.stem.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 53.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–99. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, Luke T, et al. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(−) fraction. Blood. 2010;115:1976–84. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]