Abstract
Brain networks have been of long-standing interest to neurodegeneration researchers, including but not limited to investigators focusing on conventional prion diseases, which are known to propagate along neural pathways. Tools for human network mapping, however, remained inadequate, limiting our understanding of human brain network architecture and preventing clinical research applications. Until recently, neuropathological studies were the only viable approach to mapping disease onset and progression in humans but required large autopsy cohorts and laborious methods for whole-brain sectioning and staining. Despite important advantages, postmortem studies cannot address in vivo, physiological, or longitudinal questions and have limited potential to explore early-stage disease except for the most common disorders. Emerging in vivo network-based neuroimaging strategies have begun to address these issues, providing data that complement the neuropathological tradition. Overall, findings to date highlight several fundamental principles of neurodegenerative disease anatomy and pathogenesis, as well as some enduring mysteries. These principles and mysteries provide a road map for future research.
Traditional postmortem studies cannot address in vivo, physiological, or longitudinal questions nor can they examine early-stage disease. In vivo network-based neuroimaging strategies are now addressing these issues.
Neurodegenerative diseases are united by the inexorable and targeted spread of misfolded disease protein inclusions, gliosis, and synaptic and neuronal loss. Clinical symptoms and deficits, which coalesce into recognizable syndromes, reflect the topography of neurodegeneration rather than the identity of the aggregating disease protein. Indeed, each protein is associated with a handful of distinct clinical syndromes. Uncertainty surrounds which specific aspects of each proteinopathy (i.e., “disease”) drive that protein to select its unique anatomy in an individual patient. It has become clear, however, that the ultimate spatial patterning of disease is linked to the healthy brain's connectional architecture or “connectome.”
When discussing neurodegenerative conditions, it is critical to disambiguate terms that refer to the clinical syndrome from terms that describe the underlying neuropathological entity giving rise to that syndrome. Throughout this review, I use “syndrome” when describing a named constellation of symptoms and deficits. Examples include “behavioral variant frontotemporal dementia” (bvFTD), “Alzheimer’s disease (AD)-type dementia” or “corticobasal syndrome.” In contrast, I use “disease” to refer to a histopathological entity that might be found at autopsy in a patient showing a neurodegenerative syndrome during life. Examples of disease terms include frontotemporal lobar degeneration (FTLD) with TAR DNA-binding protein 43 (TDP-43) immunoreactive inclusions (FTLD-TDP) Type A, Alzheimer's disease, or corticobasal degeneration, a subtype of FTLD with tau immunoreactive inclusions (FTLD-tau). In short, syndromes reflect where the damaging pathological process is, whereas disease terms describe what the pathological process is. Table 1 details the clinical syndromes used to illustrate key principles throughout this review. Because the frontotemporal dementia (FTD), Alzheimer-type dementia, and amyotrophic lateral sclerosis (ALS) syndromes have been particularly well-studied from a human network perspective, these disorders receive more attention here than several other equally important and related disorders.
Table 1.
Neuroanatomy of onset and progression in the major neurodegenerative syndromes
| Syndrome | Prodromal symptom(s) | Presenting symptom(s) | Onset neuron(s) | Onset region(s) | Target network | Known or candidate downstream networks | Most likely neuropathological diagnosis | Spreading protein | Differential diagnosis |
|---|---|---|---|---|---|---|---|---|---|
| AD-type dementia | Depression, sleep disturbance, anxiety | Episodic memory loss | NE 5HT Layer II stellate pyramidal |
LC DRN ERC |
DMN | ECN Language DAN |
AD | Tau Aβ* |
LBD FTLD-TDP HS Mixed |
| lvPPA | Word-finding difficulty | Hesitant speech Word finding difficulties |
Unknown | Dominant AG, TPJ | Posterior language | DMN | AD | Tau Aβ* |
FTLD-TDP |
| PCA | Unknown | Visual changes Navigation |
Meynert cell | CalcCtx Lat occipital |
1° and 2° visual | DMN | AD | Tau Aβ* |
LBD |
| bvFTD | Job loss “Midlife crisis” |
Apathy Disinhibition Compulsivity |
VEN Fork cell |
ACC, FI | Salience | ECN SAN |
FTLD-TDP FTLD-tau |
TDP-43* Tau |
FTLD-FUS AD |
| svPPA | Word-finding difficulty | Loss of word/object meaning | Unknown | Tpole | SAN | Salience | FTLD-TDP | TDP-43* | FTLD-tau |
| nfvPPA | Speech difficulty | Nonfluent aphasia | Unknown | IFGo | Anterior language | Posterior language | FTLD-tau | Tau | FTLD-TDP |
| CBS | Unknown | Loss of limb control | Unknown | Premotor cortex | Dorsal perirolandic | ECN Language |
FTLD-tau | Tau | AD FTLD-TDP |
| PSP-S | Apathy, mental rigidity | Falls | Unknown | rMT | rMT network | Dorsal perirolandic | PSP | Tau | CBD |
| ALS | Fasiculations, atrophy | Weakness | UMN LMN |
1° MC AH |
Pyramidal motor | ECN Salience |
ALS-TDP | TDP-43* | ALS-FUS ALS-SOD |
| CJD | Variable | Variable | Unknown | Variable | Variable | Variable | CJD | PrPSc | - |
Asterisk acknowledges lingering uncertainty about whether the indicated protein spreads along axonal pathways.
1° MC, primary motor cortex; 5-HT, 5-hydroxytryptamine; ACC, anterior cingulate cortex; AD, Alzheimer's disease; AH, anterior horn; ALS, amyotrophic lateral sclerosis; AG, angular gyrus; bvFTD, behavioral variant frontotemporal dementia; CalcCtx; calcarine cortex; CBS, corticobasal syndrome; CJD, Creutzfeldt–Jacob Disease; DAN, dorsal attention network; DMN, default mode network; DRN, dorsal raphe nucleus; ECN, executive-control network; ERC, entorhinal cortex; FI, frontoinsula; FTLD, frontotemporal lobar degeneration; FUS, fused in sarcoma; IFGo, inferior frontal gyrus, opercular part; LBD, Lewy body disease; LC, locus ceruleus; LMN, lower motor neuron; NE, norepinephrine; PrPSc, prion protein scrapie; PSP, progressive supranuclear palsy; rMT, rostral midbrain tegmentum; SAN, semantic-appraisal network; SOD, superoxide dismutase; TDP-43, transactive response DNA-binding protein 43 kDa; Tpole, temporal pole; TPJ, temporoparietal junction; UMN, upper motor neuron; VEN, von Economo neuron.
The need to separate clinical syndromic from neuropathologic terms stems from how few one-to-one correspondences exist between syndrome and disease. These imperfect clinicopathological correlations give rise to two related concepts, “clinicoanatomical convergence” and “phenotypic diversity,” which receive extensive consideration in the sections that follow.
