Abstract
Sleep is homeostatically regulated with sleep pressure accumulating with the increasing duration of prior wakefulness. Yet, a clear understanding of the molecular components of the homeostat, as well as the molecular and cellular processes they sense and control to regulate sleep intensity and duration, remain a mystery. Here, we will discuss the cellular and molecular basis of sleep homeostasis, first focusing on the best homeostatic sleep marker in vertebrates, slow wave activity; second, moving to the molecular genetic analysis of sleep homeostasis in the fruit fly Drosophila; and, finally, discussing more systemic aspects of sleep homeostasis.
Although much has yet to be learned about the molecular and cellular controls of sleep intensity and duration, research on mammals and flies is revealing some conserved features (e.g., neuronal plasticity).
Sleep is a tightly regulated process, which is one of the most compelling indications that it must fulfill fundamental functions. Sleep timing is controlled by the circadian clock, which promotes the consolidation of sleep during the day in nocturnal animals like many rodents, or during the night in diurnal species such as Drosophila melanogaster and humans. Sleep is also strongly homeostatically regulated, that is, the pressure to go to sleep increases with time spent awake. As sleep homeostatic processes are dependent on the duration of prior wakefulness, they can be assessed under baseline conditions, for example, after a lengthy bout of spontaneous wakefulness, or after induced sleep deprivation, that is, artificially extending wakefulness. The homeostat is thought to be a major determinant of sleep intensity even under baseline conditions. Although there is increasing appreciation of the close link between circadian and homeostatic regulation of sleep, the two processes are separable. Here, we focus on sleep homeostasis, and briefly summarize the factors known to affect sleep need in mammals and flies.
Sleep homeostasis is traditionally measured by actively disrupting sleep, typically using mechanical stimuli. When animals recover sleep after sleep loss (sleep rebound), they do so by increasing its duration and/or by enhancing its intensity. Because of the constraints imposed by the circadian clock, changes in sleep intensity are often larger and more common than changes in sleep duration, but they may be also more difficult to assess, at least in Drosophila. They include an increase in arousal thresholds—the intensity of the stimuli required to wake up a sleeping subject—a phenomenon well documented in both flies and mammals (Faville et al. 2015). In rodents, birds, and humans, they also include changes in neuronal activity that can be measured with great accuracy using the electroencephalogram (EEG), most notably changes in the number and amplitude of the sleep slow waves (slow wave activity [SWA]) (further discussed below).
Sleep benefits both the body and the brain. Sleep loss broadly impairs cognitive functions, and there is evidence that the recovery from these detrimental effects requires sleep, not just rest (Goel et al. 2015). There is also evidence that several other physiological functions are affected by sleep deprivation, from metabolism to the immune response (further discussed below). Sleep loss may also contribute to the pathogenesis of diseases, such as Alzheimer’s disease, perhaps via clearance of neurotoxic molecules (Xie et al. 2013). Not surprisingly, many physiological processes can affect sleep homeostasis. Below, we discuss some of the most important molecules and signaling pathways that mediate these processes and are known to affect sleep homeostasis, as assessed in flies and mammals using measures of both sleep duration and sleep intensity.
A core homeostatic process should both be affected by sleep and, in turn, affect sleep. Most homeostatic models posit that wake-dependent homeostatic factors accumulate with increasing duration of prior wakefulness (Porkka-Heiskanen et al. 1997). These factors are sensed in and activate neurons that promote sleep or inhibit those that promote wake. To dissect the molecular basis of sleep homeostasis, a wide range of approaches has been used to identify pathways that contribute to sleep homeostasis, including unbiased forward genetic screens using chemical mutagens (Cirelli et al. 2005a; Wu et al. 2008) or transposable elements (Koh et al. 2008), RNA interference (RNAi), and the cloning of quantitative trait loci (Harbison and Sehgal 2008) as well as reverse or candidate genetics. In theory, a “homeostatic mutant” should impact the homeostatic response to sleep deprivation, typically evident as a change in the amount or intensity (e.g., SWA in vertebrates) during recovery sleep. As homeostasis also operates in the absence of induced sleep deprivation, an alteration of sleep levels or intensity under baseline conditions may also be observed, although changes may be subtle. Nonetheless, genetic studies suggest that induced sleep deprivation may also activate homeostatic processes that may be qualitatively distinct from those seen under baseline conditions, suggesting that there are context-dependent homeostatic pathways (e.g., see Dubowy et al. 2016). In flies in which in vivo electrophysiological markers of sleep intensity are not easily or typically assessed, a reduction in recovery sleep, typically quantified as percentage of sleep lost that is recovered over a defined time period, could mean an impaired homeostat that is unable to provide sufficient sleep or a highly efficient homeostat that is capable of restorative effects with very little sleep. These opposing interpretations could be distinguished by assaying the functional consequences of sleep loss, for example, on learning and memory (Dissel et al. 2015b).
To identify processes affected by sleep and sleep loss, unbiased transcriptomic and translatomic approaches such as microarrays (Cirelli and Tononi 1999; Cirelli et al. 2004, 2006; Mackiewicz et al. 2009; Romcy-Pereira et al. 2009), RNA-sequencing (Yelin-Bekerman et al. 2015), and translating ribosome affinity purification (Bellesi et al. 2013, 2015) have been used. In addition, candidate genetic approaches using RNA in situ hybridization and immunofluorescence targeted toward specific genes and proteins (Gilestro et al. 2009) and neurophysiological approaches (Vyazovskiy et al. 2008) have revealed molecular and cellular consequences of sleep loss. Such sleep–wake-dependent processes may be evident after natural sleep–wake cycles and/or induced sleep deprivation. Changes may be assessed throughout the brain, specific parts of the brain, or in specific neurons. Of special interest to sleep homeostasis are those changes in the many specific neuronal groups that are known to play a role in initiating and/or maintaining sleep or wakefulness. Sleep–wake-induced changes may also fall into those that reflect state-dependent markers, that is, those that acutely track behavioral state, and homeostatic markers, those that reflect sleep–wake history. The latter would be expected to accumulate more slowly (i.e., over hours). A plausible sleep homeostatic factor should, for example, be induced by sleep deprivation and, in turn, promote sleep. A factor that is induced by sleep loss but has no function in sleep regulation may mediate the effects of sleep loss on the brain or allow the organism to cope with sleep loss, but is unlikely to be mediating sleep homeostasis, at least not directly. Here, we will first discuss the molecular and cellular basis of sleep homeostasis in mammals, in which a well-established electrophysiological marker (SWA) is available, and then focus on Drosophila, stressing similarities and differences in the current knowledge of sleep regulation across species.
SLOW WAVE ACTIVITY AND SLEEP HOMEOSTASIS IN MAMMALS
Sleep in mammals includes non-rapid eye movement (NREM) sleep, which represents the bulk of sleep (∼80%), and rapid eye movement (REM) sleep. The duration of NREM and of REM sleep are modulated by the circadian system to a different extent across species, with humans showing stronger circadian influence on REM sleep and the rat on NREM sleep (Yasenkov and Deboer 2012). There is no established marker of sleep homeostasis for REM sleep. In contrast, the SWA of NREM sleep is a sensitive marker of sleep intensity and sleep need. It is defined as the EEG power in the 0.5 to 4.5 Hz during NREM sleep, and provides a quantitative measure of the amplitude and incidence (number per minute of NREM sleep) of sleep slow waves (also called delta waves) (Feinberg et al. 1978). During NREM sleep, neurons in the cortex and thalamus are bistable, oscillating every second or so between an UP state characterized by membrane depolarization and wake-like tonic firing, and a DOWN state characterized by membrane hyperpolarization and neuronal silence (Steriade et al. 2001; Chauvette et al. 2011). Because these slow oscillations occur more or less synchronously in many neurons, their summed activity can be recorded from the scalp as slow waves, with the DOWN state of thalamocortical neurons corresponding to the negative phase of the slow waves recorded from the scalp. The bistability of thalamic and cortical neurons during NREM sleep impairs cortical information transmission and cortico–cortical effective connectivity (Massimini et al. 2005; Pigorini et al. 2015). Perhaps not surprisingly, therefore, SWA is linked to the depth of NREM sleep: the higher the SWA, the higher is the intensity of the stimulus required to wake up from NREM sleep (Blake and Gerard 1937; Williams et al. 1964; Rosa and Bonnet 1985; Neckelmann and Ursin 1993; Ferrara et al. 1999; Ermis et al. 2010). Thus, there is a close link between two measures of sleep intensity, arousal threshold, and SWA. Of note, recent evidence shows that isolated slow waves also occur during REM sleep, but only in primary sensory and motor areas, and mainly in layer 4, the primary recipient of thalamic inputs, and layer 3 (Funk et al. 2016). This local SWA may partly account for the sensory disconnection during REM sleep, when the EEG is tonically activated.
SWA is also a marker of sleep propensity and has been used as the main correlate for process “S,” the homeostatic factor that determines the timing of sleep in combination with the circadian factor “C” (Borbély 1982; Achermann and Borbély 2003; Tobler 2005; Deboer 2015). This is because SWA increases with wake duration, declines in the course of sleep, and it is even higher after sleep deprivation, suggesting that it reflects the accumulation of sleep pressure during wake and its discharge during sleep. It is worth pointing out that, although the focus of this review is on flies and mammals, the slow waves of NREM sleep are also homeostatically regulated in birds, with SWA increasing after short periods of sleep deprivation in pigeons and sparrows (Rattenborg et al. 2009). It should also be noted that after sleep deprivation the increase in EEG power is not restricted to SWA, but often spans all or most frequencies below 10–11 Hz, and often occurs in both NREM and REM sleep, as well as in wake (Borbély et al. 1981, 1984; Dijk et al. 1987). However, after sleep loss, the most consistent and largest increase is seen below 4.5 Hz during NREM sleep, hence the focus on NREM SWA.
