Abstract
The human endogenous retroviruses (HERV)-K of the HML-2 group include full-length or near full-length elements encoding functional proteins, and are classified as type-1 or type-2 (type-1 has a deletion in the 5′ end of the env gene).
Because proteins of different retroviruses can interact, we hypothesized that HERV-K envelope (Env) could influence HIV-1 replication. Here we describe the negative effect of envelope expression of certain type-2 HERV-Ks on HIV-1 production.
All HIV-1 and SIV strains tested were susceptible to various degrees to inhibition by the HERV-K108 envelope. We identified four residues within HERV-K108 Env as being critical to inhibit HIV-1 production. No inhibition was observed on EGFP expression, indicating that HERV-K Env does not affect general protein production.
These findings demonstrate that envelope proteins from some endogenous retroviruses can limit production of exogenous lentiviruses such as HIV-1. Future studies will elucidate the mechanism mediating HIV-1 inhibition by HERV Envs.
Keywords: Endogenous Retroviruses, HERV-K, Envelope, HIV-1
Introduction
Human endogenous retroviruses (HERVs) occupy about 8% of the human genome [1]. The human MMTV-like-2 (HML-2) group of the HERV-K subfamily includes retroviruses that entered the germ-line relatively late during human evolution [2]. HML-2 members are, therefore, more likely to maintain intact open reading frames and express functional proteins. There are about one thousand HML-2 proviruses disseminated within the human genome, of which about 90% are solo LTRs. The remaining 90 or so full-length or near full-length elements have recently been identified and their genomic localization has been established [2]. In addition to the HERV-K elements that are common to all humans, a certain number are insertionally polymorphic, suggesting that their introduction into the germ-line might have happened after the beginning of human migration out of the African continent [3,4].
Human Immunodeficiency Virus (HIV) and Human T-Lymphotropic Virus (HTLV) [5] are the two known disease-causing groups of retroviruses in humans. In recent years, the interaction between HIV and the endogenous retroviruses that populate the human genome has been investigated [6–9]. Indeed, retroviruses’ ability to functionally interact with each other, although not universal, is nonetheless well documented [10]. Gag and Env proteins from endogenous retroviruses were among the first “restriction factors“ discovered [11,12]. The protein Fv1, derived from the gag region of MuERV-L (a murine endogenous retrovirus with high similarity to the human HERV-L) [13,14], interferes with the formation of exogenous Murine Leukemia Virus (MLV) proviruses in the target cell [15]. Furthermore, the HERV-K Gag consensus sequence protein co-assembles with HIV-1 Gag and negatively affects the late phase of HIV-1 replication, indicating that co-expression of HERV-K with HIV-1 could impact the HIV lifecycle [16]. ERV envelopes also can hinder retroviral infections. For example, when expressed on the surface of the target cell, Fv4 and the enJSRV Env cause receptor interference and thereby inhibit their respective exogenous counterparts, murine leukemia virus (MLV) and Jaagsiekte sheep retrovirus (JSRV) [17,18]. Here we asked whether expression of HERV-K Env proteins can affect HIV-1.
HERV-Ks of the HML-2 group are divided in: type-1, bearing a 292 bp deletion in the 5′ region of env, and type-2, which have an intact envelope gene [19–21]. Envelope sequences belonging to members of both types retain their ability to generate a processed protein, although type-1 Envs often show inefficient glycosylation [22], and some (both type-1 and type-2) can be incorporated into HIV particles [22–24]. HERV-K108 is among the best studied HERV-Ks and its expression is prevalent in transformed tissues such as melanomas [25]. The Env of HERV-K108 has been functionally characterized, and its hydrophobicity profile has been described [23,26].
We find that the expression of Env of certain HERV-Ks has a negative effect on HIV particle production. We also identify the specific Env residues necessary and sufficient to block viral production. This study indicates the possibility that, by limiting viral production, envelopes of certain endogenous retroviruses inhibit HIV-1 replication. Further studies are needed, to investigate the cellular circumstances under which these potentially protective elements are expressed.
Materials and Methods
Cell culture
Human embryonic kidney HEK 293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) in the presence of 10% fetal calf serum (FCS; GemCell) 100 units/ml penicillin/streptomycin, and 2 mM L-glutamine at 37°C and 5% CO2.
Plasmids
All HERV-K envs, except HERV-Kcon env, were generated by PCR using genomic DNA or cDNA from primary PBMCs. All env PCR products were cloned using HinDIII-SmaI restriction sites and either T4 ligase or In-fusion technology (Clontech Laboratories, Inc.) into a derivative of vector pTR600 [27] containing the HERV-K RcRe (Rec response element) segment, for RNA nuclear export [22]. All HERV-K env mutants where obtained by PCR mediated site directed mutagenesis, using as template HERV-Kcon env encoding plasmid pCRV1/env and verified by sequencing. pCRV1/env pcRV1 K-rev, and pNL4-3 Luc HXB3, were a kind gift from Dr. Paul Bieniasz (Aaron Diamond AIDS Research Center, The Rockefeller University).
pTR600 rec-HA was generated by PCR using pcRV1 K-rev as template, and cloned using HinDIII-SmaI restriction sites and In-fusion technology (Clontech Laboratories, Inc.) into a derivative of vector pTR600.
The EGFP expressing plasmid used is pEGFP-N1 obtained from Clontech.