NETWORK DEGENERATION: HISTORICAL METHODS, OBSERVATIONS, AND LIMITATIONS
The stereotypical patterns of neurodegenerative disease onset and progression have long stimulated ideas about a link to neuronal networks (Pearson et al. 1985; Saper et al. 1987; Braak and Braak 1991; Weintraub and Mesulam 1996). The connectedness among degenerating regions was inferred from axonal tracer studies performed in laboratory mammals, chiefly rodents and primates, and engendered diverse mechanistic hypotheses ranging from spreading prions (Prusiner 1984) to transported toxins (Saper et al. 1987), disrupted growth factors (Salehi et al. 2006), and unknown pathogens (Braak et al. 2003b). Despite these seminal perspectives, for decades neurodegenerative disease anatomy was viewed through oversimplified frameworks, divided into focal versus diffuse or subcortical versus cortical. The notion that each disorder represents a network-based degeneration flows naturally, however, from careful, comprehensive postmortem neuropathological localization and staging studies (Steele et al. 1964; Brun and Gustafson 1978; Braak and Braak 1991). The great advantage of these approaches, which often used whole-brain or whole-hemisphere sectioning and staining, was and remains their capacity to resolve cellular details in patients with defined molecular pathological lesions. Early neuropathological hallmarks could therefore be identified in asymptomatic or prodromal individuals to render a detailed picture of onset and progression. The need to collect and process many brains, each requiring substantial resources, limited the use of these methods to a few laboratories. And, although the approach proved spectacularly successful for prevalent aging-related diseases like AD and Lewy body disease (LBD), it can rarely capture preclinical stages of FTLD, ALS, and other diseases too rare to be encountered by chance even in large autopsy series.
The dawn of human brain mapping in the late 1980s, made possible by brain-wide, voxel-wise statistical methods, slowly gave rise to an era in which neurodegeneration researchers could determine disease topographies in vivo without narrow a priori hypotheses. This shift enabled validation of established patterns (such as that in AD) but was most impactful for less common disorders, such as FTD (Rosen et al. 2002), for which ideas about anatomical onset and progression had been difficult to derive from postmortem data. Network-sensitive imaging approaches emerged in the mid-1990s (Biswal et al. 1995; Fox and Raichle 2007) and provided a means for visualizing network organization and degeneration in living humans (Greicius et al. 2003, 2004; Seeley et al. 2009). Around that time, complementary in vitro and animal model studies had begun to explore mechanisms of network-based dysfunction and disease protein spread. A strong tide of empirical data now supports the notion that misfolded disease protein conformers undergo prion-like spread within and between neurons and across synapses (Frost and Diamond 2010; Goedert et al. 2010; Prusiner 2012). Most recently, structural, functional, and molecular neuroimaging studies have been combined to replicate the stereotypical spread of AD pathological hallmarks (Choo et al. 2007; Whitwell et al. 2007; Thal et al. 2014; Johnson et al. 2016). Emerging positron emission tomography (PET) ligands for pathological forms of the tau protein may enable similar characterizations for the non-AD tauopathies, such as FTLD-tau, at least during their symptomatic phases. Molecular probes for α-synuclein, TDP-43, and other disease proteins remain an important target for development.
HUMAN BRAIN NETWORK MAPPING: THE METHODS
Structural and functional connectivity analyses noninvasively map healthy large-scale networks in vivo (Greicius et al. 2003; Damoiseaux et al. 2006; Fox and Raichle 2007; Biswal et al. 2010) and can detect network connectivity changes in living patients (Greicius et al. 2004; Zhou et al. 2010). The following paragraphs provide a brief overview of the major network-sensitive structural and functional magnetic resonance–based neuroimaging methods.
Intrinsic Connectivity
With task-free functional magnetic resonance imaging (tf-fMRI), researchers can now identify functional intrinsic connectivity networks (ICNs) derived from temporally synchronous, spatially distributed, spontaneous low-frequency (<0.1 Hz) blood oxygen level–dependent (BOLD) signal fluctuations (Biswal et al. 1995; Raichle et al. 2001; Fox et al. 2005; Fox and Raichle 2007). These ICNs, which may represent functional connections spanning multiple synapses, represent a conserved and robust form of organized macroscopic brain activity. Compared to conventional task-based fMRI studies, tf-fMRI is free of performance confounds, making it easier to apply and interpret in cognitively impaired populations. To derive ICNs, seed-based analyses determine correlations among low-frequency BOLD fluctuations of a seed region with the rest of the brain (Biswal et al. 1995). Other approaches, such as independent component analysis and clustering methods, take advantage of multiple simultaneous brain interactions to identify brain networks (Beckmann et al. 2005; Yeo et al. 2011). Ongoing efforts seek to characterize temporal dynamics of ICNs and elucidate possible causal relationships (see reviews by Krajcovicova et al. 2014 and Dennis and Thompson 2014). Synchronization across neuronal assemblies can likewise be computed from task-free electro- or magneto-encephalography data.
Structural Covariance
Coordinated variations in brain structure across subjects have been used as measures of the association between regions to construct large-scale “structural covariance networks” (Mechelli et al. 2005; Lerch et al. 2006; He et al. 2008; Seeley et al. 2009). This approach, which may use gray matter volume or cortical thickness data, relies on the assumption that structural covariance reflects a shared trophic influence during development or ongoing co-trophism conferred by synaptic coupling across regions. Mean gray matter volume or thickness of a region of interest is used to conduct a whole-brain voxel-wise regression across subjects to identify those voxels (or regions or vertices) whose magnitude is correlated with the region of interest. Other analytic approaches, such as independent component analysis and clustering, can likewise be used to derive structural covariance networks.
Structural Connectivity
The term “structural connectivity” most strictly refers to the axonal connections between neurons or brain regions. Although axonal connectivity remains beyond the resolution of current neuroimaging techniques, the integrity of medium to large fiber tracts can be assessed in vivo using diffusion-weighted imaging methods, which map the diffusion of water molecules and rely on the principle that diffusion is restricted by tissue structure (Le Bihan et al. 1992), especially within highly ordered white matter tracts. Region of interest analysis or data-driven voxel-based analysis allows estimation of group differences in fiber tract integrity or associations with cognitive functioning. Fiber tracking between specific region pairs can further be performed (Mori et al. 1999; Mori and Zhang 2006).
Connectomics
The term “connectome” refers to a comprehensive map of the brain's neural connections (Sporns et al. 2005), whether the connections are defined on structural (MRI/diffusion) or functional (fMRI, electroencephalogram [EEG], magnetoencephalography [MEG]) grounds. By modeling networks as graphs (brain regions as nodes and node-to-node connections as edges), graph theoretical analyses offer a flexible and quantitative approach for characterizing brain network topology. Several graph theoretical metrics quantify brain network “hubs” (i.e., regions with high degree centrality) (Sporns et al. 2007; van den Heuvel and Sporns 2011; Zuo et al. 2012; Crossley et al. 2013), whereas other metrics, such as clustering coefficient and path length, emphasize modularity or efficiency of communication. “Connectomics,” then, refers to the science of brain connectivity.
NEURODEGENERATIVE DISEASE: UNIFYING ANATOMICAL PRINCIPLES
This section introduces the key concepts of neurodegenerative disease onset and progression. In my view, the most critical unanswered questions in neurodegenerative disease research regard these two issues. In addition, I discuss two interrelated neurodegenerative disease phenomena: clinicoanatomical convergence and phenotypic diversity. Any comprehensive model of disease onset and progression must account for these observations, which cut across this class of human illness.
Onset
Patients with each neurodegenerative syndrome emerge from an incipient preclinical stage during which symptoms remain absent or subtle and the lesion remains restricted to just one or few brain regions and only to the most susceptible cells and microcircuits within the affected regions. This focal onset manifests as cell-type-specific disease protein aggregation followed by quantifiable neuronal dropout (Hyman et al. 1984; Graveland et al. 1985; Seeley et al. 2006; Kim et al. 2012).
Progression
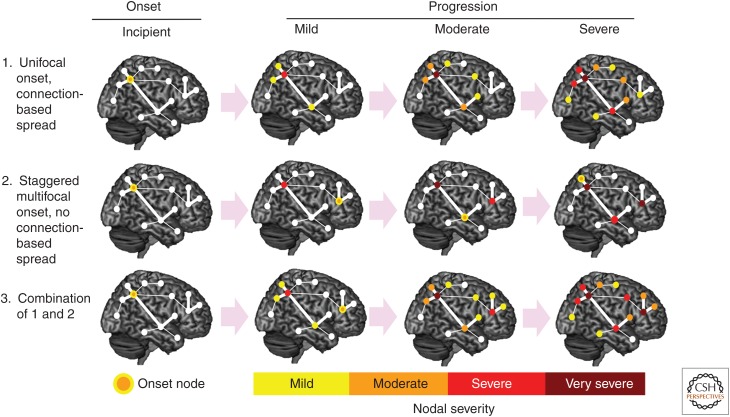
What anatomical principles govern the relentless spatiotemporal progression of each disease? Postmortem and in vivo neuroimaging studies suggest that the pattern of regional injury reflects a network-based landscape (Fig. 1), arguing against the notion that disease spreads across the cortical mantle via spatial contiguity (Steele et al. 1964; Brun and Gustafson 1978; Saper et al. 1987; Braak and Braak 1991; Greicius et al. 2004; Buckner et al. 2005; Seeley et al. 2009). But what factors govern how disease spreads from the onset node(s) to downstream regions within and beyond the target network? At least three onset-progression scenarios should be considered (Fig. 2).
Unifocal (or simultaneous oligofocal) onset with connectional spread. In this scenario, the later-affected regions are determined entirely by the axonal connections of the most vulnerable cells within the onset region(s).
Staggered multifocal onset without connectional spread. Here, anatomical progression reflects independent, temporally staggered eruptions of disease within multiple (not necessarily interconnected) regions. In this way, progression is connectivity-independent and generated by a graded hierarchy of regional and/or cellular vulnerabilities to some diffusely expressed pathogenic process.
Combined unifocal and staggered multifocal onset with connectional spread. In this model, which blends aspects of the previous two, disease progression reflects not only the connectivity of the initial onset regions but also the emergence of later but independent onset sites and the connections of affected neurons within those later-affected sites.
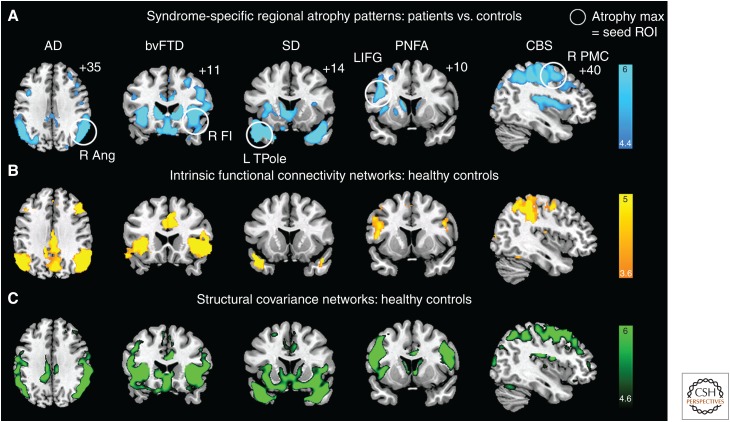
Figure 1.
Neurodegenerative syndromes reflect degeneration within large-scale networks. (A) Five clinical neurodegeneration syndromes showed distinct atrophy patterns, with atrophy maxima highlighted with white circles. Regions circled in A were used as seed regions of interest (ROIs) for task-free functional magnetic resonance imaging (fMRI) analysis (B) and structural covariance mapping (C) in healthy controls. Both approaches showed that the connectivity of the healthy brain mirrored the five atrophy patterns. These data showed that each syndrome was anatomically linked to a specific large-scale network that could be detailed in the healthy brain with connectivity-based methods. (Reproduced, with permission, from Seeley 2016, © 2016 Oxford University Press; www.oup.com.)
Figure 2.
Neurodegenerative disease onset and progression. What is the relationship between disease onset and progression? After a first locus of onset, progression to other regions could involve (1) connectivity-based spread alone, (2) secondary sites of onset within or outside the target network, or (3) a combination of these models. (Reproduced, with permission, from Seeley 2016, © 2016 Oxford University Press; www.oup.com.)
Clinicoanatomical Convergence
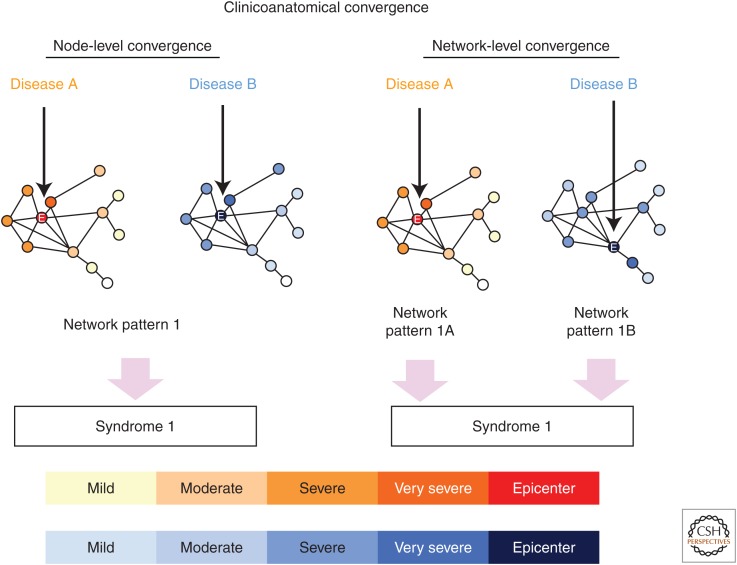
Clinicoanatomical convergence describes the observation that most clinical syndromes can be caused by at least a few and often several underlying pathological entities. For example, patients with bvFTD may be found to have any one of at least 15 different underlying pathological diagnoses, spanning three FTLD major molecular classes (FTLD with tau, TDP-43, or FUS immunoreactive inclusions) and AD. The key question is whether convergence occurs at the network, regional, or neuronal level (Fig. 3). In other words, distinct proteinopathies could converge at the network level by targeting disease-specific nodes within the same syndrome-associated network. In this scenario, neuroimaging studies might improve antemortem pathological predictions by detecting disease-specific atrophy signatures (within the syndromic network). Alternatively, convergence could occur at the regional or even neuronal level, in which case methods capturing brain structure or function would fail to discriminate between diseases, and alternative approaches, such as molecular imaging or fluid biomarkers linked to the disease proteins themselves, would be required.
Figure 3.
Clinicoanatomical convergence may occur at the neuronal, nodal, or network levels. Diseases that cause each syndrome may converge at multiple levels to create the syndrome. Convergence at the level of specific neuronal types (not shown) or even specific network nodes (left) would be expected to create nearly identical patterns of network impairment. Alternatively, convergence could occur at the level of the overall network (right), with each disease targeting different nodes but, nonetheless, manifesting as the same (or nearly the same) syndrome. Circles represent network nodes (brain regions), lines represent edges (connections between two nodes), and shorter edges indicate tighter connections between node pairs. Color shading indicates the severity of predicted regional impairment based on the onset nodes (“epicenters”) indicated by arrows. (Reproduced, with permission, from Seeley 2016, © 2016 Oxford University Press; www.oup.com.)
Phenotypic Diversity
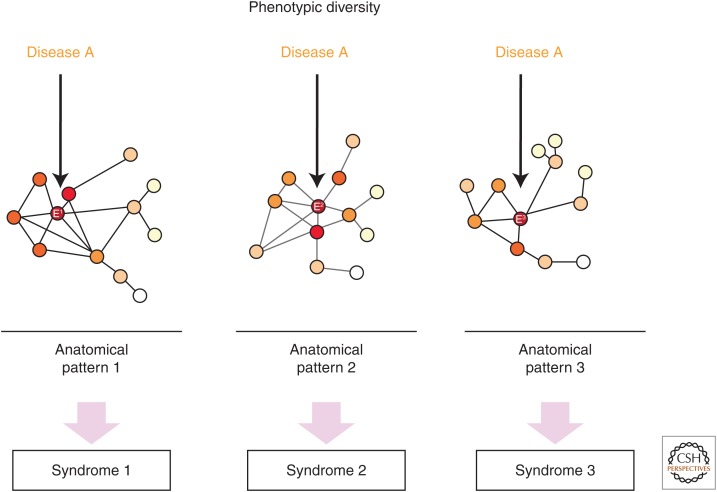
Phenotypic diversity refers to the observation that the same histopathological entity (i.e., disease) may be associated with several distinct clinical syndromes, reflecting distinct regional degeneration patterns (Fig. 4). For example, Pick's disease, a subtype of FTLD-tau, may present with bvFTD, semantic variant primary progressive aphasia, nonfluent variant primary progressive aphasia, or corticobasal syndrome, based on the targeted regional epicenter and its network-based affiliations. This observation suggests either that (1) each disease protein maintains a certain nonrandom variability with regard to where it first aggregates in an individual brain or that (2) neuropathological taxonomy remains inadequately specified and that further characterization (i.e., “splitting”) of the tau protein found in Pick's disease will, extending the example, reveal different forms of posttranslationally modified or misfolded tau in each of the syndromic presentations of Pick's disease. Such hypothetical subtypes of a given single disease protein are often conceptualized as conformer “strains” (Sanders et al. 2014). To explain phenotypic diversity entirely, the tau protein strain recognized pathologically as Pick's disease would have to be further divided into Pick-type “substrains,” one for each of the syndromic presentations of Pick's disease.
Figure 4.
Phenotypic diversity suggests that most diseases can produce multiple clinical syndromes, reflecting a small portfolio of candidate onset regions (“epicenters,” E). The heterogeneity of clinical manifestations for each disease is illustrated here at the network level, where onset within epicenters (E1, E2, or E3) that anchor distinct networks gives rise to three different clinicoanatomical presentations. Network depictions follow Figure 3.
Based on these unifying neurodegeneration principles, I will discuss disease onset regions and cell types in more detail. I will review neuroimaging data that inform competing models of disease progression. I will relate competing concepts of onset and spread to clinicoanatomical convergence and phenotypic diversity. Finally, I will consider the most important frontiers in selective vulnerability and network imaging.
MODELING ONSET: WHERE AND HOW DOES DISEASE BEGIN?
Evidence to Date
How does each neurodegenerative disease select its initial target or targets? This question remains an enduring mystery for every illness, and merely identifying the early targets has proven challenging enough. For AD and LBD, early neuronal targets have been identified through cross-sectional postmortem studies that have included patients at all stages of the disease process, from asymptomatic to prodromal to full-blown symptomatic and even end-stage (Braak et al. 1993, 2004). Regional-level observations made with this approach have been well-supported by longitudinal imaging studies in living individuals. For example, studies following older individuals from health to mild memory impairment and later AD-type dementia show early tau deposition and atrophy in the entorhinal cortex (Killiany et al. 2002; Jack et al. 2004; Varon et al. 2011; Johnson et al. 2016), consistent with classical postmortem studies (Braak and Braak 1991; Braak et al. 1993). Johnson et al. (2016) found that tau deposition extends beyond the medial temporal lobe only in patients with cortical β-amyloid deposition. On the other hand, in vivo brain imaging lacks the regional subnuclear and neuronal subtype resolution required to provide a complete picture. This limitation is well-illustrated by AD and Parkinson's disease, in which the earliest brain neuronal protein aggregates are now understood to emerge in brainstem nuclei that are difficult to resolve with conventional MRI or PET: the locus coeruleus and dorsal raphe in AD (Bondareff et al. 1981; Grinberg et al. 2009; Braak and Del Tredici 2012) and the dorsal motor nucleus of the vagus nerve in LBD (Braak et al. 2003a). In LBD, the process may begin even more peripherally, in the olfactory mucosa and enteric nervous system. For less common diseases, like FTLD, determining early neuronal subtype selectivity has been even more difficult because of the diversity of FTD syndromes and the scarcity of postmortem materials from patients with asymptomatic or prodromal disease. The few laudable attempts to derive distinct stages using cross-sectional materials have not been able to include individuals with presymptomatic disease (Brettschneider et al. 2014; Irwin et al. 2016). Furthermore, because each FTLD pathological subtype produces diverse clinical phenotypes, it would be difficult to interpret presymptomatic FTLD materials even if they became available. One remarkable exception comes from a patient who died of brainstem lymphoma but was astutely noted to harbor premanifest Pick's disease (Miki et al. 2014), with Pick bodies and other Pick-type tau inclusions in an anterior cingulate-frontoinsular pattern that almost perfectly matches the early bvFTD regional vulnerability profile (Seeley et al. 2008).
How, then, can brain imaging studies in symptomatic patients inform our understanding of disease onset? Regions showing the greatest atrophy during symptomatic disease may or may not represent the sites of initial injury, but recent neuroimaging studies support an emerging model for generating hypotheses about where each syndrome begins before it spreads. Having established that each neurodegenerative syndrome is linked to a specific network (Fig. 3) (Seeley et al. 2009), my colleagues and I, led by Juan (Helen) Zhou, showed that each syndrome-associated brain network contains a vulnerable “epicenter” (or epicenters), whose connectivity in health mirrors—and may template—the spatial patterning of each syndrome (Zhou et al. 2012). These epicenters bear close relationships to the early clinical and anatomical deficits that define each syndrome. For instance, in bvFTD the identified epicenters in the right frontoinsula and pregenual anterior cingulate cortex are known for their co-activation as part of a “salience network” for homeostatic behavioral guidance (Seeley et al. 2011) and harbor a unique class of large, bipolar projection neurons, called von Economo neurons, that may represent the initial target in bvFTD (Seeley et al. 2006; Kim et al. 2012). Identifying an epicenter, as defined above, does not prove that this epicenter represents the site of initial injury; nonetheless, there is a striking overlap between regions of peak atrophy and those that serve as epicenters (Zhou et al. 2012).
Despite the scarcity of postmortem materials representing presymptomatic FTLD, structural and functional imaging has begun to provide insights into presymptomatic inherited FTD. In the first large study of this kind, carriers of FTLD-causing microtubule-associated protein tau (MAPT) or progranulin (GRN) mutations showed fractional anisotropy reductions in the right uncinate fasciculus and decreased functional connectivity between key salience network hubs, the anterior mid-cingulate cortex and frontoinsula, compared with noncarriers (Dopper et al. 2013). More recently, using region of interest–based structural MRI, researchers have identified sites presumed to reflect incipient atrophy in each of the three major FTD-causing mutations (MAPT, GRN, and C9ORF72) (Rohrer et al. 2015). Converging with findings from patients with symptomatic bvFTD and with the bvFTD epicenters identified by Zhou and colleagues (Zhou et al. 2012), Rohrer et al. identified the insula as a region showing atrophy among the youngest mutation carriers when examining all three genetic subgroups together. Although the insula appeared to degenerate first in a GRN mutation carrier subset analysis, other regions showed even earlier deficits in MAPT (hippocampus) and C9ORF72 (thalamus) carriers, as predicted by the atrophy seen in symptomatic mutation carriers (Whitwell et al. 2009a,b; Mahoney et al. 2012; Sha et al. 2012). These important studies, however, share several methodological limitations. In presymptomatic FTD gene carriers, we have no way to predict which of the several associated clinical syndromes will later emerge; in this way, group-level results likely represent a blend of preclinical syndromic patterns, as well as the known anatomical heterogeneity within each syndrome. Studies of preclinical inherited FTD may also generalize weakly to sporadic FTD, considering the diversity of genetic mechanisms and the known anatomical differences seen in patients with inherited versus sporadic FTD. Finally, it remains uncertain whether the observed gray matter volume deficits represent incipient degeneration in early adulthood or an abnormal developmental trajectory that has yet to be traced back to its origins.
Relationship to Clinicoanatomical Convergence and Phenotypic Diversity
Does clinicoanatomical convergence reflect onset within the same vulnerable neuron population or within different neuronal constituents of the same region or network? To address this question requires that we study all relevant levels, in a single syndrome, as caused by multiple diseases. For example, does bvFTD begin in the von Economo neurons whether the syndrome is caused by FTLD-tau, TDP-43, or FUS? Some studies have provided clues toward this cell-type-level convergence on the von Economo neurons (Seeley et al. 2006; Kim et al. 2012; Santillo et al. 2013; Santillo and Englund 2014), but the studies needed to fully resolve the issue have yet to be performed.
Principles of disease onset should also be viewed in light of phenotypic diversity. For example, although most patients with underlying AD present with early memory loss, a significant minority presents with a nonamnestic syndrome (Snowden et al. 2007). Patients with nonfamilial early-onset AD (EOAD, defined as onset <65 years in most studies) show a mix of cognitive deficits, often beginning with attentional or executive impairment (Frisoni et al. 2007; Koedam et al. 2010). Focal syndromes such as posterior cortical atrophy (PCA), characterized by predominant visuospatial and visuoperceptual deficits (Crutch et al. 2012) and the logopenic variant of primary progressive aphasia (lvPPA), a progressive disorder of language (Gorno-Tempini et al. 2008), are also strongly linked to AD pathology. The factors driving this phenotypic diversity are not well understood but could reflect an internal hierarchy or “pecking order” of vulnerability that differs between individual patients based on their genetic backgrounds, life experiences, region-specific stressors (trauma, seizures, vascular malformations, etc.), or developmental anomalies (Rogalski et al. 2013).
Lingering Questions and Uncertainties
Many key questions remain within the general concept of disease onset. How many cell types and/or brain regions undergo independent (sometimes referred to as “cell autonomous”) onsets? What is the hierarchy of neuron-type vulnerabilities for each disease? Does this order vary across individuals? Does onset occur within neurons, glia, or both? Can cells undergo a “reversible onset,” such as protein aggregation and dysfunction, but then revert to a healthy state? Does protein misfolding and aggregation begin only within a select and finite group of cell types/brain regions for each protein, or, alternatively, does this homeostatically controlled process pervade the aging brain but remain in check in all but that protein's short list of onset cells/regions, which are somehow ill-equipped to manage the quality control process?
MODELING PROGRESSION: HOW DOES DISEASE MOVE BEYOND THE CELLS AND REGIONS WHERE IT BEGINS?
Evidence to Date
That each neurodegenerative syndrome reflects a large-scale network breakdown has now been established through data that converge across diseases, methods, and research groups. Early network-based imaging support for this principle came from studies of AD-type dementia, which features an anatomical profile strongly linked to the default mode network (Greicius et al. 2003, 2004; Buckner et al. 2008). Next, it was shown that AD and four distinct FTD syndromes are each associated with atrophy reflecting a healthy human intrinsic connectivity and structural covariance network (Fig. 1) (Seeley et al. 2009). But how does disease progress from the onset stage to render a network-based spatial pattern? At least four disease-general hypotheses have been put forth and can be summarized as (1) “nodal stress,” in which regions subject to heavy network traffic (i.e., “hubs”) undergo activity-related “wear and tear” that gives rise to or worsens disease (Buckner et al. 2009; Saxena and Caroni 2011); (2) “transneuronal spread,” in which some toxic agent propagates along network connections, perhaps through “prion-like” templated conformational change (Prusiner 1984; Baker et al. 1994; Ridley et al. 2006; Walker et al. 2006; Frost et al. 2009; Frost and Diamond 2010; Lee et al. 2010; Jucker and Walker 2011); (3) “trophic failure,” in which disruption of network connectivity undermines internodal trophic factor support, accelerating disease within nodes lacking collateral trophic sources (Appel 1981; Salehi et al. 2006); and (4) “shared vulnerability,” in which networked regions feature a common gene or protein expression signature (Richiardi et al. 2015) that confers relatively disease-specific susceptibility, evenly distributed throughout the network. These nonmutually exclusive candidate network degeneration mechanisms make competing predictions about how healthy network architecture should influence disease-associated regional vulnerability. Although “network degeneration” is often understood to mean “network-based spread,” only the “transneuronal spread” model proposes that progression represents physical spreading of a pathological process along axons connecting individual neurons.
To date, most efforts to investigate mechanisms of disease progression have relied on cross-sectional data. In the study by Zhou and coworkers (2012), we identified epicenters whose normal connectivity profiles most resembled the syndrome-associated atrophy patterns, as described above. We then used graph theoretical analyses in healthy subjects to show that regions with higher total connectional flow and, more consistently, shorter functional paths to the epicenters showed greater syndrome-associated vulnerability. The relationship between regional network “traffic” and vulnerability suggests that activity-dependent mechanisms, such as oxidative stress, local extracellular milieu fluctuations, or glia-dependent phenomena might influence regional vulnerability; this influence might be a key factor in determining sites of initial or secondary onset. Because nodes with shorter connectional paths to an epicenter also showed greater vulnerability, it appears that “connectional closeness” is another key vulnerability factor, an observation most parsimoniously explained by physical, transsynaptic spreading of a toxic agent. Epicenter infiltration by disease may provide privileged but graded and connectivity-driven access across the network that determines where the disease will arrive next. Predictions made by the trophic factor insufficiency hypothesis were not consistent with our data. Although a shared gene or protein expression profile across networked regions may influence sites of onset, our findings were difficult to reconcile with predictions made by the “shared expression” model. We further examined connectivity-vulnerability relationships within the “off-target” networks to determine how nodal characteristics influence downstream vulnerability. Here, overwhelmingly, the evidence supported the transneuronal spread model. In summary, the findings best fit a model in which initial vulnerability may in part reflect a node's centrality (i.e., “hubness”) within the target network, whereas downstream vulnerability within and beyond the target network more closely relates to a node's connectional proximity to the most vulnerable epicenters.
In AD, innovative studies have begun to link regional connectivity profiles to hallmark AD molecular lesions, which can now be localized in vivo with molecular PET imaging, and disease progression. In a study describing an “epidemic spreading model,” the investigators considered axonal propagation of amyloid protein along the healthy structural connectome and regional clearance mechanisms. The model was able to explain roughly 50% of the variance in measured amyloid deposition on amyloid PET (Iturria-Medina et al. 2014) based on the connectional model, supporting the general hypothesis that regional amyloid deposition in part reflects the connectional distance from specific outbreak regions, which may lie in the anterior paramedian and posterior cingulate cortices. In AD, clearly, progression models need to account for two stages of the illness, one in which amyloid-β deposition is a key factor and another in which intraneuronal tau spreading takes over and drives the clinical and anatomical deficit pattern. In a recent longitudinal study of prodromal AD and AD-type dementia, the healthy brain's structural connectome was used to predict the progression of regional atrophy by modeling progression as simple diffusion along fiber tracts (Raj et al. 2015). This model makes no assumptions about where the diffusive process begins, a feature that may allow the model to accommodate the known heterogeneity in onset sites across patients.
Relationship to Clinicoanatomical Convergence and Phenotypic Diversity
How do emerging principles of disease progression relate to clinicoanatomical convergence? If progression is driven by connectional spread, then brain-wide anatomical convergence could merely reflect a shared population of onset neurons. Alternatively, distinct onset sites within the same network could, via connectional spread, produce convergent involvement of the overall network. In other words, there may be alternative anatomical pathways to the same syndrome. A particularly clear example of this notion comes from bvFTD. In the subset of patients who carry the C9ORF72 hexanucleotide repeat expansion, salience network dysfunction resembles that seen in sporadic bvFTD, but the loss of network integrity is linked to a strategic lesion of the medial pulvinar thalamus (Lee et al. 2014). This mechanism of network breakdown differs from that seen in sporadic bvFTD, where the salience network is disrupted by early involvement of anterior cingulate and frontoinsular cortices. Thus, in bvFTD, the clinical deficits may reflect disruption of the same network by damage to distinct onset nodes.
The phenotypic diversity produced by AD raises the question of whether each clinical AD variant can be linked to a distinct large-scale network or onset site. A recent study tested this hypothesis by assessing intrinsic functional connectivity in healthy subjects, seeding regions commonly or specifically atrophied in early-onset AD, lvPPA, or posterior cortical atrophy (Lehmann et al. 2013). The investigators found that the connectivity maps derived from commonly atrophied regions of interest resembled the default mode network, which was affected in all AD variants, whereas seeding regions specifically atrophied in each AD variant revealed distinct, syndrome-specific connectivity patterns in the healthy brain. These findings indicate that the syndrome-specific neurodegenerative patterns in AD variants are driven by the involvement of specific networks outside the default mode network. One might predict that spread into these distinct networks reflects differences in the precise localization of onset in the three variants; where exactly (in which regions and neuronal subtypes) these syndromes begin remains uncertain, but meticulous neuroanatomical studies suggest that PCA may begin with neurofibrillary tangle formation and neuronal loss within large, long-range projection neurons in the primary visual cortex, such as the layer 5 Meynert cells (Hof et al. 1997).
Lingering Questions and Uncertainties
Many questions about the mechanisms of disease progression remain unanswered, and many of those questions are daunting. Considering the three hypothetical progression scenarios (Fig. 2), what is the balance between connection-based spread versus secondary sites of onset? Does spread within the local microcircuitry occur via contiguity (such as release of disease protein by dying cells and uptake by others), or is it governed by axo-dendritic (or dendro-dendritic) synapses? What better predicts disease progression: a patient's current, “personalized” (i.e., diseased) connectome, that patient's premorbid connectome, or a normative connectome from young or older subjects? How do genetic risk factors interact with the connectome to influence disease progression? Resolving these questions may help to facilitate development of individualized treatment and prevention trials.
FUTURE DIRECTIONS
To aid in the search for treatments, connectivity-based neuroimaging methods will need to detect early disease in individuals or track progression over time. For most sporadic diseases, presymptomatic detection remains a distant reality because either the right tools are lacking or the disease is too infrequent to facilitate large-scale population screening without a sensitive and affordable test. Efforts to monitor disease with connectivity metrics have, so far, been limited, with most evidence coming from cross-sectional correlations with disease severity (Zhang et al. 2010; Zhou et al. 2010). One longitudinal study showed reduced intrinsic connectivity in the posterior default mode network and increased connectivity in anterior and ventral default mode subnetworks in AD compared to healthy controls at baseline (Damoiseaux et al. 2012). At follow-up, patients showed worsening connectivity across all default mode subsystems, consistent with a network-based degeneration model in which disease first spreads from its “epicenters” to interconnected nodes within the target network (Zhou et al. 2012). An alternative model based on diffusion within the white matter architecture (i.e., structural connectome) showed that the model could predict progression in subjects with mild cognitive impairment and AD-type dementia (Raj et al. 2015). Systematic collection and analysis of multicenter multimodal imaging and biomarker data, including functional and structural connectivity metrics, will be required to assess the value of imaging biomarkers for diagnosis, prognosis, and disease monitoring.
ACKNOWLEDGMENTS
I thank Juan (Helen) Zhou for helpful discussions and for contributing to the illustrations. I also thank the study participants at the University of California, San Francisco, for their invaluable contributions to neurodegeneration research.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Appel SH. 1981. A unifying hypothesis for the cause of amyotrophic lateral sclerosis, parkinsonism, and Alzheimer disease. Ann Neurol 10: 499–505. [DOI] [PubMed] [Google Scholar]
- Baker HF, Ridley RM, Duchen LW, Crow TJ, Bruton CJ. 1994. Induction of β (A4)-amyloid in primates by injection of Alzheimer's disease brain homogenate. Comparison with transmission of spongiform encephalopathy. Mol Neurobiol 8: 25–39. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. 2005. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci 360: 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34: 537–541. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, et al. 2010. Toward discovery science of human brain function. Proc Natl Acad Sci 107: 4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondareff W, Mountjoy CQ, Roth M. 1981. Selective loss of neurones of origin of adrenergic projection to cerebral cortex (nucleus locus coeruleus) in senile dementia. Lancet 1: 783–784. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. 1991. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol 82: 239–259. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. 2012. Where, when, and in what form does sporadic Alzheimer's disease begin? Curr Opin Neurol 25: 708–714. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, Bohl J. 1993. Staging of Alzheimer-related cortical destruction. Eur Neurol 33: 403–408. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. 2003a. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging 24: 197–211. [DOI] [PubMed] [Google Scholar]
- Braak H, Rub U, Gai WP, Del Tredici K. 2003b. Idiopathic Parkinson's disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J Neural Transm 110: 517–536. [DOI] [PubMed] [Google Scholar]
- Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. 2004. Stages in the development of Parkinson's disease-related pathology. Cell Tissue Res 318: 121–134. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Irwin DJ, Grossman M, Robinson JL, Toledo JB, Lee EB, Fang L, Van Deerlin VM, Ludolph AC, et al. 2014. Sequential distribution of pTDP-43 pathology in behavioral variant frontotemporal dementia (bvFTD). Acta Neuropathol 127: 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun A, Gustafson L. 1978. Limbic lobe involvement in presenile dementia. Arch Psychiatr Nervenkr 226: 79–93. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. 2008. The brain's default network: Anatomy, function, and relevance to disease. Ann N Y Acad Sci 1124: 1–38. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, et al. 2005. Molecular, structural, and functional characterization of Alzheimer's disease: Evidence for a relationship between default activity, amyloid, and memory. J Neurosci 25: 7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. 2009. Cortical Hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci 29: 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo IH, Lee DY, Youn JC, Jhoo JH, Kim KW, Lee DS, Lee JS, Woo JI. 2007. Topographic patterns of brain functional impairment progression according to clinical severity staging in 116 Alzheimer disease patients: FDG-PET study. Alzheimer Dis Assoc Disord 21: 77–84. [DOI] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Vertes PE, Winton-Brown TT, Patel AX, Ginestet CE, McGuire P, Bullmore ET. 2013. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc Natl Acad Sci 110: 11583–11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crutch SJ, Lehmann M, Schott JM, Rabinovici GD, Rossor MN, Fox NC. 2012. Posterior cortical atrophy. Lancet Neurol 11: 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Prater KE, Miller BL, Greicius MD. 2012. Functional connectivity tracks clinical deterioration in Alzheimer's disease. Neurobiol Aging 33: 828.e19–828.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. 2006. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci 103: 13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Thompson PM. 2014. Functional brain connectivity using fMRI in aging and Alzheimer's disease. Neuropsychol Rev 24: 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopper EG, Rombouts SA, Jiskoot LC, Heijer T, de Graaf JR, Koning I, Hammerschlag AR, Seelaar H, Seeley WW, Veer IM, et al. 2013. Structural and functional brain connectivity in presymptomatic familial frontotemporal dementia. Neurology 80: 814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8: 700–711. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci 102: 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB, Pievani M, Testa C, Sabattoli F, Bresciani L, Bonetti M, Beltramello A, Hayashi KM, Toga AW, Thompson PM. 2007. The topography of grey matter involvement in early and late onset Alzheimer's disease. Brain 130: 720–730. [DOI] [PubMed] [Google Scholar]
- Frost B, Diamond MI. 2010. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci 11: 155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Ollesch J, Wille H, Diamond MI. 2009. Conformational diversity of wild-type tau fibrils specified by templated conformation change. J Biol Chem 284: 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Clavaguera F, Tolnay M. 2010. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci 33: 317–325. [DOI] [PubMed] [Google Scholar]
- Gorno-Tempini ML, Brambati SM, Ginex V, Ogar J, Dronkers NF, Marcone A, Perani D, Garibotto V, Cappa SF, Miller BL. 2008. The logopenic/phonological variant of primary progressive aphasia. Neurology 71: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveland GA, Williams RS, DiFiglia M. 1985. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington's disease. Science 227: 770–773. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. 2003. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci 100: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. 2004. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci 101: 4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg LT, Rub U, Ferretti RE, Nitrini R, Farfel JM, Polichiso L, Gierga K, Jacob-Filho W, Heinsen H, Brazilian Brain Bank Study G. 2009. The dorsal raphe nucleus shows phospho-tau neurofibrillary changes before the transentorhinal region in Alzheimer's disease. A precocious onset? Neuropathol Appl Neurobiol 35: 406–416. [DOI] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. 2008. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J Neurosci 28: 4756–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Vogt BA, Bouras C, Morrison JH. 1997. Atypical form of Alzheimer's disease with prominent posterior cortical atrophy: A review of lesion distribution and circuit disconnection in cortical visual pathways. Vision Res 37: 3609–3625. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Damasio AR, Van Hoesen GW, Barnes CL. 1984. Alzheimer's disease: Cell-specific pathology isolates the hippocampal formation. Science 298: 83–95. [DOI] [PubMed] [Google Scholar]
- Irwin DJ, Brettschneider J, McMillan CT, Cooper F, Olm C, Arnold SE, Van Deerlin VM, Seeley WW, Miller BL, Lee EB, et al. 2016. Deep clinical and neuropathological phenotyping of Pick disease. Ann Neurol 79: 272–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Toussaint PJ, Evans AC. 2014. Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders. PLoS Comput Biol 10: e1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Shiung MM, Gunter JL, O'Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, et al. 2004. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology 62: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Schultz A, Betensky RA, Becker JA, Sepulcre J, Rentz D, Mormino E, Chhatwal J, Amariglio R, Papp K, et al. 2016. Tau positron emission tomographic imaging in aging and early Alzheimer disease. Ann Neurol 79: 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M, Walker LC. 2011. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann Neurol 70: 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killiany RJ, Hyman BT, Gomez-Isla T, Moss MB, Kikinis R, Jolesz F, Tanzi R, Jones K, Albert MS. 2002. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology 58: 1188–1196. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Sidhu M, Gaus SE, Huang EJ, Hof PR, Miller BL, DeArmond SJ, Seeley WW. 2012. Selective frontoinsular von Economo neuron and fork cell loss in early behavioral variant frontotemporal dementia. Cereb Cortex 22: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedam EL, Lauffer V, van der Vlies AE, van der Flier WM, Scheltens P, Pijnenburg YA. 2010. Early-versus late-onset Alzheimer's disease: More than age alone. J Alzheimers Dis 19: 1401–1408. [DOI] [PubMed] [Google Scholar]
- Krajcovicova L, Marecek R, Mikl M, Rektorova I. 2014. Disruption of resting functional connectivity in Alzheimer's patients and at-risk subjects. Curr Neurol Neurosci Rep 14: 491. [DOI] [PubMed] [Google Scholar]
- Le Bihan D, Turner R, Douek P, Patronas N. 1992. Diffusion MR imaging: Clinical applications. AJR Am J Roentgenol 159: 591–599. [DOI] [PubMed] [Google Scholar]
- Lee JK, Jin HK, Endo S, Schuchman EH, Carter JE, Bae JS. 2010. Intracerebral transplantation of bone marrow-derived mesenchymal stem cells reduces amyloid-beta deposition and rescues memory deficits in Alzheimer's disease mice by modulation of immune responses. Stem Cells 28: 329–343. [DOI] [PubMed] [Google Scholar]
- Lee SE, Khazenzon AM, Trujillo AJ, Guo CC, Yokoyama JS, Sha SJ, Takada LT, Karydas AM, Block NR, Coppola G, et al. 2014. Altered network connectivity in frontotemporal dementia with C9orf72 hexanucleotide repeat expansion. Brain 137: 3047–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Madison CM, Ghosh PM, Seeley WW, Mormino E, Greicius MD, Gorno-Tempini ML, Kramer JH, Miller BL, Jagust WJ, et al. 2013. Intrinsic connectivity networks in healthy subjects explain clinical variability in Alzheimer's disease. Proc Natl Acad Sci 110: 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Worsley K, Shaw WP, Greenstein DK, Lenroot RK, Giedd J, Evans AC. 2006. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage 31: 993–1003. [DOI] [PubMed] [Google Scholar]
- Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC, et al. 2012. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: Clinical, neuroanatomical and neuropathological features. Brain 135: 736–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli A, Friston KJ, Frackowiak RS, Price CJ. 2005. Structural covariance in the human cortex. J Neurosci 25: 8303–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Mori F, Tanji K, Kurotaki H, Kakita A, Takahashi H, Wakabayashi K. 2014. An autopsy case of incipient Pick's disease: Immunohistochemical profile of early-stage Pick body formation. Neuropathology 34: 386–391. [DOI] [PubMed] [Google Scholar]
- Mori S, Zhang J. 2006. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51: 527–539. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, Van Zijl PCM. 1999. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45: 265–269. [DOI] [PubMed] [Google Scholar]
- Pearson RC, Esiri MM, Hiorns RW, Wilcock GK, Powell TP. 1985. Anatomical correlates of the distribution of the pathological changes in the neocortex in Alzheimer disease. Proc Natl Acad Sci 82: 4531–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 1984. Some speculations about prions, amyloid, and Alzheimer's disease. N Engl J Med 310: 661–663. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. 2012. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 336: 1511–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. 2001. A default mode of brain function. Proc Natl Acad Sci 98: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, LoCastro E, Kuceyeski A, Tosun D, Relkin N, Weiner M. 2015. Network diffusion model of progression predicts longitudinal patterns of atrophy and metabolism in Alzheimer's disease. Cell Rep 10: 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T, Barker GJ, Bokde AL, Bromberg U, Buchel C, et al. 2015. BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science 348: 1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley RM, Baker HF, Windle CP, Cummings RM. 2006. Very long term studies of the seeding of beta-amyloidosis in primates. J Neural Transm (Vienna) 113: 1243–1251. [DOI] [PubMed] [Google Scholar]
- Rogalski E, Weintraub S, Mesulam MM. 2013. Are there susceptibility factors for primary progressive aphasia? Brain Lang 127: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD, Nicholas JM, Cash DM, van Swieten J, Dopper E, Jiskoot L, van Minkelen R, Rombouts SA, Cardoso MJ, Clegg S, et al. 2015. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal Dementia Initiative (GENFI) study: A cross-sectional analysis. Lancet Neurol 14: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen HJ, Gorno-Tempini ML, Goldman WP, Perry RJ, Schuff N, Weiner M, Feiwell R, Kramer JH, Miller BL. 2002. Patterns of brain atrophy in frontotemporal dementia and semantic dementia. Neurology 58: 198–208. [DOI] [PubMed] [Google Scholar]
- Salehi A, Delcroix JD, Belichenko PV, Zhan K, Wu C, Valletta JS, Takimoto-Kimura R, Kleschevnikov AM, Sambamurti K, Chung PP, et al. 2006. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron 51: 29–42. [DOI] [PubMed] [Google Scholar]
- Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, et al. 2014. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82: 1271–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santillo AF, Englund E. 2014. Greater loss of von Economo neurons than loss of layer II and III neurons in behavioral variant frontotemporal dementia. Am J Neurodegener Dis 3: 64–71. [PMC free article] [PubMed] [Google Scholar]
- Santillo AF, Nilsson C, Englund E. 2013. von Economo neurones are selectively targeted in frontotemporal dementia. Neuropathol Appl Neurobiol 39: 572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB, Wainer BH, German DC. 1987. Axonal and transneuronal transport in the transmission of neurological disease: Potential role in system degenerations, including Alzheimer's disease. Neuroscience 23: 389–398. [DOI] [PubMed] [Google Scholar]
- Saxena S, Caroni P. 2011. Selective neuronal vulnerability in neurodegenerative diseases: From stressor thresholds to degeneration. Neuron 71: 35–48. [DOI] [PubMed] [Google Scholar]
- Seeley WS. 2016. Brain circuits: Pathway for NDD. In Neurodegenerative diseases: Unifying principles, