Abstract
The ten-eleven translocation (TET) family of enzymes were originally cloned from the translocation breakpoint of t(10;11) in infant acute myeloid leukemia (AML) with subsequent genomic analyses revealing somatic mutations and suppressed expression of TET family members across a range of malignancies, particularly enriched in hematological neoplasms. The TET family of enzymes is responsible for the hydroxylation of 5-methylcytosines (5-mC) to 5-hydroxymethylcytosine (5-hmC), followed by active and passive mechanisms leading to DNA demethylation. Given the complexity and importance of DNA methylation events in cellular proliferation and differentiation, it comes as no surprise that the TET family of enzymes is intricately regulated by both small molecules and regulatory cooperating proteins. Here, we review the structure and function of TET2, its interactions with cooperating mutations and small molecules, and its role in aberrant hematopoiesis.
The TET2 enzyme is a critical regulator of DNA methylation, which is important in cellular proliferation and differentiation. It is aberrantly expressed in various malignancies, particularly hematological neoplasms.
Although often thought to be a stable epigenetic mark, recent research has revealed DNA methylation to be dynamic modification capable of regulating critical features of cellular proliferation, differentiation, and gene expression. Integral to this regulatory function are the enzymes necessary for both addition of the DNA-methyl mark and subsequent removal. Amongst these enzymes, the ten-eleven translocation (TET) family of proteins has emerged as critical regulators of the oxidation of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC). Since its recent identification in 2009, an explosion of studies has interrogated the roles of TET2 in malignancies of the blood and brain, developmental processes, and roles in inflammation. Genetic and biochemical studies in both human tumor specimen and animal models of disease have revealed TET2 as a critical node linking alterations in tumor metabolism to alterations in DNA methylation and modified chromatin. These features require a refined understanding of how to classify a cancer-associated gene that fits neither rigid definitions of an oncogene nor a tumor suppressor.
In this review, we will review the initial studies in hematologic malignancy that led to the discovery of TET2, the function and structure of the enzyme, its interactions with cooperating mutations and small molecules, and a perspective into other diseases in which TET2 mutations have been identified.
MUTATIONS IN HEMATOLOGIC DISEASE
In 2009, a series of papers identified somatic mutations in TET2 in multiple hematologic malignancies (Delhommeau et al. 2009; Jankowska et al. 2009; Langemeijer et al. 2009; Tefferi et al. 2009a,b). Mapping of minimal regions of deletion in the 4q24 cytoband revealed TET2 loss of heterozygosity (LOH) and somatic mutations in as many as 30%–50% of myelodysplastic syndrome (MDS) and myeloproliferative neoplasia (MPN) patients, whereas 32% of secondary acute myeloid leukemia (AML) patients harbored TET2 mutations (Jankowska et al. 2009). Further genomic studies in MPN patients revealed the presence of TET2 mutations in both JAK2-V617F-positive and -negative patients, with relatively equal distribution across essential thrombocytosis (ET), polycythemia vera (PV), and myelofibrosis (MF) (Tefferi et al. 2009b). In each of these studies, deletions as well as nonsense and missense mutations were found across multiple exons. Interestingly, most AML patients with TET2 mutations retain expression of the wild-type allele with only 10% of patients possessing biallelic mutations (Delhommeau et al. 2009). Although the function of TET2 was not known at the time, these data were suggestive of a tumor suppressor role and potentially haploinsufficient loss-of-function role in TET2 mutants.