The plasmid encoding Moloney MLV gag-pol, pHCMV–intron gag-pol was a gift from Dr. François-Loic Cosset (LVRTG, ENS de Lyon–U412 INSERM, Lyon, France).
pCAGGS ZEBOV VP40 FLAG encoding a FLAG tagged version of Ebola VP40 was a gift from Dr. Christopher Basler (Center for Microbial Pathogenesis, Institute for Biomedical Sciences, Georgia State University, Atlanta).
Lab adapted HIV-1 expressing plasmids pNL4-3 [28], pLAI.2 [29] and transmitter/founder plasmids pWITO.c/2474, and pCH040.c/2625 [30] were obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. HIV-2 encoding plasmid pROD10 FL2 [31,32] was a kind gift from Dr. Klaus Strebel, National Institutes of Health. The SIVcpz LB715 [33] encoding plasmid was a kind gift from Dr. Beatrice Hahn, Perelman School of Medicine and SIVmac239 [34] was a gift from Dr. Benjamin Chen, Icahn School of Medicine at Mount Sinai.
Transfections and Western blots
All transfections were performed in 24-well plates containing 1.8 × 105 HEK 293T cells per well (plated the day before) using 3 μg/ml polyethyleneimine (Polysciences Inc.). If not otherwise specified 250ng/well of HIV-1 HIV-2 SIV and MLV-gag-pol plasmid were transfected. Plasmid amounts for HERV-K env varied and are specified for each experiment. One hundred ng/well pEGFP-N1 and Ebola VP40 encoding plasmid was used for figure 1D and 1E respectively.
Figure 1.
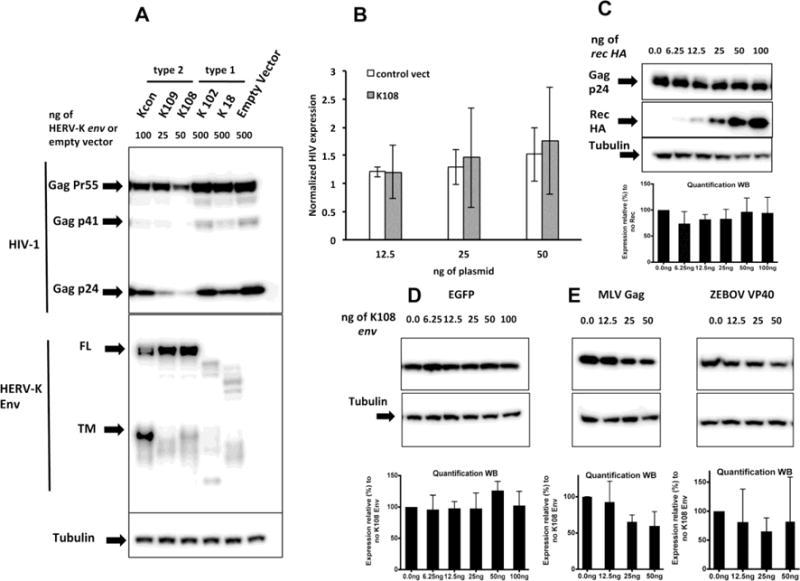
A) Effect of different HERV-K envelopes on HIV protein production. Western blot of HEK 293T transfected with 3 different HERV-K envs belonging to the type-2 group (Kcon, K109 and K108) and 2 belonging to the type-1 group (K102 and K18) transfected alongside HIV-1 NL4-3. Because sequences belonging to the different envs have different protein expression efficiencies, we adjusted plasmid amounts in order to obtain comparable amounts of protein. The anti-HERV-K Env antibody HERM 1811-5 used throughout the paper recognizes the transmembrane domain (TM) on the C-terminus of HERV-K Env, which explains the detection of full length (FL) as well as of the processed TM portion of the Env protein. B) HERV-K108 Env does not interfere with HIV-1 transcription. 32 hours after transfection, quantitative RT-PCR was performed on RNA extracted from cells transfected with different amounts of either HERV-K108 env or empty plasmid control alongside 250ng of NL4-3 expressing plasmid. Data are normalized with the expression of house keeping gene Ribosomal Protein S11 (RPS11). The results are the averages of 3 experiments and error bars represent standard deviations. P values of 2-tailed T-Tests for each amount HERV-K108 or control are all not significant, confirming that there are no differences between the values obtained. C) No effect of HERV-K Rec on HIV-1 expression. Western blot of lysates of HEK293T transfected with increasing amounts of HERV-K rec HA alongside plasmids encoding HIV-1 NL4-3. Quantifications represent the average of 3 experiments and the error bars represent the standard deviation from the mean. D) No effect of HERV-K 108 Env expression on EGFP protein production. Western blot of HEK 293T cells transfected with different amounts of HERV-K108 alongside 100ng of EGFP plasmid. Quantifications represent the average of 3 experiments and the error bars represent the standard deviation from the mean. E) Effect of HERV-K 108 Env expression on MLV Gag and Ebola VP40 production. Western blot of HEK 293T cells transfected with different amounts of HERV-K108 env alongside MLVgag-pol and ZEBOV VP40 matrix expressing plasmids. Quantifications represent the average of 3 experiments for MLV and 5 experiments for ZEBOV, and the error bars represent the standard deviation from the mean.
