Abstract
Despite the great progress in our understanding of the molecular basis of human cancer, the heterogeneity of individual tumors and the evolutionary pressures imposed by therapy have hampered our ability to effectively eradicate and control this disease. How, therefore, do cancers evolve under the selective pressures of cancer therapy? Recent studies have linked both primary (or de novo) and acquired treatment resistance to intratumor heterogeneity and clonal evolution. Resistance to targeted therapies often includes mutation of the drug target itself and aberrations of pathways upstream of, downstream from, or parallel to the drug target. For systemic chemotherapies, discrete and recurrent resistance-conferring genetic aberrations have eluded the community, due in part to their wide-ranging mutagenic effects. In this review, we discuss different patterns of clonal evolution during treatment-specific selective pressures and focus on the genetic mechanisms of treatment resistance that have emerged to both targeted therapies and chemotherapies.
Targeted therapies and chemotherapies alter the genetic makeup of a tumor and influence its evolution. Characterizing and understanding these evolutionary patterns may lead to more effective cancer treatments.
Genomic instability is considered an enabling characteristic that promotes the acquisition of other hallmarks of cancer (Hanahan and Weinberg 2011) and furthermore creates genetic variation from cell to cell. This variation can be observed as genetic differences within the same tumor, known as intratumor heterogeneity. Although intratumor heterogeneity has been recognized for many years (Nowell 1976; Hansemann 1890), until recently, we lacked the tools to fully characterize the extent and different forms of genomic instability at single-base resolution in cancer and to determine both exogenous factors and endogenous mutational processes that shape cancer genomes. The advent of broad-based next-generation sequencing (NGS), coupled with advances in computational methodologies, has made it possible to identify and interpret the processes that leave a mutational footprint in the cancer genome (Helleday et al. 2014; Hollstein et al. 2016).
In the past decade, studies have begun to apply these novel computational tools to gain further insight into intratumor heterogeneity in a range of different cancer types (Andor et al. 2015; Greaves 2015). Most studies, however, have sampled a given patient’s disease once, which provides only a snapshot of subclonal heterogeneity in treatment-naïve human cancers. Only recently has the combination of lower sequencing costs and new approaches to sample acquisition, such as liquid biopsies, enabled longitudinal analysis of cancer genomes under treatment-selective pressures. Many studies have explored the evolution of a tumor from an ancestral cell to diagnosis using a variety of both theoretical and experimental approaches. Although contributing greatly to our basic understanding of cancer pathogenesis, it is recurrent and metastatic disease that is responsible for the majority of cancer mortality. The evolution these tumors undergo after diagnosis, surgery, and/or adjuvant therapies can drastically alter the initial clonal composition of the treatment-naïve tumor and impact its biological and clinical course. To design the most effective therapeutic approaches that improve the survival of cancer patients, we must therefore study tumors longitudinally after diagnosis and throughout the course of the disease. In this review, we discuss how the selective pressures of therapy can sculpt cancer genome evolution.
RESISTANCE TO DIFFERENT TYPES OF ANTICANCER TREATMENTS
Therapy resistance is a major problem within clinical oncology, and can present itself along different points during treatment. If the treated cancer does not show an initial response and is effectively resistant upfront to the therapy, we refer to this as primary or de novo resistance. To overcome this challenge, a more tailored or precision approach is often undertaken, which is guided in part by histopathology and, at present, limited prospective genomic testing and sequencing. However, even if the patient is carefully selected based on validated biomarkers, the response is often quite variable and relapse in initial responders is common. This form of therapeutic resistance, called acquired resistance, can have a significant impact on the evolutionary course of the disease. Here, we will discuss how targeted therapies and chemotherapies directly and indirectly alter the genetic makeup of the recurrent tumor and drive the patient’s tumor into a disease state distinct from diagnosis.
Genetic Resistance Mechanisms to Targeted Therapy
Depending on the context of the selective pressure, a particular genetic aberration may or may not increase cellular fitness of a subclone (Greaves 2015), respectively contributing to either Darwinian selection and accompanying clonal outgrowth (Nowell 1976; Greaves and Maley 2012) or neutral evolution (Siegmund et al. 2009, 2011; Humphries et al. 2013; Sottoriva et al. 2015; Williams et al. 2016). In the setting of treatment-specific selective pressures, tumor subclones, which have acquired a resistance-conferring aberration but were outcompeted in the absence of therapy, will eventually be selected for and grow into the incumbent clone (Schmitt et al. 2015). Over the past two decades, studies have revealed a large number of discrete genetic aberrations that confer resistance to a myriad of anticancer therapies. In general, these resistance mechanisms can be categorized as (1) genetic aberrations directly affecting the drug target, and (2) aberrations bypassing the drug target through compensatory activation of upstream, downstream, or parallel pathways. In the first case, the drug target itself is mutated to thwart drug binding or to counterbalance target-inhibition by increasing its activity.