TET2 mutations are present in multiple lymphoid and myeloid lineages, as well as CD34+ progenitor cells, suggestive of an early clonal mutation in the stem cell compartment (Smith et al. 2010). In line with this early mutation designation, TET2 mutants have continually been found at high allele frequency indicating that they are often the “first hit” in the multihit model of leukemogenesis (Smith et al. 2010; Papaemmanuil et al. 2016). These findings are reinforced by genetic studies identifying somatic TET2 mutations in asymptomatic, healthy adults with clonal hematopoiesis (Smith et al. 2010; Busque et al. 2012). This, however, does not appear to always be the case, as two studies have shown that in MPN patients with JAK2-V617F mutations TET2 can either present as the first hit or the second hit based on mutant allele frequency (Abdel-Wahab et al. 2010; Ortmann et al. 2015). Interestingly, “JAK2-first” patients presented with significantly worse overall survival compared with “TET2-first” patients (Ortmann et al. 2015). In addition to the co-occurrence with JAK2 mutations in MPN mentioned above, mutations in TET2 have been shown to co-occur with mutations in ASXL1, SRSF2, SF3B1, U2AF1, and CALR (Rampal et al. 2014a). In AML, TET2 shows comutational patterns with NPM1, FLT3 and DNMT3a (Papaemmanuil et al. 2016). How these mutations cooperate in leukemogenesis remains an area of intense investigation and will be discussed later in this review.
Prognosis
TET2 mutational status has been found to be a variable prognostic indicator. One of the earliest studies of a cohort of 48 patients with systemic mastocytosis found no prognostic association with TET2 mutational status (Tefferi et al. 2009a). Similarly, no survival association was found in a cohort of 63 patients with AML, chronic myelomonocytic leukemia (CMML), or MPN/MDS (Jankowska et al. 2009). Meanwhile, other studies showed a significant association with poor prognosis in AML (Abdel-Wahab et al. 2009) and a favorable prognostic association in MDS (Kosmider et al. 2009). In the largest cohorts to date by The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research Network 2013) and by a group at the Sanger Institute (Papaemmanuil et al. 2016), there was no independent association with survival for TET2 mutations. Interestingly, in a cohort of 211 MDS patients, there was an association in response to the hypomethylating agents decitabine and azacitidine, in which patients with TET2 mutant AML were more likely to respond to therapy than those without the mutation, a finding that was more pronounced when the comutational partner ASXL1 was not mutated (Bejar et al. 2014). Further studies into the functional role of TET2 in disease progression and response to different therapeutic regiments may help clarify the prognostic value of TET2 mutations in various hematologic malignancies.
TET2 FUNCTION AND STRUCTURE
When mutations in TET2 were first discovered through mapping of the 4q24 region of loss/LOH, the functions of this protein remained unknown. Shortly after, homology searches for the trypanosome proteins JBP1 and JBP2, enzymes known to oxidize methyl-thymine, identified the mammalian TET family as 2-oxoglutarate (2-OG) and Fe(II)-dependent enzymes (Tahiliani et al. 2009). These studies revealed that TET1 possessed enzymatic activity for converting 5-mC to 5-hmC, and follow-up studies soon confirmed similar enzymatic activity for TET2 and TET3 (Ito et al. 2010; Ko et al. 2010). Subsequent studies would reveal that TET proteins are capable of generating iterative cytosine alterations leading to the formation of 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC) (Fig. 1) (Ito et al. 2011). These intermediates were further shown to be substrates for thymine-DNA glycosylase (TDG)-mediated base excision repair (BER), converting the modified cytosine residue back to the unmethylated cytosine base (He et al. 2011; Maiti and Drohat 2011). Alternative demethylating mechanisms involve the APOBEC family members deaminating 5-hmC into 5-hydroxymethyluracil (5-hmU), which presents as a target for TDG and selective monofunctional uracil-DNA glycosylase 1 (SMUG1)-mediated BER (Bhutani et al. 2011). In addition to these active processes of DNA demehtylation, TET2 has been implicated in passive DNA demethylation, as 5-hmC serves as a poor substrate for the cell cycle–regulated DNMT1 leading to dilution of the 5-mC mark with each round of DNA replication and cell division (Hashimoto et al. 2012). It is important to note that the relative role of these different pathways in the ultimate removal of DNA modifications back to unmethylated cytosine remains to be fully delineated.
Figure 1.
Reactions involved in TET-mediated oxidation of 5-methylcytosine (5-mC). Depicted here is cytosine-mediated methylation by the family of DNA methyltransferases (DNMT) with the substrate S-adenosyl methionine (SAM) leading to the formation of 5-mC. TET family members are then capable of mediating the iterative oxidation of 5-mC to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC), and 5-carboxylcytosine (5-caC) in an Fe(II), O2, and α-ketoglutarate (α-KG)-dependent reaction. These α-KG-dependent reactions can be inhibited by the oncometabolite 2-hydroxyglutarate (2-HG), which is a neomorphic by-product of mutant IDH1 and IDH2. Each downstream product (5-hmC, 5-fC, and 5-caC) can serve as substrates for thymine DNA glycosylase (TDG) leading to base excision repair (BER) and eventual return to unmodified cytosine.
Recent biochemical studies have identified the structural components of TET2 that mediate these catalytic functions (Hu et al. 2013). Human TET2 encodes a 2002–amino acid, 223-kDa protein with a carboxy-terminal catalytic domain and poorly conserved amino-terminal domain. Biochemical studies on truncation variants were capable of reconstituting the enzymatic functions of TET2 within an 807–amino acid stretch (1129–1936) containing cysteine rich regions and a double-stranded β helix (DSBH) separated by an unstructured linker. This fragment was subsequently crystalized in complex with methylated DNA revealing coordination of a catalytic core by two zinc finger domains. These studies further revealed a cavity allowing for recognition of various modifications on the 5-mC base in the catalytic core. Subsequent studies showed that 5-hmC and 5-fC substrates possessed enzymatically unfavorable coordination of hydrogen bonds in the catalytic cavity, offering a potential explanation for the preferred substrate specificity of TET2 for 5-mC over that of 5-hmC and 5-fC (Hu et al. 2015), as well as the stability of 5-hmC in vivo (Ito et al. 2011).
REGULATION OF TET2 FUNCTION
Inhibition by IDH1/2 Mutant-Derived 2-Hydroxyglutarate
In addition to the genetic and biochemical studies above, one of the most important clues to understanding TET2 biology was the identification of mutually exclusive mutations in the metabolic enzymes isocitrate dehydrogenase 1 (IDH1) and IDH2, placing these enzymes in a putative genetic pathway (Abdel-Wahab et al. 2010). In a broader biological context, the identification of mutation in IDH1 and IDH2 was of fundamental importance in linking of altered cellular metabolism to the genomic age of cancer research. Although the altered glycolysis was a long appreciated hallmark of tumorigenesis (Hanahan and Weinberg 2011), it was not immediately clear how mutations in enzymes canonically involved in the citric acid cycle might impact tumorigenesis. The mechanistic role of these metabolic mutations began to take shape on the discovery that R132H mutant IDH1 was capable of producing 2-HG through an NADPH-dependent reduction of α-ketoglutarate (α-KG) (Dang et al. 2009). Soon after, these findings would be extended to the more common leukemic mutations of IDH2-R172K and IDH2-R140K (Ward et al. 2010). As IDH1/2 mutations were enriched in diseases with a relatively undifferentiated phenotype, low-grade glioma and leukemia, it was hypothesized that 2-HG might block differentiation albeit through unknown mechanisms. Definitive evidence would come later that year with the discovery that IDH1/2 mutant-derived 2-HG was capable of blocking differentiation and inhibiting the α-KG dependent enzyme TET2 (Figueroa et al. 2010). Critically, these studies revealed that IDH1/2 mutant AML patients displayed a hypermethylated phenotype (Figueroa et al. 2010), linking TET2 inhibition by 2-HG with the demethylating functions of TET proteins in development (Ito et al. 2011; Ko et al. 2011). The inhibitory capacity of 2-HG would later be extended to most α-KG-dependent enzymes (Xu et al. 2011), suggesting that IDH1/2 mutations might possess TET2-independent functions. Indeed, IDH1 mutant mice have been shown to down-regulate the DNA damage sensor ATM through altered histone methylation (Inoue et al. 2016). Further work will aim to identify therapeutic vulnerabilities that are shared between TET2 and IDH1/2 mutant AML, as well as those that are specifically relevant to the pleiotropic features of IDH1/2 mutant disease.