What are the mechanisms underlying the increase of SWA with wake and its decline during sleep? In other words, why does SWA work well as a marker of sleep pressure? The first studies that linked amplitude and number of slow waves to sleep pressure speculated that sleep need may reflect the occurrence of “the metabolic processes which reverse the effects of wakefulness on the brain” (Feinberg et al. 1978), or that “it is related to the level of an endogenous sleep compound which is eliminated or inactivated during sleep” (Borbély et al. 1981). More recently, it was suggested that SWA ultimately reflects the progressive depletion of glycogen during wake, which triggers the build-up of adenosine in the cerebral cortex and the subsequent further potentiation of the potassium currents that mediate the hyperpolarization of cortical neurons during the DOWN state (Benington and Heller 1995). Sleep deprivation studies, however, have shown that changes in glycogen levels vary depending on wake duration, brain region, and mouse strain (Scharf et al. 2008). Nonetheless, there is evidence that the build-up of adenosine in the basal forebrain and other brain regions is at least one of the mechanisms by which the brain senses wake duration (Scharf et al. 2008). The arousal- and sleep-promoting systems identified in the mammalian brain are widespread and redundant, most likely because the maintenance of an ordered sleep/wake cycle is crucial to survival (Luppi et al. 2016; Weber and Dan 2016). Similarly, it is likely that several, if not many, neuronal groups can transmit the homeostatic sleep signal that is crucial to control sleep need and may do so using different signals.
Another recent hypothesis suggests that what is being accumulated during wake and reduced during sleep is actually not a molecule, but the overall synaptic strength of many neural circuits, which increases during wake because of learning and needs to be renormalized during sleep to avoid the high cost of plasticity from the increased need for energy and cellular supplies to the saturation of the ability to learn (Tononi and Cirelli 2014). According to this view, SWA increases after extended wake mainly because neurons are more strongly connected to each other, thus firing in higher synchrony. Indeed, the onset of ON and OFF periods is more synchronous among neural populations after wake, suggesting stronger neuronal coupling (Vyazovskiy et al. 2009). Higher SWA after long wake may also result from a larger number of neurons simultaneously recruited into the slow oscillation, consistent with the fact that slow waves are largest and more global (i.e., they can be simultaneously detected by more EEG electrodes) in early sleep than in late sleep (Nir et al. 2011). During sleep, instead, SWA decreases because synaptic renormalization leads to weaker connections (Tononi and Cirelli 2014). Computer simulations also showed that, in the course of sleep, the progressive decline in SWA and in the amplitude/slope of slow waves can be accounted for by a gradual increase in neuromodulatory tone, which would make neurons less bistable (Esser et al. 2007). However, so far, there is no strong evidence supporting this hypothesis, at least for the noradrenergic system (Bellesi et al. 2016). Note that the mechanisms discussed above that could account for increased SWA after sleep loss—increased potassium conductances, increased synaptic strength, and decreased arousal tone—are not mutually exclusive, because changes in synaptic strength and in neuronal excitability often go together, and neuromodulators such as acetylcholine and noradrenaline affect both of them. Note also that energy metabolism and neural plasticity are strongly linked, because synaptic potentiation requires neural activity. As discussed below, changes in SWA can be triggered by changes in neural activity, although the latter is likely not the sole, or even the most crucial, determinant of SWA.
Forced desychrony experiments in humans, as well as sleep deprivation in mice performed at different times of day, show that a circadian influence on SWA exists, but is generally small and depends on the specific cortical area (Dijk and Czeisler 1995; Cajochen et al. 2002; Hanlon et al. 2009; Curie et al. 2013; Lazar et al. 2015). NREM SWA has high hereditability, but this is true for most EEG frequencies in both wake and sleep (Cirelli 2009). More crucially, however, studies in mice show that the increase in NREM SWA after sleep deprivation is also under genetic control (Franken et al. 2001). It was found that Dps1, a quantitative trait locus (QTL) on mouse chromosome 13, accounts for 49% of the genetic variance in this trait (Franken et al. 2001; Andretic et al. 2008a), and the immediate early gene Homer1a was singled out as a likely candidate gene to carry the homeostatic signal within Dps1 (Maret et al. 2007; Mackiewicz et al. 2008; Curie et al. 2013). Homeostasis is believed to be mainly reflected not in the levels of SWA per se but in the speed of wake-dependent accumulation and sleep-dependent dissipation of SWA. The time constants of the buildup and decline of SWA have been described by exponential functions, a saturating exponential increase in wake, and an exponential decline in sleep. Time constants show strong interindividual differences and both increase and decrease show similarly wide ranges. For instance, in humans, the time constant of the SWA increase ranges between ∼14 and 26 h, and that of the decrease between 1.2 to 2.9 h (Rusterholz et al. 2010; Rusterholz and Achermann 2011). The time constant of the SWA decline also varies across cortical regions and, although different studies disagree on the specific area showing the fastest decline (Zavada et al. 2009; Rusterholz and Achermann 2011), this regional specificity suggests a use-dependent component.
In recent years, many studies have revealed that sleep SWA reflects not only wake duration, but also wake “intensity,” as first shown in humans using repeated vibration of the right hand, which activated left somatosensory cortex and resulted in a shift in the low frequency EEG power, including SWA, in the same area (Kattler et al. 1994). In rats kept awake for the same amount of time, SWA is positively correlated with the time spent exploring but not with the time spent in automatic behaviors such as grooming or eating (Huber et al. 2007). Within the same animal, the amount of exploratory behavior during wake could predict the extent to which the gene BDNF was induced in the cerebral cortex at the end of the sleep-deprivation period, as well as the extent of the homeostatic SWA response during subsequent sleep (Huber et al. 2007). Because brain-derived neurotrophic factor (BDNF) is causally related to the induction of synaptic potentiation, this finding suggests that the amount of learning and synaptic plasticity during wake is a major determinant of sleep need. Many other studies, in both animals and humans, have shown that SWA can increase locally in the cortical areas involved in task performance, for instance, right parietal cortex after a visuomotor task (Huber et al. 2004a; Landsness et al. 2009), motor cortex after training in a reaching task (Hanlon et al. 2009), or visual areas after visual tasks (Goel et al. 2014). There is also evidence in pigeons, albeit limited, that SWA homeostasis reflects local brain use and/or synaptic potentiation (Lesku et al. 2011). Several of the experiments discussed above controlled for the amount of movement used to perform the task, suggesting that changes in SWA are sensitive to neural plasticity above and beyond any effect of neuronal activity per se. Moreover, the extent of the SWA increase depends on the complexity of the task, and thus presumably on the extent of the plasticity that it triggers (Wilhelm et al. 2014). There is also evidence that the changes in SWA that follow increased neuronal activity and plasticity do not simply reflect metabolic changes, because the SWA time course is longer than that of metabolic markers such as glucose and lactate (Vyazovskiy et al. 2004; Rempe and Wisor 2014). Finally, a recent study in which cortical neurons were forced to fire at high wake-like levels for several hours during sleep found that it is unlikely that neuronal activity alone can account for the changes in SWA observed after extended wake (Rodriguez et al. 2016).
In summary, there is good evidence in mammals that sleep homeostasis may be sensed in different regions through different signaling molecules, including adenosine whose build-up in wake likely reflects increased synaptic transmission and neuronal activity. Moreover, neuronal plasticity is a major determinant of sleep homeostasis as measured by SWA, and can explain why sleep slow waves can also be regulated locally.
MOLECULAR AND NEURONAL BASIS OF SLEEP HOMEOSTASIS IN DROSOPHILA
Drosophila has been a powerful model for identifying specific genetic pathways important for sleep homeostasis. Several sleep parameters are similarly affected by sleep loss in flies and mammals, and are used to define sleep homeostasis in Drosophila. Thus, sleep deprivation in flies is followed by longer and more consolidated sleep, characterized by reduced number of brief awakenings, longer sleep episodes, and higher arousal thresholds (Hendricks et al. 2000; Shaw et al. 2000; Huber et al. 2004b; van Alphen et al. 2013; Seidner et al. 2015). These parameters serve as markers of sleep homeostasis in lieu of SWA, which requires electrophysiological measurements that are possible but technically challenging in flies. However, as in mammals, disruption of homeostasis affects processes dependent on neural plasticity. For instance, sleep deprivation impairs learning and memory (Li et al. 2009; Seugnet et al. 2009; Donlea et al. 2011) and reduces aggression, mating success (Kayser et al. 2015), and neural development (Kayser et al. 2014). In flies, the chronic recording of neuronal activity during sleep is possible but remains challenging (Nitz et al. 2002; van Swinderen et al. 2004; van Alphen et al. 2013; Bushey et al. 2015). Like in mammals, wake duration is an important factor affecting the sleep rebound in flies (Huber et al. 2004b), but wake “intensity,” for instance, the exposure to an enriched environment, also increases sleep need (Ganguly-Fitzgerald et al. 2006; Donlea et al. 2009; Bushey et al. 2011), and learning mutants are deficient for experience-dependent increases in sleep (Donlea et al. 2009). Overall, studies in flies confirm the presence of a strong link between sleep need, learning, and neuronal plasticity, at least in part through the activation of extracellular signal-regulated kinase (ERK) signaling (Foltenyi et al. 2007; Vanderheyden et al. 2013).
Acetylcholine
Acetylcholine (ACh) is the major excitatory neurotransmitter in the Drosophila brain and is an important mediator of sleep homeostasis. Activation of cholinergic neurons is sufficient to induce wakefulness and subsequent sleep rebound (Seidner et al. 2015). Sleep homeostasis defects in the short sleeping mutant insomniac (inc) could be rescued by inc expression in cholinergic neurons (Pfeiffenberger and Allada 2012). Mutations in the Shaker potassium channel (Cirelli et al. 2005a) and its activators Hyperkinetic (Bushey et al. 2007) and sleepless (sss) (Koh et al. 2008; Wu et al. 2010) are predicted to increase excitability dramatically and reduce sleep. Notably reduced sleep in sss mutants can be rescued by expression in cholinergic neurons (Wu et al. 2014). Interestingly, sss, but not Sh mutants (Cirelli et al. 2005a), show impaired rebound sleep (Koh et al. 2008), suggesting that sss may regulate other factors. SSS also interacts with and suppresses the activity of nicotinic acetylcholine receptors to promote sleep (Wu et al. 2014). On the other hand, the nicotinic acetylcholine receptor α subunit redeye (rye) interacts with SSS to promote sleep (Shi et al. 2014), suggesting that cholinergic signaling can impact sleep in both directions. Levels of the RYE receptor are induced by both spontaneous and induced wakefulness (Shi et al. 2014).