Gleevec (or Imatinib, STI571) was the first tyrosine kinase inhibitor to be approved for the treatment of cancer and targets the BCR-ABL fusion oncoprotein in chronic myeloid leukemia (CML) (Capdeville et al. 2002). CML becomes genetically more complex and aggressive as it progresses through three distinct phases, namely, the chronic phase (CML-CP), blast crisis (CML-BC), and accelerated phase (CML-AP). Although the hematological response rates to Gleevec (normalization of blood count) for CML-CP are ∼90%, this number steadily declines for CML-BC (70%) and CML-AP (30%) (Capdeville et al. 2002). Furthermore, the duration of response is often limited to months because of treatment resistance. A panel of drug target mutations, such as BCR-ABL amplification (le Coutre et al. 2000; Weisberg and Griffin 2000) and ABL kinase domain mutations (Gorre et al. 2001; Branford et al. 2003), were identified early on in (pre-) clinical studies.
Besides BCR-ABL, Gleevec’s major targets include c-KIT and PDGFR, both of which were found to be mutually exclusively mutated in gastrointestinal stromal tumors (GISTs) in 80% and 10% of cases, respectively (Hirota et al. 1998; Heinrich et al. 2003b; Rubin et al. 2007). These mutations lead to the constitutive activation of c-KIT and PDGFR-α, and within GIST signify oncogene dependency. Similar to BCR-ABL-driven CML, c-KIT- and PDGFR-α-driven GIST are sensitive to Gleevec therapy, but eventually relapse. The patients who have primary resistant tumors are enriched with c-KIT mutations in exon 9, D842V mutations in PDGFRA or are both wild-type in c-KIT and PDGFRA (Heinrich et al. 2003a). Acquired resistance is largely driven by the acquisition of secondary c-KIT mutations and c-KIT amplifications (Debiec-Rychter et al. 2005).
Another success within targeted therapy includes EGFR inhibition of EGFR-mutated (mostly including exon 19 deletions and L858R mutations) lung adenocarcinomas. Patients with EGFR-mutant lung adenocarcinoma treated with first- and second-generation EGFR inhibitors can experience dramatic responses, but usually progress within 1 or 2 years. Secondary mutations of EGFR occur in more than half of the patients and involve the selection of the EGFR T790M mutation during treatment (Kobayashi et al. 2005; Pao et al. 2005). This has led to the development of third-generation EGFR inhibitors (including rociletinib, AZD9291, and EAI045), which have shown potent activity against EGFR T790M-mutant cells (Walter et al. 2013; Cross et al. 2014; Jia et al. 2016). These drugs have enabled the sequential application of first- and second-generation EGFR inhibitors followed by third-generation EGFR inhibitors (Politi et al. 2015). The strategy of sequential drug therapy has not been limited to EGFR inhibition, but has also been used to counteract resistance of other drug targets, such as BCR-ABL (Cortes et al. 2007, 2012) and ALK (Gainor et al. 2016; Shaw et al. 2016). A complication of sequential therapy is that multiple different drug target mutations can be selected for, also referred to as compound mutations (Shah et al. 2007). Compound mutant cells contain two or more drug-target mutations within the same gene of an individual cell and, hence, may preclude the option of switching back to the initial drug. A potential strategy to prevent compound mutants from arising in the first place might be to provide combination therapy upfront (Misale et al. 2015).
Apart from drug target mutations, resistance mechanisms also include activation of parallel pathways and activation of upstream or downstream effectors in the same pathway. For example, in the setting of monoclonal EGFR-targeting antibodies for the treatment of metastatic colorectal cancer cells, KRAS mutations (downstream effector) (Amado et al. 2008; Karapetis et al. 2008) and MET amplifications (parallel pathway) (Bardelli et al. 2013) are characterized as causes of bypass resistance mechanisms. These data allude to a cancer’s dependency on activity of several key pathways, known as pathway addiction. Parallel and convergent cancer evolution, the phenomenon of different subclones originating from the same or different cancer-initiating cells acquiring genetic aberrations in similar pathways, lend support to the concept of pathway dependency (Misale et al. 2014; Shi et al. 2014; Juric et al. 2015; Pagliarini et al. 2015; Spoerke et al. 2016).
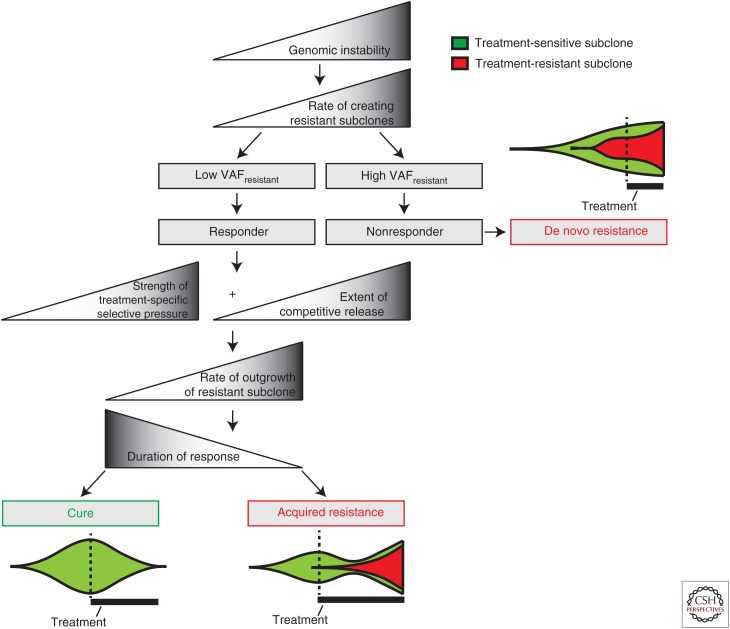
In the clinic, resistance to anticancer treatments can arise upfront (primary or de novo resistance) or after an initial response (acquired resistance). Although these two clinical presentations of resistance occur at different points of the course of disease, it appears that they might be linked (Fig. 1). A recent report suggests that primary and acquired resistance are potentially both influenced by the clonality of the resistant subclone (Laurent-Puig et al. 2015). Laurent-Puig et al. explored why almost half of all KRAS wild-type metastatic colorectal cancer patients did not respond to anti-EGFR monoclonal antibody therapy (Linardou et al. 2008). Using picodroplet digital PCR (dPCR) as a sensitive method, they reassessed the KRAS and BRAF mutation status of primary colorectal cancer samples in 136 patients, who were initially classified as KRAS, NRAS, and BRAF wild-type by qPCR. In total, 22 and two patients contained subclonal KRAS and BRAF mutations, respectively. Interestingly, they observed an inverse correlation between the mutant allele fraction and response. They found that if the mutant allele fraction was below 1.5%, patients could accurately be classified as a responder in 87% of the cases. These findings suggest that an initially minor subclone of resistant cells can be rapidly selected during anticancer treatment (Fig. 1). Hata et al. (2016) lend support for this hypothesis in an in vitro study, in which they mimic the cancer evolutionary path to drug resistance of EGFR-mutant PC9 lung cancer cells to gefitinib. They performed long-term drug exposure experiments on initially sensitive PC9 cells and found significant differences in the rate in which an EGFR T790M-mutant cell line emerged. This difference ultimately relied on the presence of preexisting EGFR T790M-mutant cells within the largely dominating EGFR T790M wild-type population. When they exposed a single-cell-cloned EGFR T790M wild-type culture to gefitinib, it could take up to 40 weeks to derive an EGFR T790M-mutant cell line. However, this process was accelerated to 2 weeks in the presence of only one EGFR T790M-mutant cell (Hata et al. 2016). These findings emphasize why it may be difficult to cure cancers with targeted therapy. First, resistant subclones are selected by treatment-specific selective pressures (Fig. 1). Second, even if the bulk of the drug-sensitive cancer cells is successfully killed, a portion of the EGFR T790M wild-type cells tolerates the drug (drug-tolerant persister cells) and may form a reservoir to produce bona fide genetically resistant cancer cells (also see Sharma et al. 2010; Ramirez et al. 2016).
Figure 1.
General rules governing response to cancer therapies. The subclonality of the resistant subclone partially determines whether the patient is a responder (resistant subclone = minor subclone) or nonresponder (resistant subclone = major subclone) to therapy. Patients who do not experience an initial response to therapy experience “de novo resistance.” In contrast, if a patient initially responds to therapy, the pretreated cancer consists of mostly treatment-sensitive cancer cells. If the cancer consists of only treatment-sensitive cancer cells, the patient can theoretically be cured. However, if residual disease consists of treatment-resistant cancer cells, treatment-induced competitive release, together with treatment-specific selective pressures, confer increased cell fitness to the treatment-resistant subclone. This will partially determine the rate at which the resistant subclone is selected, and consequently will also affect the duration of response.
Chemotherapy-Induced Competitive Release
Clinical oncology continues to be transformed by the use of increasingly mechanism-based targeted medicine; nevertheless, chemotherapy remains the mainstay of many current clinically approved anticancer regimens. Unlike most targeted therapies, chemotherapies alkylate the DNA, inhibit DNA replication, interfere with microtubule function or mitosis in general and can consequently damage the genome through broadly acting mechanisms, and as a result may leave a mutational footprint. Owing to their wide-ranging mutagenic effects, it has been more difficult to find recurrent resistance-conferring genetic aberrations, although a few have been identified (Aas et al. 1996; Li et al. 2010). Recent studies comparing pre- and postchemotherapy-treated clinical samples, have explored how (1) chemotherapy-induced elimination of sensitive subclones, and (2) how chemotherapy-induced mutagenesis influences the course of cancer evolution.
Intratumor genetic heterogeneity can arise by multiple means, either stochastically or via discrete biological mechanism, which can spawn treatment-sensitive and -resistant subclones. The elimination of sensitive subclones reduces the competitive forces imposed by cancer dominant subclones on minor subclones. This so-called “competitive release” allows treatment-resistant minor subclones to repopulate and drive the relapsed tumor, which may be distinct from the treatment-naïve tumor (Enriquez-Navas et al. 2016). To explore this question, Landau et al. (2013) used whole-exome sequencing of paired, longitudinally collected samples for 18 cases of chronic lymphocytic leukemia (CLL). Of these 18 patients, 12 received chemotherapy and six patients remained untreated. The investigators found that chemotherapy induced a shift in the clonal composition of the relapsed disease in 10 of 12 treated cases, whereas only one of six untreated cases showed a shift in the clonal composition at relapse. In general, Landau et al. and others studying haematological malignancies have observed two patterns of clonal repopulation: (1) After therapy, the dominant clone gains more aberrations and evolves into the relapse clone; and (2) a minor subclone at diagnosis later dominates the relapsed disease (Ding et al. 2012; Landau et al. 2013; Garg et al. 2015). Furthermore, Landau et al. (2013) found that patients with a subclonal driver had significantly faster progression of their CLL. These findings were later recapitulated in an expanded study by the same group (Landau et al. 2015), and also by a pan-cancer study involving 12 different cancer types (Andor et al. 2015). A possible interpretation of these findings is that tumor subclones containing cancer driver genes have a fitness advantage over the rest of the tumor but cannot drive a clonal sweep (i.e., leading to clonal dominance within the tumor) if the driver gene is acquired late in tumorigenesis. The potential for the subclone to repopulate the tumor only surfaces after treatment-induced competitive release. It is still unclear to which these observations made from genetically simpler tumors such as CLL (Landau et al. 2013) can be extrapolated to other tumors. For example, in glioma the oncogene, BRAF V600E, was found in an initial tumor, but was not detected in the recurrent tumor after treatment with chemotherapy (temozolomide) (Johnson et al. 2014). Regardless of the precise relevance of subclonal drivers in certain solid tumors, Janiszweska et al. (2015) have provided evidence that chemotherapy-induced competitive release may be a clinically relevant phenomenon in breast cancer. They performed STAR-FISH (allele-specific in situ PCR combined with FISH) for PIK3CA mutations and HER2 amplifications, which are known to respectively confer resistance (Berns et al. 2007) and sensitivity to trastuzumab, in breast cancer treated with neoadjuvant chemotherapy. They found that the PIK3CA-mutant population was low (<8%) before neoadjuvant treatment, in contrast to a relatively high HER2-amplified population (∼30%). These tumors could possibly benefit from anti-HER2 targeted therapy (such as trastuzumab), because the trastuzumab-resistant, PIC3CA-mutant population is only present as a minor subclone. Interestingly, after chemotherapy the PIK3CA-mutant population increased considerably (to ∼20%), whereas the HER2-amplified population slightly decreased (to ∼23%). These tumors are, therefore, less likely to respond to sequential anti-HER2 treatment. This study suggests that the sequence of chemotherapy could select for a population that could later induce the nonresponse to anti-HER2 therapy, possibly sculpting an anti-HER2 responsive tumor into a de novo resistant tumor.
Fortunately, our increasing understanding of a phenomenon like competitive release may provide a potential solution to impede the selection of resistant subclones, namely, by modulating the treatment-specific selective pressure. This is a form of “adaptive therapy,” which has been defined by Gatenby et al. (2009) as a “treatment-for-stability strategy” in which the goal is to maintain a stable treatment-sensitive subclone, which will repress the emergence of the treatment-resistance subclone. The fitness of these two competing subclones differs according to the strength of the treatment-specific selective pressures (Gatenby et al. 2009; Das Thakur et al. 2013; Enriquez-Navas et al. 2016). The treatment-sensitive subclone dominates in the absence of the treatment and represses the treatment-resistant subclone. In contrast, the resistant subclone has an increased fitness in the presence of treatment and furthermore experiences less competition by treatment-induced elimination of the sensitive subclone, with competitive release as a consequence (Fig. 1). The aim of adaptive therapy is to stabilize the cancer as opposed to curing the cancer, by maintaining a balance between clonal interference and treatment-induced competitive release.
Chemotherapy-Induced Mutagenesis
Besides inducing competitive release, chemotherapies can also drive distinct evolutionary trajectories in individual tumors through their mutagenic effects. In a series of seminal papers, Alexandrov et al. (2013) detailed methodology to deconstruct the mutational processes that have contributed to genetic aberrations during the development of cancers. They subdivided the six classes of nucleotide base substitutions into 16 subgroups, each according to their 5′ and 3′ bases, resulting in 96 different possible mutations. They catalogued each of these 96 different types of base substitutions in 30 different cancer types, and accordingly described more than 30 different mutational signatures (Petljak and Alexandrov 2016). Of these signatures, they were able to identify a potential biological cause for 18 of them. Although it was previously known that different (chemo)therapeutics were mutagenic (Gupta et al. 1987; Lemaire et al. 1991; Pillaire et al. 1995), NGS together with novel bioinformatics analysis methods enables the quantification of how different therapies (in)activate cellular pathways, which in turn can sculpt cancer evolution. One of these signatures includes the previously described signature of the alkylating chemotherapeutic drug, temozolomide (TMZ). Besides TMZ, the mutational signatures of additional chemotherapies have been elucidated, such as that of cisplatin and cyclophosphamide (Murugaesu et al. 2015; Szikriszt et al. 2016). Furthermore, several chemotherapies can activate APOBEC3, and can potentially promote APOBEC3 mutagenesis (Kanu et al. 2016).