Although 2-HG has received much attention as a mutant IDH1/2 neometabolite, its production is not limited to these mutations. Importantly, 2-HG is a chiral molecule with the d-enantiomer being produced by mutant IDH1/2. Recent work has identified that under hypoxic conditions the l enantiomer of 2-HG is produced as a promiscuous bioproduct of lactate dehydrogenase A (LDHA)-mediated and malate dehydrogenase 1 (MDH1)-mediated reduction of α-KG (Intlekofer et al. 2015; Oldham et al. 2015). L2-HG was shown to function as a competitive inhibitor of the EGLN prolyl hydroxylase promoting hypoxia-inducible factor 1-α (HIF1-α) stability, whereas D2-HG served a substrate leading to HIF1-α degradation (Intlekofer et al. 2015). Meanwhile, both enantiomers are capable of inhibiting TET2 (Figueroa et al. 2010; Shim et al. 2014). These studies lead to the possibility that physiological production of L2-HG might play a role in modulating TET2 function in homeostasis, especially in the context of the hypoxic hematopoietic stem cell niche (Spencer et al. 2014).
TET2 Binding Proteins
The amino terminus of TET1 and TET3 contain a well-conserved CXXC domain that has been shown to mediate binding to unmethylated CpG residues (Xu et al. 2012); however, no such domain is present in TET2. Interestingly, a CXXC domain–containing protein, IDAX (CXXC4), is encoded 5′ of the TET2 genomic locus, suggestive of evolutionary splitting of the original TET2-CXXC gene into two separate genes (Ko et al. 2013). Biochemical studies revealed that IDAX is capable of binding both the amino terminus and catalytic domain of TET2, in which binding was associated with caspase-mediated TET2 degradation. This negative regulation of TET2 may present an additional mechanism for affecting 5-mC levels in malignancy, independent of genomic alterations to TET2 or mutations in IDH1/2. Indeed, IDAX has found to be overexpressed in villous adenomas in the colon (Nguyen et al. 2010). In addition to its interaction with TET2, IDAX is a known inhibitor of WNT signaling (Hino et al. 2001), suggesting a potential source of cross talk between these pathways. Another interacting partner with WNT signaling, WT1, has also been shown to bind TET2 and TET3 (Rampal et al. 2014b), acting as a guide for TET2 to specific genomic loci associated with proliferation (Wang et al. 2015). In support of this observation, WT1 is mutated in AML, in a mutually exclusive pattern with TET2, and WT1 loss was further shown to phenocopy TET2 loss in hematopoiesis (Rampal et al. 2014b). In addition to these factors, the CRL4-VprBP complex has been shown to stabilize TET family members through monoubiquitination, increasing TET family members binding to DNA (Yu et al. 2013). Mutation at, or around, the TET2 monoubiquitination site at residue K1299 have been identified in several leukemia cell lines, offering another plausible mechanism for TET2 dysfunction in cancer (Nakagawa et al. 2015).
Vitamin C
Vitamin C has been shown to induce TET activity in embryonic stem (ES) cells, and to induce a global increase in 5-hmC content (Blaschke et al. 2013). Although this activity appeared to be specific to vitamin C and no other reducing agents, vitamin C affected both TET1 and TET2, the only TET family members expressed in ES cells. Interestingly, in this study, the investigators found that not all methylation marks were equally sensitive to subsequent demethylation. Indeed 5-hmC levels were most robustly affected at the promoters of genes, whereas methylation of retro elements remained unchanged. Vitamin C has previously been shown to regulate the activity of several iron-dependent dioxygenases; however, in this setting its effects did not appear to depend on either iron availability or α-KG concentration. In contrast, Hore et al. (2016) found in ES cells that vitamin C increased iron recycling and did not function as a cofactor. Although the details of the regulation may diverge, the capacity for vitamin C to induce TET2 activity is robust, with consistent effects in ES cells, mouse embryonic fibroblasts, T regulatory cells (Nair et al. 2016; Yue et al. 2016), and melanoma cells (Gustafson et al. 2015). Interestingly, in 2009, a single-arm clinical trial on 16 AML patients revealed a subset of patients that showed a clinical response following vitamin C deprivation (Park et al. 2009), highlighting the clinical relevance of vitamin C to leukemia biology. In addition to vitamin C, vitamin A has also been shown to play a role in inducing TET activity through the direct transcriptional regulation of both TET2 and TET3 (Hore et al. 2016). Understanding the mechanisms of both vitamin A and vitamin C activities on TET function could provide key insights into therapeutic options for both IDH and DNMT mutant cancers.
MECHANISMS OF CONTRIBUTION TO LEUKEMOGENESIS
The cancer genetics and biochemical studies discussed above provided substantial insight into the function of TET2 in DNA methylation, yet understanding the cellular manifestations of these activities was made possible through the development of genetic mouse models. Conditional loss of TET2 activity by Vav:Cre-mediated removal of exon 3 led to an expansion of the lineage negative Sca.1+ cKit+ (LSK) cells in vivo and an increase in replating potential in a colony forming unit assay in vitro (Moran-Crusio et al. 2011). These studies further showed that TET2KO/KO bone marrow was capable of outcompeting TET2WT/WT bone marrow in competitive transplant assays and showed increased stem cell function and self-renewal. Finally, aged TET2KO/KO mice developed a CMML-like syndrome with expansion of the monocytes, increased spleen weight, and proliferative growth in the bone marrow, spleen, liver, and lung. Multiple studies published in the same year confirmed these findings (Ko et al. 2011; Li et al. 2011; Quivoron et al. 2011; Shide et al. 2012), with many studies revealing decreased 5-hmC levels in the LSK population. The expansion of the LSK and hematopoietic stem cell (HSC) populations in these mice mirror the findings in patient samples in which clonal TET2 mutations were found in healthy individuals with clonal hematopoiesis (Busque et al. 2012).
In AML and myeloproliferative disease, TET2 mutations are typically present in concert with other mutations. Mutations in the fms related tyrosine kinase 3 (FLT3) are among the most common events in AML with point mutations in the tyrosine kinase domain, and internal tandem duplications (ITDs) near the juxtamembrane domain, leading to autoactivation of the kinase (Levis and Small 2003). Interestingly, when TET2 loss was combined with a FLT3-ITD mutation there was a distinct set of genomic loci that underwent hypermethylation compared with either mutation alone (Shih et al. 2015). Among these loci, hypermethylation of the GATA2 promoter led to a reduction in expression, blockade in differentiation and the development of a transplantable leukemia derived from the LSK progenitor compartment. Interestingly, in addition to the hypermethylated regions, there were more than 500 hypomethylated regions in the combined TET2-FLT3-ITD mutants that were not present in either mutant alone. These, at first paradoxical, findings may be partially explained by FLT3’s role in the commitment to the myeloid lineage and thus hypomethylation of genes necessary for that engagement. Future studies on DNA methylation and 5-hmC will be of interest to determine if the loci-specific effects are indeed specific to this model or representative of a more general leukemic transformation phenotype. Additional models of mutational cooperation with TET2 loss include expression of a mutant c-KIT in mast cells (Soucie et al. 2012), expression of AML-ETO (Hatlen et al. 2016), and loss of Notch signaling (Lobry et al. 2013).
MUTATIONS IN OTHER MALIGNANCIES
T-Cell Lymphoma
In addition to myeloid malignancies detailed above, TET2 mutations have also been identified in patients with T-cell lymphoma (Quivoron et al. 2011). One study found TET2 mutations in 47% of angioimmunoblastic T-cell lymphomas (ATLs) and in 38% of peripheral T-cell lymphomas not otherwise specified (PCTL-NOS) (Lemonnier et al. 2012). Subsequent studies have identified TET2 mutations in upward of 75% of ATL patients (Odejide et al. 2014). Interestingly, this study found multiple subclonal mutations in TET2 within individual patients, all of which resulted in truncation or disruption of the final gene product. Unlike the myeloid leukemia setting, few mutations were found in CD34+ progenitors or subsequent myeloid colonies derived from this population (Odejide et al. 2014). Collectively, these results place TET2 loss as a recurrent driver in ATL with acquisition of the mutation in a lineage committed stage, contrasting sharply with the myeloid malignancies. A related contrast was identified when patients presented with both TET2 and IDH1 mutations (Odejide et al. 2014), events that are largely mutually exclusive in myeloid malignancy. This may reflect a different role for subclonal TET2 loss of function in ATL versus the presumed expansion of a preleukemic clone in AML.
Melanoma
TET2 mutations have been predominantly associated with hematologic malignancies; however, whole-genome analyses through TCGA have identified additional mutations in melanoma and cutaneous squamous cell carcinoma (Cancer Genome Atlas Network 2015). Consistent with these genomic findings, loss of 5-hmC has been proposed to be a prevalent, epigenetic hallmark, of melanoma (Lian et al. 2012). Indeed, epigenetic silencing of TET2 and TET3 has been shown to drive TGF-β-dependent invasion and acquisition of EMT-like features (Gong et al. 2016). Subsequent in vivo studies showed that overexpression of TET2 blunted tumor growth and metastasis. Given the prominent role of dedifferentiation in metastatic melanoma, it will be interesting to investigate the potentially parallel roles of TET2 in hematopoietic and melanocyte differentiation.
Glioma
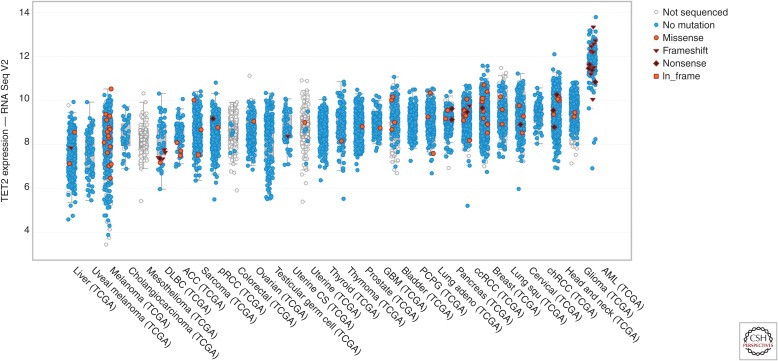
CpG-island hypermethylator phenotypes (CIMPs) have also been identified in low-grade gliomas, as well as some glioblastoma patients. Although TET2 promoter methylation has been identified in glioma (Kim et al. 2011), the predominant mechanism appears to be driven by mutations in IDH1, with few loss-of-function mutations present in TET2 (Kraus et al. 2015). This is in stark contrast to MPN and AML studies in which TET2, IDH1, and IDH2 mutations are all present. These studies support a growing literature showing mutant IDH1 and IDH2 elicit functions outside of 2-HG-mediated TET2 inhibition, including inhibition of histone demethylases (Lu et al. 2012), alteration of DNA damage repair (Inoue et al. 2016), and alterations in branched chain amino acid metabolism (Tonjes et al. 2013). This apparent tissue-specific distinction in mutational patterns may also be the result of tissue-specific gene expression levels of TET2, which is substantially more highly expressed in AML than in the gliomas (Fig. 2).
Figure 2.
TET2 expression across malignancy. Normalized RNA-Seq counts (log2) are shown for the indicated malignancies ranked from lowest to highest mean expression of TET2. Samples with frameshift mutations are denoted with an inverted triangle, nonsense mutations are denoted as a diamond, and in-frame mutations are shown with a square. Data was collected and graphed using the cBioPortal (see cbioportal.org/) (Gao et al. 2013).
Other Diseases
In addition to melanoma, TET2 has been shown to be down-regulated in an androgen-dependent manner in prostate cancer, with lower expression conferring worse prognosis for patients (Nickerson et al. 2016). In colorectal cancer, TET2 has been shown to be excluded from the nucleus (Huang et al. 2016), with similar findings for TET1 in glioma (Waha et al. 2012).
CONCLUDING REMARKS
In sum, TET2 is a critical regulator of DNA methylation in development and malignancy. Genomic alterations of TET2, in addition to modulation of binding partners, lead to alterations in 5-hmC levels and downstream outputs on proliferation and maintenance of stem cells. Although mutations are enriched in hematologic neoplasms, TET2 loss of function has been observed in solid tumors as well. Collectively, these studies have provided insight into how TET2 contributes to disease and may provide clues for identifying specific therapeutic avenues for patients harboring these mutations.
ACKNOWLEDGMENTS
We thank members of the Levine laboratory for their discussion and critical insights.