Potential loci of acetylcholine function are specific mushroom body output neurons (MBONs) (Sitaraman et al. 2015a). The Drosophila mushroom bodies (MBs) are central to associative learning and are analogous to the mammalian cerebral cortex (Tomer et al. 2010). Early unbiased neurogenetic screens identified the MBs as a major sleep regulatory center capable of both sleep and wake promotion (Joiner et al. 2006; Pitman et al. 2006). Distinct subsets of intrinsic MB neurons (Kenyon cells [KCs]) promote sleep via activation of cholinergic MBONs (Sitaraman et al. 2015a). Sleep deprivation increases the activity of this KC–MBON circuit and this activity is also essential for normal recovery sleep, indicating that this circuit is important for conveying homeostatic sleep drive (Sitaraman et al. 2015a). Notably, blocking this circuit did not affect baseline sleep, further supporting the idea of separable homeostatic mechanisms (Sitaraman et al. 2015a). The role of ACh in flies may be akin to that of another arousal system, the noradrenergic locus coeruleus in mammals, whose lesions impair the induction of plasticity-related genes during wake and blunt the sleep homeostatic response after sleep deprivation (Cirelli et al. 1996, 2005b; Cirelli and Tononi 2004). Given the extensive use of ACh in the fly brain, a remaining challenge is to identify the key sites of action and the underlying molecular mechanisms at those sites.
Dopamine
Dopaminergic (DA) neuromodulation, likely via cyclic AMP (cAMP) signal transduction, plays a key role in arousal and sleep homeostasis. In flies, enhancement of dopaminergic neurotransmission genetically or pharmacologically can dramatically reduce sleep, arousal threshold, and increase locomotor activity (Hendricks et al. 2003; Andretic et al. 2005; Kume et al. 2005; Wu et al. 2008) in line with the wake-promoting effects of dopamine in mammals (Eban-Rothschild et al. 2016). Dopaminergic signaling in the MBs mediates the wake promoting effects of caffeine (Andretic et al. 2008b). Dopamine receptor activation in the MBs can also reduce the effects of sleep loss on learning (Seugnet et al. 2008). On the other hand, a subset of MB-projecting DA neurons is specifically active during spontaneous and induced wake and this activity can stimulate forgetting, providing a circuit basis for sleep-dependent memory (Berry et al. 2015). Wake-promoting effects of dopaminergic neurons are mediated by activation of glutamatergic wake-promoting MB output neurons (Sitaraman et al. 2015b). In addition, elevated dopamine can also reduce rebound sleep, suggesting a contribution to sleep homeostasis (Kume et al. 2005). The E3 ubiquitin ligase Cullin-3 (Cul3) and its substrate adaptor, inc, have potent roles in both baseline and recovery sleep (Stavropoulos and Young 2011; Pfeiffenberger and Allada 2012). These effects depend on DA but do not occur in DA neurons (Pfeiffenberger and Allada 2012). The molecular and cellular targets of Cul3/INC remain to be identified, although they do appear to function in cholinergic neurons.
The dorsal fan-shaped body (dFB) of the central complex is another brain center important for mediating homeostatic sleep drive. Induction of dFB activity induces sleep states that can even enhance memory consolidation (Donlea et al. 2009). Sleep deprivation further enhances the activity of dFB neurons (Donlea et al. 2014). The effects of sleep loss depend on the RhoGTPase-activating protein, CROSSVEINLESS-C (CV-C) whose function also strongly affects baseline sleep (Donlea et al. 2014). dFB neurons are inhibited by key arousal promoting dopaminergic neurons via activation of the potassium “leak” current carried by Sandman, a two-pore potassium channel, and the suppression of voltage-gated A-type potassium current likely encoded in part by Sh (Pimentel et al. 2016). Although these studies reveal the mechanistic basis for dopaminergic arousal, it remains unclear whether these dopamine-dependent changes are important for homeostatic responses to sleep deprivation. It appears likely that the dFB lies downstream from sites where sleep drive is sensed and encoded. Interestingly, the DA-dFB circuit also mediates developmental regulation of sleep (Kayser et al. 2014) and is important for the response to inhaled general anesthetics (Kottler et al. 2013).
Dopamine sleep effects may also act via cAMP and the cAMP-response element(CRE)-binding protein (CREB). In Drosophila, genetic manipulations of the cAMP signal transduction cascade alter sleep–wake (Hendricks et al. 2001; Joiner et al. 2006). Sleep deprivation induces the activity of a CRE reporter (Hendricks et al. 2001). Impairment of the CREB transcription factor increases sleep rebound, further supporting the connection to homeostatic mechanisms (Hendricks et al. 2001). Comparable functions for cAMP have been observed in other invertebrates as well as mammals (Graves et al. 2003; Raizen et al. 2008). Notably, the cAMP pathway has been intimately linked to synaptic plasticity as well as learning and memory. In fact, sleep–wake-induced alterations of cAMP are crucial for the adverse effects of sleep deprivation on memory processing in mice (Vecsey et al. 2009). Anaplastic lymphoma kinase, which interacts with neurofibromatosis-1 to regulate plasticity/memory function, also functions in the MBs to regulate sleep (Bai and Sehgal 2015). Although cAMP is a key player in synaptic plasticity, it remains unclear whether cAMP effects on sleep homeostasis are mediated through its effects on wake-dependent synaptic plasticity. Interestingly, promoting sleep pharmacologically or genetically can rescue learning and memory dysfunction in cAMP-based mutants, suggesting that sleep may also promote learning and memory independently of cAMP (Dissel et al. 2015a).
Glutamate
Enhancement of glutamatergic neurotransmission caused by elevated expression of the vesicular glutamate transporter results in increased sleep (Robinson et al. 2016), whereas pan-neuronal knockdown of an NMDA receptor (dNR1) reduces sleep (Tomita et al. 2015). Glutamate neurotransmission among specific circadian clock neurons, on the other hand, has a timed sleep-promoting effect (Guo et al. 2016). Broad manipulations of glutamatergic neurotransmission indicate that, on net, glutamate signaling promotes wakefulness especially at night (Zimmerman et al. 2016). Homer proteins, adaptors for metabotropic glutamate receptors, also contribute to sleep homeostasis in both flies and mammals. Homer is up-regulated during sleep in Drosophila, whereas the mammalian ortholog, H1a, is up-regulated during wake (Naidoo et al. 2012). Loss of homer/H1a results in fragmented sleep and wake, respectively (Naidoo et al. 2012).
Wake-dependent synaptic plasticity may be a key mediator of sleep homeostasis. Spontaneous, induced, and/or enriched waking experience can increase synapse number, size, and/or synaptic protein levels in multiple brain regions, which can be reversed by sleep (Donlea et al. 2009; Gilestro et al. 2009; Hanlon et al. 2009; Bushey et al. 2011; de Vivo et al. 2016, 2017). Which of these synaptic changes mediate homeostatic sleep drive? In both flies and mammals, many neuronal groups that are able to initiate and/or maintain wake or sleep have been identified (Allada and Wu 2015; Luppi et al. 2016; Weber and Dan 2016). Similarly, it is likely that several, if not many, neuronal groups can transmit the homeostatic sleep signal that is crucial to control sleep need. In flies, modulation of glutamatergic synaptic activity in specific neurons, the R2 ring neurons of the ellipsoid body, may be one of the molecular mechanisms by which sleep homeostasis and synaptic plasticity are coupled. Transient activation of R2 neurons using the heat-activated cation channel, TrpA1, induces sleep rebound after activation is relieved with only minimal wake-promoting effects during activation, suggesting that R2 activation can result in persistent sleep drive (Liu et al. 2016). Sleep deprivation induces burst firing, intracellular calcium, and the presynaptic protein BRUCHPILOT (BRP) in R2 neurons (Liu et al. 2016). In addition, sleep deprivation also induced the dNR1 transcript that encodes an NMDA receptor (Liu et al. 2016). BRP changes require the inositol triphosphate receptor (IP3R) and can be induced by TrpA1 activation, each of which regulates intracellular calcium (Liu et al. 2016). Knockdown of IP3R as well as dNR1 reduces, but does not eliminate, recovery sleep (Liu et al. 2016). Moreover, tetanus toxin-induced blockade of R2 neuron synaptic output has little effect on baseline sleep levels (Liu et al. 2016). Thus, other homeostatic circuits are likely important, especially under baseline conditions. The finding that genetic manipulations that impair wake-dependent synaptic changes also affect recovery sleep provides important evidence that synaptic changes in specific neurons can mediate sleep drive.
GABA
γ-Aminobutyric acid (GABA) plays a major role in promoting sleep in both flies and mammals. Most, if not all, sleep-promoting neurons identified so far in the rodent brain, from the basal forebrain to the brainstem, are GABAergic (Luppi et al. 2016) and most of the commonly used sleep medications, including benzodiazepines and the so-called Z-drugs, target and enhance ionotropic GABA receptor signaling. In mammals, these drugs increase sleep continuity and the lighter stages of NREM sleep, but decrease SWA and REM sleep. Of note, these drugs prevent the SWA rebound normally seen after sleep deprivation, but preserve the homeostatic decline in SWA in the course of the night (Landolt et al. 2000), again suggesting a possible distinction between homeostatic mechanisms during baseline and after extended wake. In flies, GABAergic neurons promote sleep. GABA effects are mediated in part via the Rdl GABA receptor, which both promotes sleep but is especially important for regulating sleep latency, particularly in PDF+ circadian clock neurons (Agosto et al. 2008; Parisky et al. 2008; Chung et al. 2009; Liu et al. 2014). GABAergic dorsal paired median neurons that target the MBs to promote sleep provide at least one site of action (Haynes et al. 2015). Mutations in the glial GABA-metabolizing enzyme, GABA transaminase, increase sleep presumably by increasing GABA levels (Chen et al. 2015), providing a potential link between GABA and sleep homeostasis in Drosophila.
Other Pathways: Circadian Clock, Protein Misfolding Stress, and Growth/Differentiation
Although the circadian and homeostatic processes for sleep regulation are separable, an accumulating body of evidence indicates that the two processes are more deeply intertwined than previously thought (Borbély et al. 2016). In fact, theoretical modeling studies have proposed that the circadian clock may regulate the set point of the sleep homeostat, although experimental evidence for a direct connection is currently lacking (Achermann and Borbély 2003; Borbély et al. 2016). Genetic disruption of core circadian clock components often, but not always, modestly alters baseline and recovery sleep in both Drosophila (Shaw et al. 2002) and mice (Naylor et al. 2000; Wisor et al. 2002; Dudley et al. 2003; Laposky et al. 2005; Franken et al. 2006). In some cases, mutant effects on the clock do not parallel those on sleep, suggesting that these genes may be acting in a clock-independent manner (Franken et al. 2007). Mutations in cyclin A and its regulators reveal a postmitotic role in neurons targeted by clock neurons in regulating both baseline and rebound sleep, further supporting the clock–homeostasis interaction (Rogulja and Young 2012). Sleep–wake state or history also feeds back onto circadian pacemaker neuron physiology (Deboer et al. 2007) and neuropeptidergic outputs. Sleep deprivation can phase shift circadian behavior and notably induces per1 and per2 expression in the mammalian cortex (Wisor et al. 2008). Thus, the clock and homeostatic processes appear to be bidirectionally linked.
Wake-induced stress, especially during sleep deprivation, may act via heat shock (HS) and the endoplasmic reticulum (ER) unfolded protein response (UPR) pathways to mediate rebound sleep. HS increases sleep in flies (Lenz et al. 2015) as it does in worms (Hill et al. 2014; Nelson et al. 2014), and HS proteins mediate the lethal effects of sleep deprivation observed in mutants of the circadian clock gene cyc (Shaw et al. 2002). BiP, an ER chaperone component of the UPR, is up-regulated by spontaneous and induced wake in both flies (Shaw et al. 2000; Naidoo et al. 2007) and rodents (Cirelli and Tononi 2000; Naidoo et al. 2005). BiP, in turn, regulates the amount of recovery sleep (Naidoo et al. 2007). The role of the UPR appears to be conserved in mammals. As discussed below, stress signals affecting sleep can also arise from the periphery, suggesting system-wide regulation of sleep.
Canonical growth and differentiation pathways with more traditional roles in development may also impact sleep homeostasis. Epidermal growth factor (EGF) signaling strongly promotes sleep (Foltenyi et al. 2007). The Delta ligand/Notch receptor mediates sleep rebound in flies in which both Notch gain-of-function mutations and Delta overexpression in the MBs can specifically reduce sleep rebound (Seugnet et al. 2011). Notably, sleep deprivation soon after eclosion can impact neural development (Kayser et al. 2014).
INTERACTION BETWEEN SLEEP AND OTHER PHYSIOLOGICAL SYSTEMS
A brain-centric view of sleep still dominates, but we now have increasing recognition of the systemic benefits of sleep or, more commonly, of widespread physiological consequences of sleep loss. In addition, evidence for regulation of sleep by nonneural signals continues to accumulate. Of these, the signals that have been studied in the context of their association with sleep derive from the immune system or from metabolic pathways.
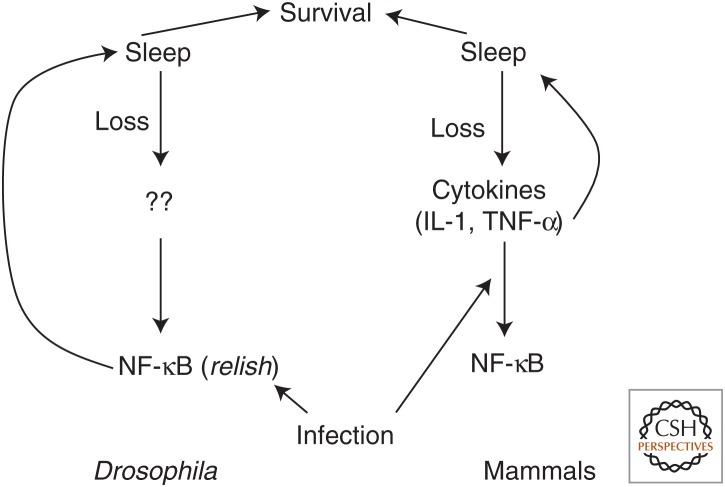
Perhaps one of the first indications of peripheral effects of sleep loss came from examination of rats that died following prolonged sleep deprivation. Pathological symptoms included fluid in the lungs and trachea, stomach ulcers, internal hemorrhage, and excessive edema, many of which could result from elevated proinflammatory signaling (Rechtschaffen et al. 1983; Rechtschaffen and Bergmann 2002). The presence of lesions in the stomach was also shown in another study, implicating inflammation and maybe also metabolic dysfunction (Murison et al. 1982). As described below, studies in recent years have bolstered the significance of these findings and also highlighted the reciprocity of the relationship between sleep and these other physiological systems (Fig. 1) (Everson et al. 2014).
Figure 1.
Model for the relationship between sleep and the immune system. In both flies and mammals, loss of sleep increases expression of nuclear factor κB (NF-κB) in mammals by increasing cytokine expression and in Drosophila through unknown mechanisms. The same immune/inflammation molecules are increased by infection. Cytokines or NF-κB, in turn, increase sleep in both systems, and increased sleep is associated with increased survival. IL-1, Interleukin 1; TNF-α, tumor necrosis factor α.
Sleep and Immune Function
There is no lack of anecdotal evidence for a connection between sleep and immunity, but the effects are complex and may differ for innate and adaptive immunity (Besedovsky et al. 2012). We typically sleep more when we are sick, and whereas a mechanistic explanation is still awaited, research already conducted does present viable hypotheses. We discuss below the bidirectional nature of the sleep–immune association and prevailing ideas in the field.
Immune/Inflammation Molecules as Sleep Modulators
Experiments conducted more than a century ago in dogs (Opp and Krueger 2015) and almost 50 years ago in sheep and goats (Pappenheimer et al. 1967) indicated that sleep deprivation leads to the buildup of one or more humoral factors that promote sleep. The actual experiment involved extraction of cerebrospinal fluid (CSF) from sleep-deprived animals and injection into other animals that had been allowed to sleep. Sleep was induced in the injected animals, prompting the idea of the sleep homeostat signaling through secreted molecules (somnogens). Among the first such secreted molecules identified was muramyl peptide, which also stimulates the immune system and elicits a febrile response (Krueger et al. 1982). Interestingly, muramyl peptide isolated from urine was shown to induce sleep in rats, rabbits, and cats, suggesting somnogenic effects of peripheral factors. Sleep-promoting effects of muramyl could be dissociated from fever induction, as the latter was blocked by antipyretics such as acetaminophen, whereas sleep effects persisted.
Subsequent studies supported effects of sleep loss on inflammation regulators, specifically cytokines, and even showed elevated interleukin 1 (IL-1) in plasma of sleep-deprived humans (Moldofsky et al. 1989). Indeed, activity of nuclear factor κB (NF-κB), a major transcription factor targeted by cytokines, is elevated following sleep deprivation in the cortex of rodents and in peripheral lymphocytes of human subjects (Obal and Krueger 2003). Effects of NF-κB on sleep have been studied, but are complicated by the existence of multiple subunits (Jhaveri et al. 2006). Of the different known cytokines, IL-1α and tumor necrosis factor 1 (TNF-1) appear to have the most significant role as sleep modulators. They both increase calcium flux in brain neurons and can increase NREM sleep together with SWA. In fact, TNF was shown to locally increase SWA in the area of application within the cortex (Taishi et al. 2007). The mechanisms of IL-1/TNF action include modulation of major neurotransmitter systems; for instance, IL-1 interacts with serotonin signaling to promote sleep (Imeri et al. 1999). Both IL-1 and TNF display diurnal rhythms in expression that occur in phase with NREM sleep (reviewed in Obal and Krueger 2003).
IL-6 is also expressed with a diurnal rhythm and appears to increase in human plasma on sleep deprivation (Bauer et al. 1994; Redwine et al. 2000) although results in humans are not always consistent (Besedovsky et al. 2012). Thus, it may be important for promoting sleep in response to sleep loss or infection, perhaps even as a downstream component from TNF and IL-1 signaling. Genetic knockouts of IL-6 have subtle phenotypes, indicating that this particular cytokine is not necessary for the regulation of daily sleep (Morrow and Opp 2005). It should be noted that cytokines are also indirectly implicated in sleep regulation. Prostaglandin D2, which is a component of cytokine pathways, may act through adenosine signaling to promote sleep (Huang et al. 2007).
Not surprisingly, unbiased screens for genes whose expression is altered by sleep loss reveal up-regulation of inflammation/stress pathways. This is true in rodents as well as in Drosophila, in which the NF-κB ortholog, Relish, is elevated by sleep loss (Cirelli et al. 2004, 2006; Williams et al. 2007). Like its mammalian counterpart, Relish is essential for Drosophila immunity (Hedengren et al. 1999). In an interesting analogy to sickness-induced sleep in mammals, bacterial infection of flies increases sleep (Kuo et al. 2010). Relish mutants do not show this increase (Kuo et al. 2010) and also have reduced daily sleep (Williams et al. 2007), suggesting an important sleep-regulating function of Relish. Importantly, sleep-promoting effects of Relish are mediated through the fat body, the Drosophila equivalent of the liver and adipose tissue as well as an important site for regulating innate immune responses, highlighting a role for a peripheral tissue (Kuo et al. 2010; Kuo and Williams 2014). Cellular stress also induces sleep in flies and worms, and it does so through a conserved mechanism involving the FMRFamide peptide (Nelson et al. 2014; Lenz et al. 2015).
Does Sleep Affect the Immune Response?
As discussed above, sleep is affected by immune challenges and by signaling pathways involved in the immune response, but the question, still open, is whether these changes in sleep are beneficial to the organism or are merely epiphenomena resulting from overlap in molecular mechanisms (Opp and Krueger 2015). In other words, does the immune-system-mediated increase in sleep enhance survival on injury/infection? Or conversely, does sleep loss impair immune function?