Johnson et al. (2014) used one mutational signature to investigate the contribution of TMZ treatment on the progression of low-grade gliomas to high-grade gliomas. They sequenced matched samples of 23 grade II, IDH1-mutant gliomas and their recurrences after tumor progression (Johnson et al. 2014). Ten patients were treated with TMZ and seven of these recurred as glioblastomas. Interestingly, six of the seven patients contained a TMZ-induced hypermutation phenotype, which was previously linked to TMZ resistance (Bodell et al. 2003; Hunter et al. 2006; Cahill et al. 2007; Yip et al. 2009). Johnson et al. investigated whether any of these TMZ-induced mutations occurred in any of previously identified driver genes in glioblastoma (Cancer Genome Atlas Research 2008; Brennan et al. 2013). Accordingly, they found mutations in RB1, CDKN2A, PIK3CA, PTEN, and MTOR within a TMZ mutational context (Johnson et al. 2014). These findings caused further concern regarding the use of TMZ in the treatment of low-grade gliomas; besides chemotherapy-induced competitive release, TMZ also appeared to induce mutations in driver genes, suggesting it might fuel a more aggressive, higher-grade glioma at recurrence. In a different study by Kim et al. (2015b), paired pre- and post-TMZ treated samples of 34 primary glioblastomas and four secondary glioblastomas were sequenced. Interestingly, they found that within IDH1 wild-type primary glioblastomas, TMZ did not induce a hypermutation phenotype in the recurrent tumor. In an independent study (Kim et al. 2015a), in which 21 matched IDH1 wild-type glioblastomas and recurrences were investigated, four recurred as hypermutated tumors. Collectively, these studies indicate that TMZ can induce a hypermutation phenotype in recurrent tumors of primary as well as secondary glioblastomas (Johnson et al. 2014; Kim et al. 2015a), albeit secondary glioblastomas appear more prone to this phenomenon (Kim et al. 2015b). It is still unclear whether the risks of TMZ outweigh its benefits, especially in IDH1-mutant low-grade gliomas (Field et al. 2016). Similar to TMZ, the mutagenic effects of other chemotherapies should be systematically assessed using pre- and post-chemotherapy-treated clinical samples. An unanswered question remains whether other chemotherapies can induce driver mutations and whether the antitumor benefits of the specific chemotherapy outweigh the risk of malignant progression (Lee et al. 2012), especially in the treatment of indolent tumors (Field et al. 2016). Apart from driver mutations, chemotherapeutic regimens may increase intratumor heterogeneity by contributing to the burden of subclonal mutations. If these mutations are within the exome, they may create neoantigens that can be presented to the immune system. McGranahan et al. (2016) observed that the extent of neoantigen intratumor heterogeneity holds predictive value for response to immune checkpoint blockade within non-small-cell lung cancer and melanoma. Two patients with the most heterogeneous tumors within a cohort of melanoma patients were found to be pretreated with the alkylating agent dacarbazine before immune checkpoint blockade. They found that the tumors of these two patients were enriched for signature 11 mutations (McGranahan et al. 2016), which is associated with the mutagenic effects of alkylating agents (Alexandrov et al. 2013). Although still speculative, because of the small sample size, their data suggest that subclonal diversification might preclude the generation of an effective immune response (McGranahan et al. 2016).
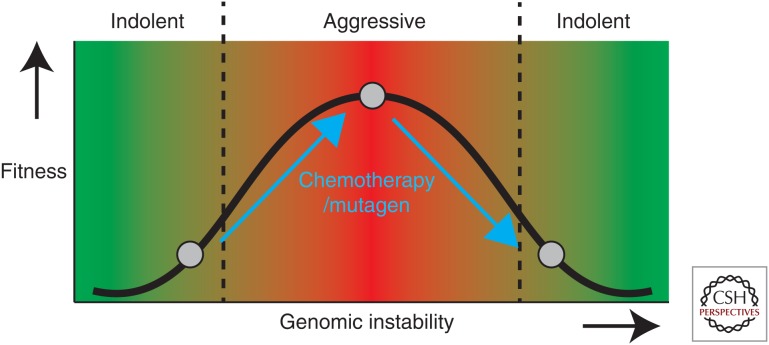
Although informative, mutational signatures only provide information regarding smaller-scaled genetic aberrations, such as single point mutations, dinucleotide mutations, and insertions and deletions (Alexandrov et al. 2013; Helleday et al. 2014), and not about chromosome level changes. This tendency of cancer cells to lose, gain, or rearrange portions of chromosomes, is also known as chromosomal instability (CIN) and is a form of genomic instability. CIN appears to contribute to drug resistance (Lee et al. 2011), tumor progression within colorectal cancer (Lengauer et al. 1998; Carter et al. 2006), and worse patient prognosis if CIN is in an optimal range (Birkbak et al. 2011; Andor et al. 2015). Recent studies have quantified the extent to which different classes of therapies induce CIN (Lee et al. 2013, 2016; Kim et al. 2016). Lee et al. (2013) have developed a nonselective human artificial chromosome (HAC) that contains kinetochores and the EGFP transgene. This enabled them to investigate the rate at which the HAC is lost during different drug treatments, which they used as a measure of CIN (Lee et al. 2013, 2016). They confirmed that different classes of chemotherapeutics, such as microtubule-stabilizing drugs and inhibitors targeting the DNA damage response/replication, were especially potent in inducing HAC-loss and thus CIN (Lee et al. 2013, 2016). It is conceivable that CIN-enhancing drugs can force indolent cancers toward a more genomic instable, aggressive state; and force highly genomic instable cancers beyond the point of tolerance and diminish cancer cell fitness (Fig. 2) (Janssen and Medema 2013).
Figure 2.
Chemotherapies and mutagens may increase genomic instability and alter the cancer cell fitness as a consequence. Genomic instability can promote tumor cell fitness, if present in an optimal range. If the level of genomic instability is too low or too high, it can diminish tumor cell fitness. Chromosomal instability (CIN)-enhancing drugs can potentially increase the level of genomic instability and can push indolent cancers toward a more genomic-instable, aggressive state and force highly genomic instable cancers beyond the point of tolerance, diminishing cancer cell fitness.
CONCLUSIONS
Both targeted therapies and chemotherapies influence the path of cancer evolution. Although we have focused our discussions around genetic correlates of cancer evolution, transcriptional, epigenetic, and posttranslational mechanisms of resistance must not be overlooked. Research over the past few decades has shown that cancer is dependent on the activity of specific pathways (Misale et al. 2014). Genetic resistance mechanisms to targeted therapy show how genomic instability enables tumors to create the appropriate subclone, which contains the (re)activated pathway. Knowledge about the mechanisms of drug resistance has enabled the scientific community to anticipate the phenomenon clinically and to develop drugs that counteract these specific resistance mechanisms. We hope that characterization of these different resistance mechanisms will enable us to formulate cancer evolutionary rulebooks. This will be especially challenging for chemotherapies. In addition to imposing treatment-specific selective pressures, they also tend to be mutagenic (Lee et al. 2013, 2016; Olivier et al. 2014; Hollstein et al. 2016; Kim et al. 2016; Szikriszt et al. 2016). Intratumor heterogeneity together with Darwinian selection can confound almost any type of cancer treatment, including new therapeutic strategies of great promise such as immunotherapy, in which immunoediting selects for less immunogenic escape subclones (Dudley and Roopenian 1996; Phillips 2002; Schreiber et al. 2011; DuPage et al. 2012; Matsushita et al. 2012; Zaretsky et al. 2016). Nevertheless, we are optimistic that developments in NGS, together with evolving bioinformatics tools, will allow us to systematically deconstruct the evolutionary history of cancers and to predict cancer’s Achilles’ heel.
ACKNOWLEDGMENTS
We thank Dr. Gerald Goh for helpful discussions of this manuscript. S.V. is funded by Stand Up 2 Cancer and a CRICK PhD studentship. C.S. is a Royal Society Napier Research Professor. This work is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001169), the UK Medical Research Council (FC001169), and the Wellcome Trust (FC001169); by the UK Medical Research Council (Grant reference MR/FC001169 /1); C.S. is funded by Cancer Research UK (TRACERx), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees Trust, NovoNordisk Foundation (ID 16584), the Prostate Cancer Foundation, the Breast Cancer Research Foundation, the European Research Council (THESEUS) and Support was provided to C.S. by the National Institute for Health Research, the University College London Hospitals Biomedical Research Centre, and the Cancer Research UK University College London Experimental Cancer Medicine Centre. B.S.T. acknowledges support from the Prostate Cancer Foundation, the Sontag Foundation, the Josie Robertson Foundation, Cycle for Survival, the American Cancer Society (127350-RSG-15-067-01-TBG), National Institutes of Health (NIH) grants R01CA207244 and U54OD202355, and the Sloan Kettering Institute for Cancer Research Cancer Center Support Grant (P30CA008748). J.F.C. is generously supported by a gift from the Dabbiere family and the Hana Jabsheh Research Initiative, and by the National Institutes of Health R01CA169316 and P01CA118816. The authors have no conflicts of interest to declare.