Cells were lysed 40 hours after transfection using 200μl of RIPA buffer (10 mM Tris-Cl pH 8.0, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 140 mM NaCl) in the presence of Complete Protease Inhibitor Cocktail (Roche). Lysates were run on 10% polyacrylamide gels (Invitrogen, Thermo Fisher Corporation) and transferred to polyvinylidene difluoride (PVDF) membranes (Pierce, Thermo Fisher Corporation). HERV-K Env proteins were probed with monoclonal antibody (mAb) HERM 1811-5 (Austral Biologicals), HIV-1 Gag was probed with α-HIV-1 p24 monoclonal antibody (183–H12-5C) (NIH AIDS Reagent Program), in Fig. 2B. HIV-1 HIV-2 and SIV Gag were probed with broadly reactive mAb AG3.0 (NIH AIDS Reagent Program) known to recognize Gag of HIV-1, HIV-2, and SIV. GFP was probed with α-GFP polyclonal antibody (sc-8334 Santa Cruz). MLV Gag was probed with α-MLV p12 mAb (a gift from Dr. Paul Bieniasz). Tubulin was assessed with monoclonal anti-α-Tubulin (T5168 Sigma). FLAG was visualized using α-FLAG antibody (F7425 Sigma) while HA using α-HA antibody (H6908 Sigma). All secondary antibodies were horseradish peroxidase-conjugated (Sigma). Membranes were developed with SuperSignal West Pico or Femto (Pierce, Thermo Fisher Corporation), and imaging was performed using the ProteinSimple FluorChem E imaging system (ProteinSimple). The Western blots shown are from independently run gels, and tubulin represents sample preparation controls.
Figure 2.
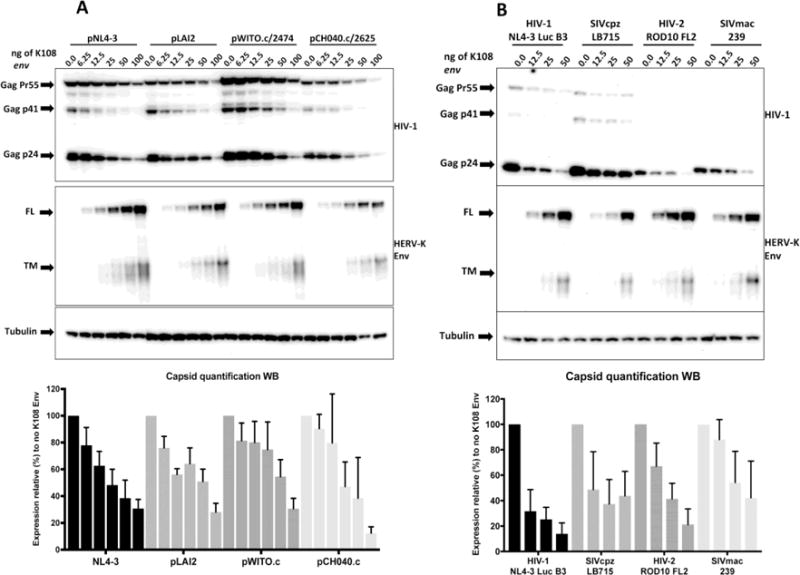
A) Both lab-adapted and transmitter-founder strains of HIV-1 are sensitive to HERV-K108 Env. Western blot of lysates of HEK293T transfected with increasing amounts of HERV K108 env alongside plasmids encoding the following strains of HIV-1: NL4-3, LAI, WITO.c/2474 and CH040.c/2625. Quantifications represent the average of 3 independent experiments and the error bars represent the standard deviation from the mean. B) Different lentiviruses are sensitive to HERV-K108 Env. Western blot of lysates of HEK293T transfected with increasing amounts of HERV K108 env alongside plasmids encoding the following lentiviruses: HIV-1 NL4-3 Luc.HXB3, SIVcpz LB715, HIV-2 ROD10, SIVmac239. Quantifications are based on the average of four independent experiments and the error bars represent the standard deviation from the mean. FL stands for full-length and TM for transmembrane domain.
Quantification of Western blots of at least three independent experiments was performed using AlphaView software (ProteinSimple) and represent relative expression levels normalized by each sample’s tubulin levels.
Quantitative Real-Time RT-PCR
Cells were lysed 32 hours after transfection, RNA was extracted using TRIzol (Invitrogen, Thermo Fisher Corporation) and following the manufacturer’s directions. Samples were DNase treated twice using the DNA-free kit (Ambion). RNA of each sample was reverse transcribed using iScript cDNA synthesis kit (Bio-Rad). HIV-1 expression was measured by qPCR using iQ SYBR green Supermix (Bio-Rad). Sequences of the primers used for this assay are the following: HIV-1 forward primer TGTGTGCCCGTCTGTTGTGT reverse primer GAGTCCTGCGTCGAGAGATC [35] spanning 143 nt transcript of the first exon from nt 102 to 244.
Expression was normalized by Ribosomal Protein S11 (RPS11) expression using primers: Rps11 Fw GCCGAGACTATCTGCACTAC and Rps11 Rv ATGTCCAGCC TCAGAACTTC [36]. qPCR conditions were: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 60 s. Expression levels for individual RNAs were calculated based on their threshold cycle (CT) (ΔΔCT values). RT minus samples were run as negative controls.
Capsid p24 ELISA
Forty-two hours after transfection supernatants were harvested and p24 was measured using HIV-1 p24 ELISA (XpressBio # XB-1000) following manufacturer’s instructions.
Measurement of viral infectivity
TZM-bl reporter cell-line (cat# 8129 NIH AIDS Reagent Program), harboring the β-galactosidase reporter gene driven by the HIV-1 long terminal repeat, was used to assess the infectivity of NL4-3 viruses produced in the presence of increasing amounts of HERV-K108 env. TZM-bl cells were infected with 2.2 ng of CA-p24 equivalents of each virus. β-galactosidase activity was quantified 44 h post-infection using chemiluminescent substrate Tropix, Galacto-Star (Applied Biosystems, Thermo Fisher Corporation).