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. 2009. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114: 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Manshouri T, Patel J, Harris K, Yao J, Hedvat C, Heguy A, Bueso-Ramos C, Kantarjian H, Levine RL, et al. 2010. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res 70: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Lord A, Stevenson K, Bar-Natan M, Pérez-Ladaga A, Zaneveld J, Wang H, Caughey B, Stojanov P, Getz G, et al. 2014. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 124: 2705–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. 2011. DNA demethylation dynamics. Cell 146: 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. 2012. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 44: 1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Guo Y, Niu Y, Jin J, Zhang X, Shi X, Zhang L, Li R, Chen L, Ma RZ. 2016. Epigenetic silencing of TET2 and TET3 induces an EMT-like process in melanoma. Oncotarget. 10.18632/oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson CB, Yang C, Dickson KM, Shao H, Van Booven D, Harbour JW, Liu ZJ, Wang G. 2015. Epigenetic reprogramming of melanoma cells by vitamin C treatment. Clin Epigenetics 7: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. 2012. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40: 4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlen MA, Arora K, Vacic V, Grabowska EA, Liao W, Riley-Gillis B, Oschwald DM, Wang L, Joergens JE, Shih AH, et al. 2016. Integrative genetic analysis of mouse and human AML identifies cooperating disease alleles. J Exp Med 213: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S, Kishida S, Michiue T, Fukui A, Sakamoto I, Takada S, Asashima M, Kikuchi A. 2001. Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol Cell Biol 21: 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hore TA, Von Meyenn F, Ravichandran M, Bachman M, Ficz G, Oxley D, Santos F, Balasubramanian S, Jurkowski TP, Reik W. 2016. Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc Natl Acad Sci 113: 12202–12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. 2013. Crystal structure of TET2–DNA complex: Insight into TET-mediated 5mC oxidation. Cell 155: 1545–1555. [DOI] [PubMed] [Google Scholar]
- Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H, Lou Z, Zhang L, Li W, Gong W, et al. 2015. Structural insight into substrate preference for TET-mediated oxidation. Nature 527: 118–122. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wang G, Liang Z, Yang Y, Cui L, Liu CY. 2016. Loss of nuclear localization of TET2 in colorectal cancer. Clin Epigenetics 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, Lemonnier F, Kron KJ, Cescon DW, Hao Z, et al. 2016. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell 30: 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Dematteo RG, Venneti S, Finley LW, Lu C, Judkins AR, Rustenburg AS, Grinaway PB, Chodera JD, Cross JR, et al. 2015. Hypoxia induces production of l-2-hydroxyglutarate. Cell Metab 22: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Dalessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O’Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP. 2009. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood 113: 6403–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Pierscianek D, Mittelbronn M, Vital A, Mariani L, Hasselblatt M, Ohgaki H. 2011. TET2 promoter methylation in low-grade diffuse gliomas lacking IDH1/2 mutations. J Clin Pathol 64: 850–852. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. 2011. Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci 108: 14566–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Äijö T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al. 2013. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497: 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, Viguie F, Quesnel B, Beyne-Rauzy O, Solary E, et al. 2009. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs). Blood 114: 3285–3291. [DOI] [PubMed] [Google Scholar]
- Kraus TF, Greiner A, Steinmaurer M, Dietinger V, Guibourt V, Kretzschmar HA. 2015. Genetic characterization of ten-eleven-translocation methylcytosine dioxygenase alterations in human glioma. J Cancer 6: 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. 2009. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838–842. [DOI] [PubMed] [Google Scholar]
- Lemonnier F, Couronné L, Parrens M, Jaïs JP, Travert M, Lamant L, Tournillac O, Rousset T, Fabiani B, Cairns RA, et al. 2012. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120: 1466–1469. [DOI] [PubMed] [Google Scholar]