Intuitively, one would assume affirmative answers to both the questions above. However, acute sleep deprivation increases expression of immune molecules, which could facilitate immunity and, thereby, survival. The mammalian literature provides examples of both scenarios—sleep-restricted workers are more prone to infection than rested individuals (de Almeida and Malheiro 2017); on the other hand, tumor growth was reportedly reduced on severe and extended sleep deprivation in rats (Bergmann et al. 1996). This issue has also been addressed in Drosophila, which may shed some light on these seemingly contradictory results. Acute sleep deprivation of flies enhances survival in response to infection, consistent with the elevated expression of immune genes (Williams et al. 2007). However, the increased survival, which is NF-κB-mediated, depends on enhanced postinfection and postdeprivation sleep (Kuo and Williams 2014). Similar experiments conducted previously with rabbits showed that sleep deprivation affected postinfection sleep, but had little effect on clinical symptoms (Toth 1995). However, survival of infected animals was generally associated with enhanced sleep (Toth et al. 1993).
Sleep and Metabolism
The interaction between sleep and metabolism needs to be considered on multiple levels. As is now widely recognized, sleep loss has systemic effects on metabolic pathways. Whether these, in turn, modulate sleep remains debatable, although energy balance can influence sleep (Collet et al. 2016). In addition, sleep–wake states alter cellular metabolism, which has also been implicated in sleep function.
Systemic Effects of Sleep Loss
Prominent studies about a decade ago drew attention to a possible link between short sleep times and the rising obesity epidemic in the United States. It was shown that in people sleeping less than 8 hours per night, sleep times are inversely correlated with body mass index (BMI) (Taheri et al. 2004). Moreover, short sleep times were also associated with lower leptin and higher ghrelin, hormonal changes that are expected to increase appetite (Taheri et al. 2004). The idea that sleep disturbances are associated with aberrant glucose metabolism, hyperglycemia, and insulin resistance, in summary with an increased risk for diabetes, has since been supported by several other studies (Hanlon and Van Cauter 2011; Reutrakul and Van Cauter 2014).
Consistent with the sleep time–obesity connection in humans, sleep deprivation of Drosophila increases triglycerides. However, a similar increase was found in flies that were subjected to the sleep-depriving stimulation, but did not lose much sleep as the stimulation was applied during a period of wake (Harbison and Sehgal 2009). This suggests that the triglyceride increase resulted from increased stress rather than sleep loss, a possibility that should also be considered for the human data. For instance, people with higher levels of stress may sleep less at night. Rats deprived of sleep usually lose weight, most likely because of the energy expenditure caused by the forced activity (Barf et al. 2012). Although human subjects may be deprived of sleep without significantly increased activity, this is not the case for animal models. Nevertheless, it is worth considering other variables—for example, circadian disruption, increased food intake during the additional waking hours—that may contribute to the obesity seen in short-sleeping humans (Arble et al. 2015).
Recent work has identified plasma metabolites altered by either total sleep deprivation or sleep restriction in human subjects (Giskeodegard et al. 2015; Weljie et al. 2015). Several metabolites were common to the two protocols; both revealed an increase in lipids and in polar metabolites, and indicated a generally oxidized environment. A comparison of data from rats and humans highlighted two metabolites that decrease on sleep restriction in both systems—oxalic acid and a specific species of diacylglycerol (36:3)—which could potentially serve as biomarkers of sleep loss (Weljie et al. 2015).
Another link between sleep and metabolism is provided by orexin, a neuropeptide whose loss causes narcolepsy. Although not as well studied or appreciated, loss of orexin also disrupts energy balance and is associated with obesity (Nixon et al. 2015). The relationship between these different effects of orexin is unclear.
Effects of Metabolic Status on Sleep
The well-known Tor/Tsc pathway affects circadian period of rhythmic behavior in flies (Zheng and Sehgal 2010), as does a high-fat diet in mice (Kohsaka et al. 2007), indicating that metabolic signals can modulate behavior. However, pathways involved in lipid, carbohydrate, and protein metabolism have not been studied for effects on sleep to the extent immune molecules have. Nevertheless, specific nutrients have been associated with sleep symptoms (Grandner et al. 2014) and some molecules involved in lipid metabolism regulate sleep in Drosophila (Thimgan et al. 2010; Gerstner et al. 2011). Interestingly, starvation suppresses sleep in Drosophila, perhaps because the animals must forage for food under these conditions (Thimgan et al. 2010). Indeed, it appears that sleep deprivation induced by starvation does not even lead to buildup of sleep drive and so does not impair performance on learning tasks (Thimgan et al. 2010). In humans, however, starvation seems to have very different effects, increasing the time spent in the deepest stage of slow wave sleep without changing total sleep time (Collet et al. 2016).
Little is known about the cellular and molecular mechanisms underlying sleep loss in response to starvation, although some relevant molecules have been identified. Thus, flies lacking the circadian clock genes, Clock or cycle, fail to suppress sleep in response to starvation (Keene et al. 2010). In general, starvation-induced sleep suppression probably represents an important homeostatic mechanism. Flies with low octopamine signaling have increased daily sleep, but they suppress sleep when starved, probably because their low endogenous triglyceride levels drive a robust foraging response despite the increased sleep drive (Erion et al. 2012). Thus, under low nutrient conditions, metabolic needs appear to override sleep need, at least in the short term.
Sleep and Cellular Metabolism
Maintenance of cellular metabolism is one of the hypothesized functions of sleep, the idea being that energy is depleted by neurons during wake, and so must be replenished during sleep. As discussed in the section on SWA, sleep loss may result in the breakdown of ATP and thereby accumulation of adenosine. Increases in adenosine have been reported in response to prolonged wake in specific regions of the brain, and are thought to contribute to the sleep drive that builds up under these conditions. In fact, adenosine is implicated in the reduced sleep homeostasis produced by blocking glial transmission, suggesting it is released from glia (Halassa et al. 2009), although these findings have been challenged (Fujita et al. 2014). Finally, wake-promoting effects of caffeine are widely believed to be mediated by antagonism of adenosine receptors.
Despite this considerable literature supporting a role for adenosine, the extent of its importance remains unclear, not least because of technical issues. Adenosine and ATP levels are typically very difficult to measure reliably as they tend to be very labile and mouse mutants of adenosine receptors have subtle, if any, sleep phenotypes, indicating that adenosine is at best one of several neuromodulators regulating sleep (Bjorness and Greene 2009). Moreover, other targets of caffeine action have probably not been accorded the attention they deserve. Caffeine has very robust wake-promoting effects in Drosophila, and these are not mediated by the only known adenosine receptor in flies but rather by dopamine signaling (Andretic et al. 2008b; Wu et al. 2009; Nall et al. 2016).
Regardless of whether the adenosine hypothesis holds up, a role for sleep in cellular metabolism should be seriously considered. Sleep loss–induced changes in mitochondrial gene expression were reported when differential gene-expression technology had just started to be applied to the study of sleep (Cirelli and Tononi 1998). GABA is one of the most potent sleep-promoting molecules, and GABA catabolism occurs in mitochondria. It is broken down by the enzyme GABA transaminase (GABAT), which is expressed predominantly in glial mitochondria in which it functions in a shunt of the tricarboxylic acid (TCA) cycle. As noted above, mutants lacking GABAT have high sleep, but they also display metabolic phenotypes (Maguire et al. 2015). Although the mechanisms underlying these two different effects of GABAT may be different, these data nevertheless support an interaction between sleep and cellular metabolism.
CONCLUSIONS
Although much is yet to be learned about the mechanisms underlying sleep homeostasis, research in this area is revealing some conserved features. Measurements of homeostasis vary greatly from flies to mammals, but both models indicate a close link of sleep homeostasis to neural plasticity. Also, across different organisms, the regulation of sleep involves redundant circuits and similar actions of the major neuromodulators. Finally, analysis of sleep is shifting from a brain-focused view to the recognition that peripheral systems are also impacted by sleep loss and may contribute to the determination of sleep need.