Results
HERV-K 108 Env interferes with HIV-1 production
To investigate whether HERV-K Env’s capacity to interact with HIV had any consequence for lentivirus production, we produced HIV-1 by transfecting a full length HIV-1 expression vector pNL4-3 [28] alongside HERV-K rec and a panel of HERV-K env encoding plasmids representing both type-1 and type-2. Type-1 included HERV-K102 (HML-2 1q22, JN675014.1) and HERV-K18 (HML-2 1q23.3, JN675013), while type-2 included HERV-K108 (HML-2 7p22.1a, JN675043.1), HERV-K109 (HML-2 6q14.1, JN675041.1) and HERV-Kcon, (a consensus sequence of 10 different type-2 HML-2 viruses, 6 of which were used to reconstitute the env sequence) [37].
We first assessed the amount of Gag produced in each condition by Western blot analysis of the lysates of transfected cells. Two of the three type-2 HERV-Ks Envs (K108 and K109) strongly inhibited retroviral Gag production (Fig. 1A), whereas the inhibitory effect was less pronounced with HERV-Kcon Env. Of note, type-1 and type-2 sequences were cloned in the same backbone vector but nonetheless resulted in different protein expression efficiencies. We adjusted therefore the amounts of transfected expression vectors to obtain comparable protein levels (Fig. 1A).
Since HERV-K 108 Env is one of the best-characterized HERV-K envelopes so far [23,26], we focused next on its mode of action. To investigate whether HERV-K108 Env expression interferes with HIV-1 transcription we measured HIV-1 RNA production in the presence of different amounts of K108 Env. Newly transcribed HIV-1 RNA was measured by quantitative RT-PCR 32h after transfection. We found that HIV-1 transcription was not affected by K108 env transfection, as compared to control vector (Fig. 1B).
Next we tested whether the disruption of RNA export could be the mechanism by which expression of HERV-K108 Env glycoprotein inhibits HIV-1 production. The rational for this experiment resides in the fact that HERV-K type II env sequences harbor the alternative ORF encoding rec, the HERV-K counterpart of HIV-1 rev, which could theoretically affect HIV-1 RNA export and thereby the equilibrium between the different HIV-1 splice forms. We co-transfected pNL4-3 alongside increasing amounts (6.25–100ng) of pTR600 rec HA (the Rec protein here has the same sequence as the one expressed by HERV-K108 [37]). Western blots of cell-lysates ran 40 hours after transfection (Fig. 1C) show that co-expression of HIV-1 with increasing amounts of Rec does not induce any change of Gag-p24 production. These data suggest that neither of the proteins encoded by the HERV-K108 env sequence, either Rec or Env, influences HIV-1 RNA metabolism.
We also assessed whether HIV-1 inhibition by K108 Env was due to a general effect on the host cell’s protein production capacity. We tested the effect of K108 on the expression of EGFP (Fig. 1D). We co-transfected EGFP expression vector (100ng) alongside increasing amounts of HERV-K108 env vector (from 6.25ng to 100ng), and found that HERV-K Env did not alter EGFP expression. Of note is that higher amounts of HERV-K108 or lower confluence of the transfected HEK 293T can result in cell detachment from the tissue culture plate, but we did not observe this effect under the experimental conditions used here.
To test whether the production of other viruses is affected by the expression of HERV-K108 Env, we measured the production of Moloney Murine Leukemia Virus (MLV) Gag and Ebola virus matrix proteins, both sufficient to produce virus-like particles, in the presence of different amounts of HERV K108 Env. In contrast to HIV-1, both MLV and Ebola matrix (ZEBOV VP-40) were less sensitive to K108 expression (Fig. 1E) although with some variability in the case of ZEBOV VP-40.
These data suggest that HIV-1 production is more heavily influenced by the co-expression of HERV-K 108 Env compared to more distant viruses such as MLV and Ebola.
Susceptibility of different lentiviruses to HERV-K108 Env expression
We next asked whether different strains of HIV-1 are similarly affected by HERV-K108 expression. We transfected four different HIV-1 molecular clones: two lab adapted strains (pNL4-3 [28], pLAI2 [29]) and two transmitter-founder viral strains (pWITO.c/2474, pCH040.c/2625 [30]. We transfected the same amount of each HIV-1 molecular clone (250ng) alongside a serial dilution of HERV-K108 env vector. Western blot analysis of the cell lysates (Fig. 2A) shows that all viral strains tested are sensitive to HERV-K Env expression, although with different dose-response dynamics.
Given that production of viruses distant from HIV-1 was minimally affected by the expression of HERV-K108, we further investigated whether other lentiviruses were influenced by the HERV-K envelope. We co-transfected the following molecular clones with HERV-K 108 env (2-fold serial dilution 50ng-12.5ng): SIVcpz LB715 [33], HIV-2 pROD10 FL2 [31,32] and SIVmac239 [34]. In all cases HERV-K108 affected viral production, with HIV-2 being inhibited to levels similar to HIV-1, retaining approx. 22% of its expression at 50ng of K108 env, while both the SIVs showing to be on average less sensitive (Fig. 2B).
Taken together, these data show that the production of both lab-adapted and transmitter founder HIV-1 strains is targeted by HERV-K108 Env, and that lentiviruses other than HIV-1 demonstrate variable levels of sensitivity to HERV-K108 Env.
Effect of HERV-K108 Env on cell free HIV-1 production and infectivity
To confirm that HERV-K108 Env interfered with the production of HIV-1 virions, we determined the p24 capsid concentration in the culture supernatants collected 42h after transfecting HIV-1 NL4-3 expressing plasmid (250ng) with increasing amounts of HERV-K108 env (6.25–100ng) (Fig. 3A). As expected, we observed a HERV-K108 Env dose-dependent reduction of HIV-1 p24 concentration, corroborating the notion that co-expression of HERV-K108 Env with HIV-1 inhibits virion production.
Figure 3.
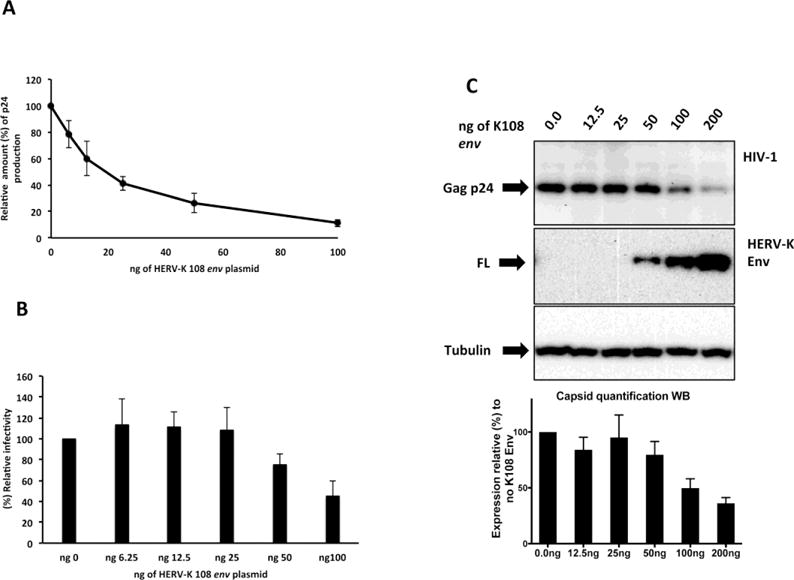
A) HERV-K108 Env expression inhibits HIV-1 virion production in cell culture supernatant. HIV-1 virion release in the tissue culture supernatant was measured 42h after transfection by ELISA for capsid p24. The results are represented as p24 production relative (%) to control, which was obtained by HIV-1 co-transfection with empty control vector. The results shown reflect the averages of three independent experiments and error bars represent standard deviations. B) HERV-K108 Env expression only minimally affects HIV-1 infectivity. TZM-bl reporter cell line was infected with 2.2ng of CA-p24 equivalents of HIV-1 produced in the presence of increasing amounts of HERV-K 108 env plasmid. Β-galactosidase was measured 44 hours after infection. The results presented are the averages of three independent experiments and error bars represent standard deviations. C) HERV-K108 Env expression in HeLa cells inhibits HIV-1 prduction. Western blot of lysates of HeLa cells transfected with increasing amounts of HERV K108 env alongside plasmids encoding HIV-1 LAI. Quantifications represent the average of 3 experiments and the error bars represent the standard deviation from the mean.
Next we tested the infectivity of the HIV-1 viruses produced in the presence of increasing amounts of HERV-K108 env (6.25–100ng). We found that HERV-K108 Env expression only minimally decreased (approx. 2 fold reduction) HIV-1 infectivity and only at the highest concentrations (Fig. 3B).
To assess the extent of HIV-1 production inhibition by HERV-K108 Env expression in cells other than HEK23T, we co-transfected HIV-1 LAI with HERV-K 108 env (2-fold serial dilution 200ng–12.5ng) in HeLa cells. Western blot analysis of the cell lysates in Fig. 3C shows a dose dependent inhibitory effect on HIV-1 Gag p24 production by HERV-K 108 Env, confirming that the K108 Env influence on HIV-1 is not cell type dependent.
Identification of the residues responsible for the HERV-K 108 Env inhibition of HIV-1
Our initial analysis revealed that the transmembrane portion of HERV-Kcon Env was far more abundant than that of the other two type 2 HERV-Ks, indicating there could be marked differences in the level of HERV-K Env processing by furin (Fig. 1A). Furin cleavage is a common process for viral glycoproteins, which is necessary to complete virion maturation and to ensure viral infectivity [38–40]. We asked, therefore, whether the level of HERV-K Env processing by furin correlated with the interference of HIV-1 production and whether inefficient HERV-K Env processing was at the root of the observed phenotype. We mutated the furin cleavage site (RSKR) in HERV-Kcon Env (to make it furin-resistant, i.e. RSKR➔QSQQ or RSKR➔AAAA) and in HERV-K 108 Env (to render it more susceptible for furin processing RSKR➔RRRRRR [41]). The mutated HERV-Kcon Env showed decreased processing by furin and inhibited HIV-1 production, in contrast to the original Kcon-Env (Fig. 4B and 4C). Conversely, the mutated HERV-K108 Env was more efficiently cleaved by furin but had similar inhibitory effect on HIV-1 production as the wild-type HERV-K108 Env (Fig. 4B and 4C). Therefore, inefficient furin-dependent processing of HERV-K Env can hinder HIV-1 production but is not the mechanism by which K108 Env interferes with HIV-1 production.
Figure 4.
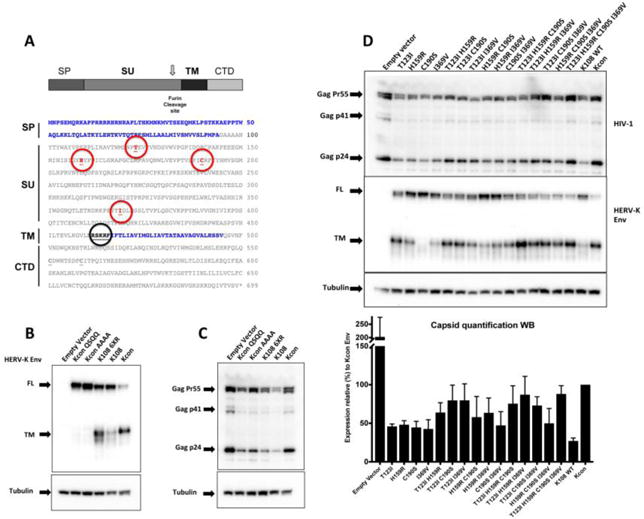
A) Schematic representation of HERV-K con and HERV-K108 envelope sequences. The positions where the residues differ between HERV-K108 and HERV-Kcon are identified by red circles. The black circle highlights the furin cleavage site. SP stands for signal peptide (shown as a blue sequence), SU stands for surface domain, TM for transmembrane (TM ectodomain shown blue sequence) and CTD stands for cyotoplasmic domain. B) and C) Effect of furin cleavage site mutagenesis on the interference on HIV by HERV-K108 Env. Disruption of furin cleavage site inhibits HERV-Kcon Env processing and increases its inhibitory effect on HIV production, while optimization of the furin cleavage site increases HERV-K108 Env processing but does not rescue HIV-1 production. FL stands for full-length and TM for transmembrane domain. D) Mutagenesis of all four residues that differ between K108 and Kcon rescues HIV-1 production. Western blot of lysates of HEK 293T co-transfected with 250ng of HIV-1 pLAI2, 100ng pcRV1-K rev and 60ng of HERV-K108 env versions harboring all possible combinations of the 4 mutant residues as well as HERV-Kcon env. Quantifications reflect the average of three independent experiments and the error bars represent the standard deviation from the mean. FL stands for full-length and TM for transmembrane domain.
Only four amino acids differ between the envelopes of HERV-K108 and HERV-Kcon (Fig. 4A). To determine which residues play a role in the inhibition of HIV-1 production by HERV-K108 Env, we mutated each of them to the corresponding residue found in HERV-Kcon (T123I, H159R, C190S and I369V), individually or in combination (16 combinations in total). We then assessed their influence on HIV-1 production. We found that different combinations, especially those including T123I, increased furin–dependent processing, and that T123I C190S, T123I I369V, and T123I H159R I369V, rescued at least 75% of HIV-1 production obtained in the presence of HERV-Kcon, with T123I H159R C190S I369V mutant restoring almost 90% (88% +/− 10%) of Kcon HIV-1 production.
Taken together, these results show that furin processing of HERV-K Env can influence HIV-1 production, as shown by the mutagenesis of the furin cleavage site in HERV-Kcon env, but it is not the primary reason by which HERV-K108 Env limits HIV-1 production. Indeed, it appears that the four positions by which HERV-K 108 Env differs from HERV-Kcon Env are required for the HERV-K108 interference of HIV-1 production.
Discussion
Within the host cell, endogenous retroviruses constitute a form of retroviral environment that exogenous retroviruses have to navigate during infection. Sometimes these interactions are lethal for the invading retrovirus, as in the case of Fv1 or consensus Gag of HERV-Kcon [15,16]. It is clear though that we have not fully grasped the extent of the impact of these resident endogenous retroviruses on the life cycle of exogenous viruses.
Here we report that the envelope of one these endogenous retroviruses, HERV-K108, has a negative effect on the production of HIV-1. Our results show that Env of HERV-K108, a type-2 HERV-K HML-2, consistently interfered with production of HIV-1. However, this is not a general feature of all type-2 HERV-Ks, as a consensus sequence type-2 Env had little effect on HIV-1 (Fig. 1A). Our experiments reveal that the four residues by which K108 differs from Kcon are required to confer the HIV-1 interference characteristic to HERV-K108 (Fig. 4D). The type-1 HERV-K Envs tested showed no effect on HIV-1 production, although it should be noted that their expression levels were lower than those seen for type-2 Envs.
Interestingly, we find that not all lentiviruses are equally sensitive to the HERV-K108 Env effect. Indeed, HIV-1 strain pWITO.c/2474 and the two SIV strains tested, SIVmac239 and SIVcpz, displayed a certain level of resistance to K108 Env.
It is important to note that the absence of strong effects on MLV Gag, Ebola matrix, GFP and the intermediate effect on pWITO.c/2474 and SIV Gag, indicate that, under the chosen experimental conditions, HERV-K108 Env inhibition of HIV production is not due to general cellular toxicity. Moreover, we have proven that there is no major impact on HIV-1 transcription by HERV-K108 Env (Fig. 1B) and that RNA binding protein Rec (the alternative ORF contained in HERV-K env sequence and the HERV-K counterpart of HIV-1 Rev) is not responsible for HIV-1 inhibition observed. However, more detailed investigations are necessary to determine how these HERV-K Envs interfere with HIV-1 production. Possible mechanisms include the induction of ER stress and unfolded protein response as well as the interference with HIV-1 protein trafficking and recycling pathways. Preliminary experiments to test the involvement of protein degradation pathways such as proteasome inhibitors (e.g., MG132 and Lactacystin) or lysosomal inhibitors (e.g., NH4Cl and Chloroquine) have failed to consistently rescue HIV-1 production in the presence of HERV-K 108 Env (data not shown), raising the possibility that more complex mechanisms are at play.