- Levis M, Small D. 2003. FLT3: It does matter in leukemia. Leukemia 17: 1738–1752. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. 2011. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118: 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. 2012. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150: 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, et al. 2013. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med 210: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem 286: 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair VS, Song MH, Oh KI. 2016. Vitamin C facilitates demethylation of the Foxp3 enhancer in a Tet-dependent manner. J Immunol 196: 2119–2131. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D’Alessio AC, Nakayama K, Fan HY, Chen X, Xiong Y. 2015. CRL4VprBP E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol Cell 57: 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AV, Albers CG, Holcombe RF. 2010. Differentiation of tubular and villous adenomas based on Wnt pathway-related gene expression profiles. Int J Mol Med 26: 121–125. [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Das S, Im KM, Turan S, Berndt SI, Li H, Lou H, Brodie SA, Billaud JN, Zhang T, et al. 2016. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene. 10.1038/onc.2016.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, Kim S, Van Bodegom D, Bolla S, Schatz JH, et al. 2014. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood 123: 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham WM, Clish CB, Yang Y, Loscalzo J. 2015. Hypoxia-mediated increases in l-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab 22: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, Baxter EJ, Massie CE, Papaemmanuil E, Menon S, et al. 2015. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med 372: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. 2016. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 374: 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Kimler BF, Yi SY, Park SH, Kim K, Jung CW, Kim SH, Lee ER, Rha M, Kim S, et al. 2009. Depletion of l-ascorbic acid alternating with its supplementation in the treatment of patients with acute myeloid leukemia or myelodysplastic syndromes. Eur J Haematol 83: 108–118. [DOI] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. 2011. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38. [DOI] [PubMed] [Google Scholar]
- Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D, Otto GA, Yelensky R, Hricik T, McKenney AS, et al. 2014a. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci 111: E5401–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. 2014b. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 9: 1841–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shide K, Kameda T, Shimoda H, Yamaji T, Abe H, Kamiunten A, Sekine M, Hidaka T, Katayose K, Kubuki Y, et al. 2012. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia 26: 2216–2223. [DOI] [PubMed] [Google Scholar]
- Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et al. 2015. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 27: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EH, Livi CB, Rakheja D, Tan J, Benson D, Parekh V, Kho EY, Ghosh AP, Kirkman R, Velu S, et al. 2014. l-2-Hydroxyglutarate: An epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov 4: 1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, Przychodzen B, Mian SA, Nasser EE, Shooter C, et al. 2010. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood 116: 3923–3932. [DOI] [PubMed] [Google Scholar]
- Soucie E, Hanssens K, Mercher T, Georgin-Lavialle S, Damaj G, Livideanu C, Chandesris MO, Acin Y, Letard S, de Sepulveda P, et al. 2012. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood 120: 4846–4849. [DOI] [PubMed] [Google Scholar]
- Spencer JA, Ferraro F, Roussakis E, Klein A, Wu J, Runnels JM, Zaher W, Mortensen LJ, Alt C, Turcotte R, et al. 2014. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508: 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen YH, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Levine RL, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Finke CM, Mullally A, Li CY, Pardanani A, et al. 2009a. Frequent TET2 mutations in systemic mastocytosis: Clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia 23: 900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Pardanani A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Gangat N, Finke CM, Schwager S, Mullally A, et al. 2009b. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 23: 905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AH, Karra D, Piro RM, et al. 2013. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med 19: 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waha A, Müller T, Gessi M, Waha A, Isselstein LJ, Luxen D, Freihoff D, Freihoff J, Becker A, Simon M, et al. 2012. Nuclear exclusion of TET1 is associated with loss of 5- hydroxymethylcytosine in IDH1 wild-type gliomas. Am J Pathol 181: 675–683. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H, et al. 2015. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 57: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. 2012. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 151: 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zhang YL, Pan WW, Li XM, Wang ZW, Ge ZJ, Zhou JJ, Cang Y, Tong C, Sun QY, et al. 2013. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science 342: 1518–1521. [DOI] [PubMed] [Google Scholar]
- Yue X, Trifari S, Äijö T, Tsagaratou A, Pastor WA, Zepeda-Martínez JA, Lio CWJ, Li X, Huang Y, Vijayanand P, et al. 2016. Control of Foxp3 stability through modulation of TET activity. J Exp Med 213: 377–397. [DOI] [PMC free article] [PubMed] [Google Scholar]