Footnotes
Editors: Paolo Sassone-Corsi, Michael W. Young, and Akhilesh B. Reddy
Additional Perspectives on Circadian Rhythms available at www.cshperspectives.org
REFERENCES
- Achermann P, Borbély AA. 2003. Mathematical models of sleep regulation. Front Biosci 8: s683–s693. [DOI] [PubMed] [Google Scholar]
- Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. 2008. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci 11: 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Wu M. 2015. Genetics and genomic basis of sleep in simple model organisms. In Principles and practice of sleep medicine (ed. Kryger M, Roth T, Dement WC), pp. 281–309. Elsevier, Amsterdam. [Google Scholar]
- Andretic R, van Swinderen B, Greenspan RJ. 2005. Dopaminergic modulation of arousal in Drosophila. Curr Biol 15: 1165–1175. [DOI] [PubMed] [Google Scholar]
- Andretic R, Franken P, Tafti M. 2008a. Genetics of sleep. Annu Rev Genet 42: 361–388. [DOI] [PubMed] [Google Scholar]
- Andretic R, Kim YC, Jones FS, Han KA, Greenspan RJ. 2008b. Drosophila D1 dopamine receptor mediates caffeine-induced arousal. Proc Natl Acad Sci 105: 20392–20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arble DM, Bass J, Behn CD, Butler MP, Challet E, Czeisler C, Depner CM, Elmquist J, Franken P, Grandner MA, et al. 2015. Impact of sleep and circadian disruption on energy balance and diabetes: A summary of workshop discussions. Sleep 38: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Sehgal A. 2015. Anaplastic lymphoma kinase acts in the Drosophila mushroom body to negatively regulate sleep. PLoS Genet 11: e1005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barf RP, Van Dijk G, Scheurink AJ, Hoffmann K, Novati A, Hulshof HJ, Fuchs E, Meerlo P. 2012. Metabolic consequences of chronic sleep restriction in rats: Changes in body weight regulation and energy expenditure. Physiol Behav 107: 322–328. [DOI] [PubMed] [Google Scholar]
- Bauer J, Hohagen F, Ebert T, Timmer J, Ganter U, Krieger S, Lis S, Postler E, Voderholzer U, Berger M. 1994. Interleukin-6 serum levels in healthy persons correspond to the sleep–wake cycle. Clin Investig 72: 315. [DOI] [PubMed] [Google Scholar]
- Bellesi M, Pfister-Genskow M, Maret S, Keles S, Tononi G, Cirelli C. 2013. Effects of sleep and wake on oligodendrocytes and their precursors. J Neurosci 33: 14288–14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellesi M, de Vivo L, Tononi G, Cirelli C. 2015. Effects of sleep and wake on astrocytes: Clues from molecular and ultrastructural studies. BMC Biol 13: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellesi M, Tononi G, Cirelli C, Serra PA. 2016. Region-specific dissociation between cortical noradrenaline levels and the sleep/wake cycle. Sleep 39: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benington JH, Heller HC. 1995. Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol 45: 347–360. [DOI] [PubMed] [Google Scholar]
- Bergmann BM, Rechtschaffen A, Gilliland MA, Quintáns J. 1996. Effect of extended sleep deprivation on tumor growth in rats. Am J Physiol 271: R1460–R1464. [DOI] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Chakraborty M, Davis RL. 2015. Sleep facilitates memory by blocking dopamine neuron-mediated forgetting. Cell 161: 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besedovsky L, Lange T, Born J. 2012. Sleep and immune function. Pflugers Arch 463: 121–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorness TE, Greene RW. 2009. Adenosine and sleep. Curr Neuropharmacol 7: 238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake H, Gerard R. 1937. Brain potentials during sleep. Am J Physiol 119: 692–703. [Google Scholar]
- Borbély AA. 1982. A two process model of sleep regulation. Human Neurobiol 1: 195–204. [PubMed] [Google Scholar]
- Borbély AA, Baumann F, Brandeis D, Strauch I, Lehmann D. 1981. Sleep deprivation: Effect on sleep stages and EEG power density in man. Electroencephalogr Clin Neurophysiol 51: 483–495. [DOI] [PubMed] [Google Scholar]
- Borbély AA, Tobler I, Hanagasioglu M. 1984. Effect of sleep deprivation on sleep and EEG power spectra in the rat. Behav Brain Res 14: 171–182. [DOI] [PubMed] [Google Scholar]
- Borbély AA, Daan S, Wirz-Justice A, Deboer T. 2016. The two-process model of sleep regulation: A reappraisal. J Sleep Res 25: 131–143. [DOI] [PubMed] [Google Scholar]
- Bushey D, Huber R, Tononi G, Cirelli C. 2007. Drosophila hyperkinetic mutants have reduced sleep and impaired memory. J Neuroscience 27: 5384–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C. 2011. Sleep and synaptic homeostasis: Structural evidence in Drosophila. Science 332: 1576–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C. 2015. Sleep- and wake-dependent changes in neuronal activity and reactivity demonstrated in fly neurons using in vivo calcium imaging. Proc Natl Acad Sci 112: 4785–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajochen C, Wyatt JK, Czeisler CA, Dijk DJ. 2002. Separation of circadian and wake duration-dependent modulation of EEG activation during wakefulness. Neuroscience 114: 1047–1060. [DOI] [PubMed] [Google Scholar]
- Chauvette S, Crochet S, Volgushev M, Timofeev I. 2011. Properties of slow oscillation during slow-wave sleep and anesthesia in cats. J Neurosci 31: 14998–15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WF, Maguire S, Sowcik M, Luo W, Koh K, Sehgal A. 2015. A neuron–glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol Psychiatry 20: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. 2009. The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 19: 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C. 2009. The genetic and molecular regulation of sleep: From fruit flies to humans. Nat Rev Neurosci 10: 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. 1998. Differences in gene expression between sleep and waking as revealed by mRNA differential display. Brain Res Mol Brain Res 56: 293–305. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. 1999. Differences in brain gene expression between sleep and waking as revealed by mRNA differential display and cDNA microarray technology. J Sleep Res 8: 44–52. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. 2000. Gene expression in the brain across the sleep-waking cycle. Brain Res 885: 303–321. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. 2004. Locus ceruleus control of state-dependent gene expression. J Neurosci 24: 5410–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Pompeiano M, Tononi G. 1996. Neuronal gene expression in the waking state: A role for the locus coeruleus. Science 274: 1211–1215. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. 2004. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41: 35–43. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill SL, Huber R, Kreber R, Ganetzky B, Tononi G. 2005a. Reduced sleep in Drosophila Shaker mutants. Nature 434: 1087–1092. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Huber R, Gopalakrishnan A, Southard TL, Tononi G. 2005b. Locus ceruleus control of slow-wave homeostasis. J Neurosci 25: 4503–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Faraguna U, Tononi G. 2006. Changes in brain gene expression after long-term sleep deprivation. J Neurochem 98: 1632–1645. [DOI] [PubMed] [Google Scholar]
- Collet TH, van der Klaauw AA, Henning E, Keogh JM, Suddaby D, et al. 2016. The sleep/wake cycle is directly modulated by changes in energy balance. Sleep 39: 1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie T, Mongrain V, Dorsaz S, Mang GM, Emmenegger Y, Franken P. 2013. Homeostatic and circadian contribution to EEG and molecular state variables of sleep regulation. Sleep 36: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida CMO, Malheiro A. 2017. Sleep, immunity and shift workers. Sleep Sci 9: 164–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboer T. 2015. Behavioral and electrophysiological correlates of sleep and sleep homeostasis. Curr Top Behav Neurosci 25: 1–24. [DOI] [PubMed] [Google Scholar]
- Deboer T, Detari L, Meijer JH. 2007. Long term effects of sleep deprivation on the mammalian circadian pacemaker. Sleep 30: 257–262. [DOI] [PubMed] [Google Scholar]
- de Vivo L, Nelson AB, Bellesi M, Noguti J, Tononi G, Cirelli C. 2016. Loss of sleep affects the ultrastructure of pyramidal neurons in the adolescent mouse frontal cortex. Sleep 39: 861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vivo L, Bellesi M, Marshall W, Bushong EA, Ellisman MH, Tononi G, Cirelli C. 2017. Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science 355: 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk DJ, Czeisler CA. 1995. Contribution of the circadian pacemaker and the sleep homeostat to sleep propensity, sleep structure, electroencephalographic slow waves, and sleep spindle activity in humans. J Neurosci 15: 3526–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk DJ, Beersma DGM, Daan S, Bloem GM, Van den Hoofdakker RH. 1987. Eur Arch Psychiatry Neurol Sci 236: 323–328. [DOI] [PubMed] [Google Scholar]
- Dissel S, Angadi V, Kirszenblat L, Suzuki Y, Donlea J, Klose M, Koch Z, English D, Winsky-Sommerer R, van Swinderen B, et al. 2015a. Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol 25: 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S, Melnattur K, Shaw PJ. 2015b. Sleep, performance, and memory in flies. Curr Sleep Med Rep 1: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Ramanan N, Shaw PJ. 2009. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. 2011. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332: 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, Miesenbock G. 2014. Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy C, Moravcevic K, Yue Z, Wan JY, Van Dongen HP, Sehgal A. 2016. Genetic dissociation of daily sleep and sleep following thermogenetic sleep deprivation in Drosophila. Sleep 39: 1083–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. 2003. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science 301: 379–383. [DOI] [PubMed] [Google Scholar]
- Eban-Rothschild A, Rothschild G, Giardino WJ, Jones JR, de Lecea L. 2016. VTA dopaminergic neurons regulate ethologically relevant sleep–wake behaviors. Nat Neurosci 19: 1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion R, DiAngelo JR, Crocker A, Sehgal A. 2012. Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. J Biol Chem 287: 32406–32414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermis U, Krakow K, Voss U. 2010. Arousal thresholds during human tonic and phasic REM sleep. J Sleep Res 19: 400–406. [DOI] [PubMed] [Google Scholar]
- Esser SK, Hill SL, Tononi G. 2007. Sleep homeostasis and cortical synchronization. I: Modeling the effects of synaptic strength on sleep slow waves. Sleep 30: 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everson CA, Henchen CJ, Szabo A, Hogg N. 2014. Cell injury and repair resulting from sleep loss and sleep recovery in laboratory rats. Sleep 37: 1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faville R, Kottler B, Goodhill GJ, Shaw PJ, van Swinderen B. 2015. How deeply does your mutant sleep? Probing arousal to better understand sleep defects in Drosophila. Sci Rep 5: 8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg I, March JD, Fein G, Floyd TC, Walker JM, Price L. 1978. Period and amplitude analysis of 0.5–3c/sec activity in NREM sleep of young adults. Electroencephalogr Clin Neurophysiol 44: 202–213. [DOI] [PubMed] [Google Scholar]
- Ferrara M, De Gennaro L, Casagrande M, Bertini M. 1999. Auditory arousal thresholds after selective slow-wave sleep deprivation. Clin Neurophysiol 110: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. 2007. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 10: 1160–1167. [DOI] [PubMed] [Google Scholar]