HIV-1 infection has been reported to up-regulate expression of HML-2 group of HERV-K [9,42] although the specific identity of the up-regulated proviruses is currently unclear. In addition, the consequences of such increased expression on the life cycle of HIV-1 remains to be explored. Our study here tries to answer the proof of principle question of whether HERV-K Env expression can interfere with HIV-1 and was therefore performed in HEK 293T cells as they are a particularly amenable model system widely used in in HIV-1 research. Since we and others have shown that cells of different origin possess different HERV-K expression profiles [22,25], future studies are needed to assess the extent by which different HERV expression profiles correlate, and perhaps influence, cellular susceptibility to infection and replication by an exogenous virus such as HIV-1, and in turn whether such exogenous viruses can regulate the expression profiles of endogenous retroviruses.
Highlights.
We find that the expression of Env of certain HERV-Ks has a negative effect on HIV particle production.
We also identify the specific Env residues necessary and sufficient to block viral production.
Acknowledgments
We like to thank: Drs. Domenico Tortorella and Ines Chen, for advice and for critically reading the manuscript, Dr. Beatrice Hahn, Perelman School of Medicine for plasmid SIVcpz LB715, Dr. Klaus Strebel, National Institutes of Health, for plasmid pROD10, Dr. Benjamin Chen, Icahn School of Medicine at Mount Sinai for plasmid SIVmac239, Dr. Christopher Basler, Georgia State University, for plasmid pCAGGS ZEBOV VP40 FLAG. Dr. François-Loic Cosset for pHCMV–intron gag-pol, Dr. Paul Bieniasz for pCRV1/env, pCRV1 K-rev pNL4-3 Luc HXB3, and α-MLV p12 monoclonal antibody. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: α-HIV-1 p24 monoclonal antibody (183–H12-5C) (cat# 3537) from Dr. Bruce Chesebro and Kathy Wehrly, monoclonal antibody to HIV-1 p24 (AG3.0) (cat# 4121), from Dr. Jonathan Allan, pNL4-3 from Dr. Malcolm Martin, pLAI 2 from Dr. Keith Peden, courtesy of the MRC AIDS Directed Program, pWITO.c/2474 (cat# 11739) and pCH040.c/2625 (cat# 11740) from Dr. John Kappes and Dr. Christina Ochsenbauer. TZM-bl reporter cell-line (cat# 8129) from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc.
LCFM is funded by NIH Grant R01 GM113886 and GM113886-01S1, VS is funded by NIH grants R01 R01AI064001 and R01AI120998.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
S.N.T., L.M., A.C., D.B., L.C.F.M. performed the experiments, L.C.F.M. designed the study, interpreted the data, and wrote the manuscript. V.S. provided reagents, interpreted the data and edited the manuscript. All authors approved the final manuscript.
Conflict of interests
The authors declare no competing financial interests.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the herv-k (hml-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 4.Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallo RC, Reitz MS., Jr The first human retroviruses: Are there others? Microbiol Sci. 1985;2:97–98. 101–104. [PubMed] [Google Scholar]
- 6.Contreras-Galindo R, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. Comparative longitudinal studies of herv-k and hiv-1 rna titers in hiv-1-infected patients receiving successful versus unsuccessful highly active antiretroviral therapy. AIDS research and human retroviruses. 2007;23:1083–1086. doi: 10.1089/aid.2007.0054. [DOI] [PubMed] [Google Scholar]
- 7.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, et al. Characterization of human endogenous retroviral elements in the blood of hiv-1-infected individuals. Journal of virology. 2012;86:262–276. doi: 10.1128/JVI.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. Detection of herv-k(hml-2) viral rna in plasma of hiv type 1-infected individuals. AIDS research and human retroviruses. 2006;22:979–984. doi: 10.1089/aid.2006.22.979. [DOI] [PubMed] [Google Scholar]
- 9.Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF, et al. Herv-k-specific t cells eliminate diverse hiv-1/2 and siv primary isolates. The Journal of clinical investigation. 2012;122:4473–4489. doi: 10.1172/JCI64560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landau NR, Page KA, Littman DR. Pseudotyping with human t-cell leukemia-virus type-i broadens the human-immunodeficiency-virus host range. Journal of virology. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odaka T. Inheritance of susceptibility to friend mouse leukemia virus. V. Introduction of a gene responsible for susceptibility in the genetic complement of resistant mice. Journal of virology. 1969;3:543–548. doi: 10.1128/jvi.3.6.543-548.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pincus T, Hartley JW, Rowe WP. A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J Exp Med. 1971;133:1219–1233. doi: 10.1084/jem.133.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 14.Benit L, De Parseval N, Casella JF, Callebaut I, Cordonnier A, Heidmann T. Cloning of a new murine endogenous retrovirus, muerv-l, with strong similarity to the human herv-l element and with a gag coding sequence closely related to the fv1 restriction gene. Journal of virology. 1997;71:5652–5657. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolicoeur P, Baltimore D. Effect of fv-1 gene product on proviral DNA formation and integration in cells infected with murine leukemia viruses. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:2236–2240. doi: 10.1073/pnas.73.7.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monde K, Contreras-Galindo R, Kaplan MH, Markovitz DM, Ono A. Human endogenous retrovirus k gag coassembles with hiv-1 gag and reduces the release efficiency and infectivity of hiv-1. Journal of virology. 2012;86:11194–11208. doi: 10.1128/JVI.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer TE, Mura M, Gray CA, Griebel PJ, Palmarini M. Receptor usage and fetal expression of ovine endogenous betaretroviruses: Implications for coevolution of endogenous and exogenous retroviruses. Journal of virology. 2003;77:749–753. doi: 10.1128/JVI.77.1.749-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda H, Sugimura H. Fv-4 resistance gene: A truncated endogenous murine leukemia virus with ecotropic interference properties. Journal of virology. 1989;63:5405–5412. doi: 10.1128/jvi.63.12.5405-5412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types a and b retrovirus genes. Journal of virology. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agoni L, Guha C, Lenz J. Detection of human endogenous retrovirus k (herv-k) transcripts in human prostate cancer cell lines. Front Oncol. 2013;3:180. doi: 10.3389/fonc.2013.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lower R, Boller K, Hasenmaier B, Korbmacher C, Mullerlantzsch N, Lower J, Kurth R. Identification of human endogenous retroviruses with complex messenger-rna expression and particle formation. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinzevich D, Young GR, Sebra R, Ayllon J, Maio SM, Deikus G, Chen BK, Fernandez-Sesma A, Simon V, Mulder LC. Hiv-1 interacts with human endogenous retrovirus k (hml-2) envelopes derived from human primary lymphocytes. Journal of virology. 2014;88:6213–6223. doi: 10.1128/JVI.00669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewannieux M, Blaise S, Heidmann T. Identification of a functional envelope protein from the herv-k family of human endogenous retroviruses. Journal of virology. 2005;79:15573–15577. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanke K, Kramer P, Seeher S, Beimforde N, Kurth R, Bannert N. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus k and modulation of its functional activity by truncation of the cytoplasmic domain. Journal of virology. 2009;83:12790–12800. doi: 10.1128/JVI.01368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group herv-k(hml-2) loci in melanoma. Genome Biol Evol. 2013;5:307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henzy JE, Coffin JM. Betaretroviral envelope subunits are noncovalently associated and restricted to the mammalian class. Journal of virology. 2013;87:1937–1946. doi: 10.1128/JVI.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green TD, Newton BR, Rota PA, Xu Y, Robinson HL, Ross TM. C3d enhancement of neutralizing antibodies to measles hemagglutinin. Vaccine. 2001;20:242–248. doi: 10.1016/s0264-410x(01)00266-3. [DOI] [PubMed] [Google Scholar]
- 28.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. Journal of virology. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of hiv-1lai, hiv-1mal, and hiv-1eli. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 30.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, et al. Generation of transmitted/founder hiv-1 infectious molecular clones and characterization of their replication capacity in cd4 t lymphocytes and monocyte-derived macrophages. Journal of virology. 2012;86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bour S, Schubert U, Peden K, Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: A vpu-like factor? Journal of virology. 1996;70:820–829. doi: 10.1128/jvi.70.2.820-829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan-Graham MA, Peden KW. Both virus and host components are important for the manifestation of a nef-phenotype in hiv-1 and hiv-2. Virology. 1995;213:158–168. doi: 10.1006/viro.1995.1556. [DOI] [PubMed] [Google Scholar]
- 33.Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S, et al. Genetic diversity and phylogeographic clustering of sivcpzptt in wild chimpanzees in cameroon. Virology. 2007;368:155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 34.Kestler HW, 3rd, Li Y, Naidu YM, Butler CV, Ochs MF, Jaenel G, King NW, Daniel MD, Desrosiers RC. Comparison of simian immunodeficiency virus isolates. Nature. 1988;331:619–622. doi: 10.1038/331619a0. [DOI] [PubMed] [Google Scholar]
- 35.Butler SL, Hansen MS, Bushman FD. A quantitative assay for hiv DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 36.Manganaro L, Pache L, Herrmann T, Marlett J, Hwang Y, Murry J, Miorin L, Ting AT, Konig R, Garcia-Sastre A, et al. Tumor suppressor cylindromatosis (cyld) controls hiv transcription in an nf-kappab-dependent manner. Journal of virology. 2014;88:7528–7540. doi: 10.1128/JVI.00239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loving R, Wu SR, Sjoberg M, Lindqvist B, Garoff H. Maturation cleavage of the murine leukemia virus env precursor separates the transmembrane subunits to prime it for receptor triggering. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7735–7740. doi: 10.1073/pnas.1118125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson LR, Whelan SP. Infectious entry pathway mediated by the human endogenous retrovirus k envelope protein. Journal of virology. 2016;90:3640–3649. doi: 10.1128/JVI.03136-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker JA, Molloy SS, Thomas G, Sakaguchi T, Yoshida T, Chambers TM, Kawaoka Y. Sequence specificity of furin, a proprotein-processing endoprotease, for the hemagglutinin of a virulent avian influenza virus. Journal of virology. 1994;68:1213–1218. doi: 10.1128/jvi.68.2.1213-1218.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, Travis B, Kuhmann S, Burton DR, Hu SL, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. Journal of virology. 2002;76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. Hiv-1 infection increases the expression of human endogenous retroviruses type k (herv-k) in vitro. AIDS research and human retroviruses. 2007;23:116–122. doi: 10.1089/aid.2006.0117. [DOI] [PubMed] [Google Scholar]