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
- Aas T, Borresen AL, Geisler S, Smith-Sorensen B, Johnsen H, Varhaug JE, Akslen LA, Lonning PE. 1996. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 2: 811–814. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. 2008. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634. [DOI] [PubMed] [Google Scholar]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. 2015. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D, et al. 2013. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 3: 658–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. 2007. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12: 395–402. [DOI] [PubMed] [Google Scholar]
- Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, Tan IB, Richardson AL, Szallasi Z, Swanton C. 2011. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res 71: 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodell WJ, Gaikwad NW, Miller D, Berger MS. 2003. Formation of DNA adducts and induction of lacI mutations in Big Blue Rat-2 cells treated with temozolomide: Implications for the treatment of low-grade adult and pediatric brain tumors. Cancer Epidemiol Biomarkers Prev 12: 545–551. [PubMed] [Google Scholar]
- Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, Taylor K, Herrmann R, Seymour JF, Arthur C, et al. 2003. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood 102: 276–283. [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. 2013. The somatic genomic landscape of glioblastoma. Cell 155: 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, et al. 2007. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res 13: 2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdeville R, Buchdunger E, Zimmermann J, Matter A. 2002. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov 1: 493–502. [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 38: 1043–1048. [DOI] [PubMed] [Google Scholar]
- Cortes J, Jabbour E, Kantarjian H, Yin CC, Shan J, O’Brien S, Garcia-Manero G, Giles F, Breeden M, Reeves N, et al. 2007. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood 110: 4005–4011. [DOI] [PubMed] [Google Scholar]
- Cortes JE, Kantarjian H, Shah NP, Bixby D, Mauro MJ, Flinn I, O’Hare T, Hu S, Narasimhan NI, Rivera VM, et al. 2012. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med 367: 2075–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ, et al. 2014. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 4: 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, Stuart DD. 2013. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debiec-Rychter M, Cools J, Dumez H, Sciot R, Stul M, Mentens N, Vranckx H, Wasag B, Prenen H, Roesel J, et al. 2005. Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology 128: 270–279. [DOI] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley ME, Roopenian DC. 1996. Loss of a unique tumor antigen by cytotoxic T lymphocyte immunoselection from a 3-methylcholanthrene-induced mouse sarcoma reveals secondary unique and shared antigens. J Exp Med 184: 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. 2012. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 482: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Navas PM, Kam Y, Das T, Hassan S, Silva A, Foroutan P, Ruiz E, Martinez G, Minton S, Gillies RJ, et al. 2016. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Transl Med 8: 327ra324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field KM, Rosenthal MA, Khasraw M, Sawkins K, Nowak AK. 2016. Evolving management of low grade glioma: No consensus amongst treating clinicians. J Clin Neurosci 23: 81–87. [DOI] [PubMed] [Google Scholar]
- Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K, Singh M, et al. 2016. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 6: 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, Zang ZJ, Okuno Y, Shiraishi Y, Chiba K, et al. 2015. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood 126: 2491–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Silva AS, Gillies RJ, Frieden BR. 2009. Adaptive therapy. Cancer Res 69: 4894–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. 2001. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293: 876–880. [DOI] [PubMed] [Google Scholar]
- Greaves M. 2015. Evolutionary determinants of cancer. Cancer Discov 5: 806–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. 2012. Clonal evolution in cancer. Nature 481: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Bromke A, Bryant DW, Gupta R, Singh B, McCalla DR. 1987. Etoposide (VP16) and teniposide (VM26): Novel anticancer drugs, strongly mutagenic in mammalian but not prokaryotic test systems. Mutagenesis 2: 179–186. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hansemann D. 1890. Ueber asymmetrische Zelltheilung in Epithelkrebsen und deren biologische Bedeutung. Virchows Archiv 119: 299–326. [Google Scholar]
- Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HC, Krishnamurthy Radhakrishna V, et al. 2016. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 22: 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, et al. 2003a. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 21: 4342–4349. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. 2003b. PDGFRA activating mutations in gastrointestinal stromal tumors. Science 299: 708–710. [DOI] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. 2014. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet 15: 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, et al. 1998. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 279: 577–580. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Alexandrov LB, Wild CP, Ardin M, Zavadil J. 2016. Base changes in tumour DNA have the power to reveal the causes and evolution of cancer. Oncogene 10.1038/onc.2016/192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries A, Cereser B, Gay LJ, Miller DS, Das B, Gutteridge A, Elia G, Nye E, Jeffery R, Poulsom R, et al. 2013. Lineage tracing reveals multipotent stem cells maintain human adenomas and the pattern of clonal expansion in tumor evolution. Proc Natl Acad Sci 110: E2490–E2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, et al. 2006. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res 66: 3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszweska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, Weigelt B, Hanker AB, Chandarlapaty S, King TA, et al. 2015. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet 47: 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Medema RH. 2013. Genetic instability: Tipping the balance. Oncogene 32: 4459–4470. [DOI] [PubMed] [Google Scholar]
- Jia Y, Yun CH, Park E, Ercan D, Manuia M, Juarez J, Xu C, Rhee K, Chen T, Zhang H, et al. 2016. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 534: 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. 2014. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343: 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et al. 2015. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 518: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanu N, Cerone MA, Goh G, Zalmas LP, Bartkova J, Dietzen M, McGranahan N, Rogers R, Law EK, Gromova I, et al. 2016. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol 17: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. 2008. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765. [DOI] [PubMed] [Google Scholar]
- Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, Grimsby J, Sougnez C, Muller F, Hu J, et al. 2015a. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res 25: 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, Nam SH, Kim BS, Johnson MD, Kong DS, et al. 2015b. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell 28: 318–328. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee HS, Lee NC, Goncharov NV, Kumeiko V, Masumoto H, Earnshaw WC, Kouprina N, Larionov V. 2016. Development of a novel HAC-based “gain of signal” quantitative assay for measuring chromosome instability (CIN) in cancer cells. Oncotarget 7: 14841–14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. 2005. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352: 786–792. [DOI] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Bottcher S, et al. 2015. Mutations driving CLL and their evolution in progression and relapse. Nature 526: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Puig P, Pekin D, Normand C, Kotsopoulos SK, Nizard P, Perez-Toralla K, Rowell R, Olson J, Srinivasan P, Le Corre D, et al. 2015. Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res 21: 1087–1097. [DOI] [PubMed] [Google Scholar]
- le Coutre P, Tassi E, Varella-Garcia M, Barni R, Mologni L, Cabrita G, Marchesi E, Supino R, Gambacorti-Passerini C. 2000. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood 95: 1758–1766. [PubMed] [Google Scholar]
- Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, Downward J, Szallasi Z, Tomlinson IP, Howell M, et al. 2011. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 71: 1858–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, et al. 2012. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 122: 2983–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lee NC, Grimes BR, Samoshkin A, Kononenko AV, Bansal R, Masumoto H, Earnshaw WC, Kouprina N, Larionov V. 2013. A new assay for measuring chromosome instability (CIN) and identification of drugs that elevate CIN in cancer cells. BMC Cancer 13: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lee NC, Kouprina N, Kim JH, Kagansky A, Bates S, Trepel JB, Pommier Y, Sackett D, Larionov V. 2016. Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res 76: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire MA, Schwartz A, Rahmouni AR, Leng M. 1991. Interstrand cross-links are preferentially formed at the d (GC) sites in the reaction between cis-diamminedichloroplatinum (II) and DNA. Proc Natl Acad Sci 88: 1982–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, Desmedt C, Sotiriou C, Szallasi Z, Iglehart JD, et al. 2010. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 16: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. 2008. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 9: 962–972. [DOI] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. 2012. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482: 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT. 2016. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351: 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. 2014. Resistance to anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov 4: 1269–1280. [DOI] [PubMed] [Google Scholar]
- Misale S, Bozic I, Tong J, Peraza-Penton A, Lallo A, Baldi F, Lin KH, Truini M, Trusolino L, Bertotti A, et al. 2015. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nat Commun 6: 8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaesu N, Wilson GA, Birkbak NJ, Watkins TB, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, et al. 2015. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 5: 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. 1976. The clonal evolution of tumor cell populations. Science 194: 23–28. [DOI] [PubMed] [Google Scholar]
- Olivier M, Weninger A, Ardin M, Huskova H, Castells X, Vallee MP, McKay J, Nedelko T, Muehlbauer KR, Marusawa H, et al. 2014. Modelling mutational landscapes of human cancers in vitro. Sci Rep 4: 4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini R, Shao W, Sellers WR. 2015. Oncogene addiction: Pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep 16: 280–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. 2005. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petljak M, Alexandrov LB. 2016. Understanding mutagenesis through delineation of mutational signatures in human cancer. Carcinogenesis 37: 531–540. [DOI] [PubMed] [Google Scholar]
- Phillips RE. 2002. Immunology taught by Darwin. Nat Immunol 3: 987–989. [DOI] [PubMed] [Google Scholar]
- Pillaire M, Hoffmann J, Defais M, Villani G. 1995. Replication of DNA containing cisplatin lesions and its mutagenic consequences. Biochimie 77: 803–807. [DOI] [PubMed] [Google Scholar]
- Politi K, Ayeni D, Lynch T. 2015. The next wave of EGFR tyrosine kinase inhibitors enter the clinic. Cancer Cell 27: 751–753. [DOI] [PubMed] [Google Scholar]
- Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu CH, Thurley K, et al. 2016. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 7: 10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Heinrich MC, Corless CL. 2007. Gastrointestinal stromal tumour. Lancet 369: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Schmitt MW, Loeb LA, Salk JJ. 2015. The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev Clin Oncol 13: 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. 2011. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 331: 1565–1570. [DOI] [PubMed] [Google Scholar]
- Shah NP, Skaggs BJ, Branford S, Hughes TP, Nicoll JM, Paquette RL, Sawyers CL. 2007. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest 117: 2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, Burke BJ, Deng YL, Liu W, Dardaei L, et al. 2016. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med 374: 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. 2014. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Marjoram P, Woo YJ, Tavare S, Shibata D. 2009. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci 106: 4828–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Marjoram P, Tavare S, Shibata D. 2011. High DNA methylation pattern intratumoral diversity implies weak selection in many human colorectal cancers. PLoS ONE 6: e21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, et al. 2015. A Big Bang model of human colorectal tumor growth. Nat Genet 47: 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, Aimi J, Derynck MK, Chen M, Chan IT, et al. 2016. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 7: 11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szikriszt B, Poti A, Pipek O, Krzystanek M, Kanu N, Molnar J, Ribli D, Szeltner Z, Tusnady GE, Csabai I, et al. 2016. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol 17: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al. 2013. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov 3: 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E, Griffin JD. 2000. Mechanism of resistance to the ABL tyrosine kinase inhibitor STI571 in BCR/ABL-transformed hematopoietic cell lines. Blood 95: 3498–3505. [PubMed] [Google Scholar]
- Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. 2016. Identification of neutral tumor evolution across cancer types. Nat Genet 48: 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN. 2009. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res 15: 4622–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. 2016. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 375: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]