- Franken P, Chollet D, Tafti M. 2001. The homeostatic regulation of sleep need is under genetic control. J Neurosci 21: 2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Dudley CA, Estill SJ, Barakat M, Thomason R, O’Hara BF, McKnight SL. 2006. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: Genotype and sex interactions. Proc Natl Acad Sci 103: 7118–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Thomason R, Heller HC, O’Hara BF. 2007. A non-circadian role for clock-genes in sleep homeostasis: A strain comparison. BMC Neurosci 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Chen MJ, Li B, Smith NA, Peng W, Sun W, Toner MJ, Kress BT, Wang L, Benraiss A, et al. 2014. Neuronal transgene expression in dominant-negative SNARE mice. J Neurosci 34: 16594–16604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk CM, Honjoh S, Rodriguez AV, Cirelli C, Tononi G. 2016. Local slow waves in superficial layers of primary cortical areas during REM sleep. Curr Biol 26: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald I, Donlea J, Shaw PJ. 2006. Waking experience affects sleep need in Drosophila. Science 313: 1775–1781. [DOI] [PubMed] [Google Scholar]
- Gerstner JR, Vanderheyden WM, Shaw PJ, Landry CF, Yin JC. 2011. Fatty-acid binding proteins modulate sleep and enhance long-term memory consolidation in Drosophila. PLoS ONE 6: e15890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro GF, Tononi G, Cirelli C. 2009. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giskeodegard GF, Davies SK, Revell VL, Keun H, Skene DJ. 2015. Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Sci Rep 5: 14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel N, Abe T, Braun ME, Dinges DF. 2014. Cognitive workload and sleep restriction interact to influence sleep homeostatic responses. Sleep 37: 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel N, Basner M, Dinges DF. 2015. Phenotyping of neurobehavioral vulnerability to circadian phase during sleep loss. Methods Enzymol 552: 285–308. [DOI] [PubMed] [Google Scholar]
- Grandner MA, Jackson N, Gerstner JR, Knutson KL. 2014. Sleep symptoms associated with intake of specific dietary nutrients. J Sleep Res 23: 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. 2003. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol 90: 1152–1159. [DOI] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M. 2016. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536: 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, Frank MG. 2009. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron 61: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon EC, Van Cauter E. 2011. Quantification of sleep behavior and of its impact on the cross-talk between the brain and peripheral metabolism. Proc Natl Acad Sci 108: 15609–15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon EC, Faraguna U, Vyazovskiy V, Tononi G, Cirelli C. 2009. Effects of skilled training on sleep slow wave activity and cortical gene expression in the rat. Sleep 32: 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Sehgal A. 2008. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics 178: 2341–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Sehgal A. 2009. Energy stores are not altered by long-term partial sleep deprivation in Drosophila melanogaster. PLoS ONE 4: e6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes PR, Christmann BL, Griffith LC. 2015. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. eLife 10.7554/eLife.03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell 4: 827–837. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. 2000. Rest in Drosophila is a sleep-like state. Neuron 25: 129–138. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. 2001. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nature Neurosci 4: 1108–1115. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI. 2003. Modafinil maintains waking in the fruit fly Drosophila melanogaster. Sleep 26: 139–146. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Mansfield R, Lopez JM, Raizen DM, Van Buskirk C. 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr Biol 24: 2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZL, Urade Y, Hayaishi O. 2007. Prostaglandins and adenosine in the regulation of sleep and wakefulness. Curr Opin Pharmacol 7: 33–38. [DOI] [PubMed] [Google Scholar]
- Huber R, Ghilardi MF, Massimini M, Tononi G. 2004a. Local sleep and learning. Nature 430: 78–81. [DOI] [PubMed] [Google Scholar]
- Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. 2004b. Sleep homeostasis in Drosophila melanogaster. Sleep 27: 628–639. [DOI] [PubMed] [Google Scholar]
- Huber R, Tononi G, Cirelli C. 2007. Exploratory behavior, cortical BDNF expression, and sleep homeostasis. Sleep 30: 129–139. [DOI] [PubMed] [Google Scholar]
- Imeri L, Mancia M, Opp MR. 1999. Blockade of 5-hydroxytryptamine (serotonin)-2 receptors alters interleukin-1-induced changes in rat sleep. Neuroscience 92: 745–749. [DOI] [PubMed] [Google Scholar]
- Jhaveri KA, Ramkumar V, Trammell RA, Toth LA. 2006. Spontaneous, homeostatic, and inflammation-induced sleep in NF-κB p50 knockout mice. Am J Physiol Regul Integr Comp Physiol 291: R1516–R1526. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. 2006. Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441: 757–760. [DOI] [PubMed] [Google Scholar]
- Kattler H, Dijk DJ, Borbely AA. 1994. Effect of unilateral somatosensory stimulation prior to sleep on the sleep EEG in humans. J Sleep Res 3: 159–164. [DOI] [PubMed] [Google Scholar]
- Kayser MS, Yue Z, Sehgal A. 2014. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344: 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, Mainwaring B, Yue Z, Sehgal A. 2015. Sleep deprivation suppresses aggression in Drosophila. eLife 4: e07643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Duboue ER, McDonald DM, Dus M, Suh GS, Waddell S, Blau J. 2010. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol 20: 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. 2008. Identification of SLEEPLESS, a sleep-promoting factor. Science 321: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. 2007. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6: 414–421. [DOI] [PubMed] [Google Scholar]
- Kottler B, Bao H, Zalucki O, Imlach W, Troup M, van Alphen B, Paulk A, Zhang B, van Swinderen B. 2013. A sleep/wake circuit controls isoflurane sensitivity in Drosophila. Curr Biol 23: 594–598. [DOI] [PubMed] [Google Scholar]
- Krueger JM, Pappenheimer JR, Karnovsky ML. 1982. Sleep-promoting effects of muramyl peptides. Proc Natl Acad Sci 79: 6102–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. 2005. Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25: 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Williams JA. 2014. Increased sleep promotes survival during a bacterial infection in Drosophila. Sleep 37: 1077–1086, 1086A–1086D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Pike DH, Beizaeipour Z, Williams JA. 2010. Sleep triggered by an immune response in Drosophila is regulated by the circadian clock and requires the NF-κB Relish. BMC Neurosci 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt HP, Finelli LA, Roth C, Buck A, Achermann P, Borbély AA. 2000. Zolpidem and sleep deprivation: Different effect on EEG power spectra. J Sleep Res 9: 175–183. [DOI] [PubMed] [Google Scholar]
- Landsness EC, Crupi D, Hulse BK, Peterson MJ, Huber R, Ansari H, Coen M, Cirelli C, Benca RM, Ghilardi MF, et al. 2009. Sleep-dependent improvement in visuomotor learning: A casual role for slow waves. Sleep 32: 1273–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. 2005. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep 28: 395–409. [DOI] [PubMed] [Google Scholar]
- Lazar AS, Lazar ZI, Dijk DJ. 2015. Circadian regulation of slow waves in human sleep: Topographical aspects. NeuroImage 116: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O, Xiong J, Nelson MD, Raizen DM, Williams JA. 2015. FMRFamide signaling promotes stress-induced sleep in Drosophila. Brain Behav Immun 47: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesku JA, Vyssotski AL, Martinez-Gonzalez D, Wilzeck C, Rattenborg NC. 2011. Local sleep homeostasis in the avian brain: Convergence of sleep function in mammals and birds? Proc Biol Sci 278: 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yu F, Guo A. 2009. Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep 32: 1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont J, Blackshaw S, Lloyd TE, et al. 2014. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82: 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Q, Tabuchi M, Wu MN. 2016. Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165: 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppi PH, Peyron C, Fort P. 2016. Not a single but multiple populations of GABAergic neurons control sleep. Sleep Med Rev 32: 85–94. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Paigen B, Naidoo N, Pack AI. 2008. Analysis of the QTL for sleep homeostasis in mice: Homer1a is a likely candidate. Physiol Genomics 33: 91–99. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Zimmerman JE, Shockley KR, Churchill GA, Pack AI. 2009. What are microarrays teaching us about sleep? Trends Mol Med 15: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire SE, Rhoades S, Chen WF, Sengupta A, Yue Z, Lim JC, Mitchell CH, Weljie AM, Sehgal A. 2015. Independent effects of γ-aminobutyric acid transaminase (GABAT) on metabolic and sleep homeostasis. J Biol Chem 290: 20407–20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret S, Dorsaz S, Gurcel L, Pradervand S, Petit B, Pfister C, Hagenbuchle O, O’Hara BF, Franken P, Tafti M. 2007. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci 104: 20090–20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimini M, Ferrarelli F, Huber R, Esser SK, Singh H, Tononi G. 2005. Breakdown of cortical effective connectivity during sleep. Science 309: 2228–2232. [DOI] [PubMed] [Google Scholar]
- Moldofsky H, Lue FA, Davidson JR, Gorczynski R. 1989. Effects of sleep deprivation on human immune functions. FASEB J 3: 1972–1977. [DOI] [PubMed] [Google Scholar]
- Morrow JD, Opp MR. 2005. Sleep–wake behavior and responses of interleukin-6-deficient mice to sleep deprivation. Brain Behav Immun 19: 28–39. [DOI] [PubMed] [Google Scholar]
- Murison R, Ursin R, Coover GD, Lien W, Ursin H. 1982. Sleep deprivation procedure produces stomach lesions in rats. Physiol Behav 29: 693–694. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Giang W, Galante RJ, Pack AI. 2005. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 92: 1150–1157. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Casiano V, Cater J, Zimmerman J, Pack AI. 2007. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep 30: 557–565. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Galante RJ, McShane B, Hu JH, Zimmerman J, Maislin G, Cater J, Wyner A, Worley P, et al. 2012. Role of Homer proteins in the maintenance of sleep–wake states. PLoS ONE 7: e35174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall AH, Shakhmantsir I, Cichewicz K, Birman S, Hirsh J, Sehgal A. 2016. Caffeine promotes wakefulness via dopamine signaling in Drosophila. Sci Rep 6: 20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, Turek FW. 2000. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci 20: 8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckelmann D, Ursin R. 1993. Sleep stages and EEG power spectrum in relation to acoustical stimulus arousal threshold in the rat. Sleep 16: 467–77. [PubMed] [Google Scholar]
- Nelson MD, Lee KH, Churgin MA, Hill AJ, Van Buskirk C, Fang-Yen C, Raizen DM. 2014. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr Biol 24: 2406–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir Y, Staba RJ, Andrillon T, Vyazovskiy VV, Cirelli C, Fried I, Tononi G. 2011. Regional slow waves and spindles in human sleep. Neuron 70: 153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. 2002. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol 12: 1934–1940. [DOI] [PubMed] [Google Scholar]
- Nixon JP, Mavanji V, Butterick TA, Billington CJ, Kotz CM, Teske JA. 2015. Sleep disorders, obesity, and aging: The role of orexin. Ageing Res Rev 20: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obal F Jr, Krueger JM. 2003. Biochemical regulation of non-rapid-eye-movement sleep. Front Biosci 8: d520–d550. [DOI] [PubMed] [Google Scholar]
- Opp MR, Krueger JM. 2015. Sleep and immunity: A growing field with clinical impact. Brain Behav Immun 47: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenheimer JR, Miller TB, Goodrich CA. 1967. Sleep-promoting effects of cerebrospinal fluid from sleep-deprived goats. Proc Natl Acad Sci 58: 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al. 2008. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C, Allada R. 2012. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet 8: e1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigorini A, Sarasso S, Proserpio P, Szymanski C, Arnulfo G, Casarotto S, Fecchio M, Rosanova M, Mariotti M, Lo Russo G, et al. 2015. Bistability breaks off deterministic responses to intracortical stimulation during non-REM sleep. NeuroImage 112: 105–113. [DOI] [PubMed] [Google Scholar]
- Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJ, Miesenbock G. 2016. Operation of a homeostatic sleep switch. Nature 536: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. 2006. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441: 753–756. [DOI] [PubMed] [Google Scholar]
- Porkka-Heiskanen T, Strecker RE, Thakkar M, Bjorkum AA, Greene RW, McCarley RW. 1997. Adenosine: A mediator of the sleep-inducing effects of prolonged wakefulness. Science 276: 1265–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572. [DOI] [PubMed] [Google Scholar]
- Rattenborg NC, Martinez-Gonzalez D, Lesku JA. 2009. Avian sleep homeostasis: Convergent evolution of complex brains, cognition and sleep functions in mammals and birds. Neurosci Biobehav Rev 33: 253–270. [DOI] [PubMed] [Google Scholar]
- Rechtschaffen A, Bergmann BM. 2002. Sleep deprivation in the rat: An update of the 1989 paper. Sleep 25: 18–24. [DOI] [PubMed] [Google Scholar]
- Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. 1983. Physiological correlates of prolonged sleep deprivation in rats. Science 221: 182–184. [DOI] [PubMed] [Google Scholar]
- Redwine L, Hauger RL, Gillin JC, Irwin M. 2000. Effects of sleep and sleep deprivation on interleukin-6, growth hormone, cortisol, and melatonin levels in humans. J Clin Endocrinol Metab 85: 3597–3603. [DOI] [PubMed] [Google Scholar]
- Rempe MJ, Wisor JP. 2014. Cerebral lactate dynamics across sleep/wake cycles. Front Comput Neurosci 8: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reutrakul S, Van Cauter E. 2014. Interactions between sleep, circadian function, and glucose metabolism: Implications for risk and severity of diabetes. Ann NY Acad Sci 1311: 151–173. [DOI] [PubMed] [Google Scholar]
- Robinson JE, Paluch J, Dickman DK, Joiner WJ. 2016. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat Commun 7: 10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AV, Funk CM, Vyazovskiy VV, Nir Y, Tononi G, Cirelli C. 2016. Why does sleep slow-wave activity increase after extended wake? Assessing the effects of increased cortical firing during wake and sleep. J Neurosci 36: 12436–12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Young MW. 2012. Control of sleep by cyclin A and its regulator. Science 335: 1617–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romcy-Pereira RN, Erraji-Benchekroun L, Smyrniotopoulos P, Ogawa S, Mello CV, Sibille E, Pavlides C. 2009. Sleep-dependent gene expression in the hippocampus and prefrontal cortex following long-term potentiation. Physiol Behav 98: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa RR, Bonnet MH. 1985. Sleep stages, auditory arousal threshold, and body temperature as predictors of behavior upon awakening. Int J Neurosci 27: 73–83. [DOI] [PubMed] [Google Scholar]
- Rusterholz T, Achermann P. 2011. Topographical aspects in the dynamics of sleep homeostasis in young men: Individual patterns. BMC Neurosci 12: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusterholz T, Durr R, Achermann P. 2010. Inter-individual differences in the dynamics of sleep homeostasis. Sleep 33: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf MT, Naidoo N, Zimmerman JE, Pack AI. 2008. The energy hypothesis of sleep revisited. Prog Neurobiol 86: 264–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidner G, Robinson JE, Wu M, Worden K, Masek P, Roberts SW, Keene AC, Joiner WJ. 2015. Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol 25: 2928–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ. 2008. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol 18: 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Galvin JE, Suzuki Y, Gottschalk L, Shaw PJ. 2009. Persistent short-term memory defects following sleep deprivation in a Drosophila model of Parkinson disease. Sleep 32: 984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, Shaw PJ. 2011. Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr Biol 21: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. 2000. Correlates of sleep and waking in Drosophila melanogaster. Science 287: 1834–1837. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. 2002. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417: 287–291. [DOI] [PubMed] [Google Scholar]
- Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A. 2014. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. eLife 3: e01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin M, Nitabach MN. 2015a. Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr Biol 25: 2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D, Aso Y, Rubin GM, Nitabach MN. 2015b. Control of sleep by dopaminergic inputs to the Drosophila mushroom body. Front Neural Circuits 9: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N, Young MW. 2011. Insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72: 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steriade M, Timofeev I, Grenier F. 2001. Natural waking and sleep states: A view from inside neocortical neurons. J Neurophysiol 85: 1969–1985. [DOI] [PubMed] [Google Scholar]
- Taheri S, Lin L, Austin D, Young T, Mignot E. 2004. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med 1: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taishi P, Churchill L, Wang M, Kay D, Davis CJ, Guan X, De A, Yasuda T, Liao F, Krueger JM. 2007. TNFα siRNA reduces brain TNF and EEG delta wave activity in rats. Brain Res 1156: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. 2010. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol 8: e1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler I. 2005. Phylogeny of sleep regulation. In Principles and practice of sleep medicine (ed. Kryger MH, Dement WC), pp. 77–90. Saunders, Philadelphia. [Google Scholar]
- Tomer R, Denes AS, Tessmar-Raible K, Arendt D. 2010. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142: 800–809. [DOI] [PubMed] [Google Scholar]
- Tomita J, Ueno T, Mitsuyoshi M, Kume S, Kume K. 2015. The NMDA receptor promotes sleep in the fruit fly, Drosophila melanogaster. PLoS ONE 10: e0128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Cirelli C. 2014. Sleep and the price of plasticity: From synaptic and cellular homeostasis to memory consolidation and integration. Neuron 81: 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth LA. 1995. Sleep, sleep deprivation and infectious disease: Studies in animals. Adv Neuroimmunol 5: 79–92. [DOI] [PubMed] [Google Scholar]
- Toth LA, Tolley EA, Krueger JM. 1993. Sleep as a prognostic indicator during infectious disease in rabbits. Proc Soc Exp Biol Med 203: 179–192. [DOI] [PubMed] [Google Scholar]
- van Alphen B, Yap MH, Kirszenblat L, Kottler B, van Swinderen B. 2013. A dynamic deep sleep stage in Drosophila. J Neurosci 33: 6917–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden WM, Gerstner JR, Tanenhaus A, Yin JC, Shaw PJ. 2013. ERK phosphorylation regulates sleep and plasticity in Drosophila. PLoS ONE 8: e81554.24244744 [Google Scholar]
- van Swinderen B, Nitz DA, Greenspan RJ. 2004. Uncoupling of brain activity from movement defines arousal states in Drosophila. Curr Biol 14: 81–87. [PubMed] [Google Scholar]
- Vecsey CG, Baillie GS, Jaganath D, Havekes R, Daniels A, Wimmer M, Huang T, Brown KM, Li XY, Descalzi G, et al. 2009. Sleep deprivation impairs cAMP signalling in the hippocampus. Nature 461: 1122–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy VV, Welker E, Fritschy JM, Tobler I. 2004. Regional pattern of metabolic activation is reflected in the sleep EEG after sleep deprivation combined with unilateral whisker stimulation in mice. Eur J Neurosci 20: 1363–1370. [DOI] [PubMed] [Google Scholar]
- Vyazovskiy V, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G. 2008. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat Neurosci 11: 200–208. [DOI] [PubMed] [Google Scholar]
- Vyazovskiy VV, Olcese U, Lazimy YM, Faraguna U, Esser SK, Williams JC, Cirelli C, Tononi G. 2009. Cortical firing and sleep homeostasis. Neuron 63: 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Dan Y. 2016. Circuit-based interrogation of sleep control. Nature 538: 51–59. [DOI] [PubMed] [Google Scholar]
- Weljie AM, Meerlo P, Goel N, Sengupta A, Kayser MS, Abel T, Birnbaum MJ, Dinges DF, Sehgal A. 2015. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci 112: 2569–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm I, Kurth S, Ringli M, Mouthon AL, Buchmann A, Geiger A, Jenni OG, Huber R. 2014. Sleep slow-wave activity reveals developmental changes in experience-dependent plasticity. J Neurosci 34: 12568–12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HL, Hammack JT, Daly RL, Dement WC, Lubin A. 1964. Responses to auditory stimulation, sleep loss and the EEG stages of sleep. Electroencephalogr Clin Neurophysiol 16: 269–279. [DOI] [PubMed] [Google Scholar]
- Williams JA, Sathyanarayanan S, Hendricks JC, Sehgal A. 2007. Interaction between sleep and the immune response in Drosophila: A role for the NF-κB relish. Sleep 30: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor JP, O’Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, Edgar DM, Franken P. 2002. A role for cryptochromes in sleep regulation. BMC Neurosci 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor JP, Pasumarthi RK, Gerashchenko D, Thompson CL, Pathak S, Sancar A, Franken P, Lein ES, Kilduff TS. 2008. Sleep deprivation effects on circadian clock gene expression in the cerebral cortex parallel electroencephalographic differences among mouse strains. J Neurosci 28: 7193–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Koh K, Yue Z, Joiner WJ, Sehgal A. 2008. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep 31: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Ho K, Crocker A, Yue Z, Koh K, Sehgal A. 2009. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J Neurosci 29: 11029–11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. 2010. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci 13: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Robinson JE, Joiner WJ. 2014. SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Curr Biol 24: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O’Donnell J, Christensen DJ, Nicholson C, Iliff JJ, et al. 2013. Sleep drives metabolite clearance from the adult brain. Science 342: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasenkov R, Deboer T. 2012. Circadian modulation of sleep in rodents. Prog Brain Res 199: 203–218. [DOI] [PubMed] [Google Scholar]
- Yelin-Bekerman L, Elbaz I, Diber A, Dahary D, Gibbs-Bar L, Alon S, Lerer-Goldshtein T, Appelbaum L. 2015. Hypocretin neuron-specific transcriptome profiling identifies the sleep modulator Kcnh4a. eLife 4: e08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavada A, Strijkstra AM, Boerema AS, Daan S, Beersma DG. 2009. Evidence for differential human slow-wave activity regulation across the brain. J Sleep Res 18: 3–10. [DOI] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. 2010. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol 20: 1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Chan MT, Lenz OT, Keenan BT, Maislin G, Pack AI. 2016. Glutamate is a wake active neurotransmitter in Drosophila melanogaster. Sleep 40: zsw046. [DOI] [PMC free article] [PubMed] [Google Scholar]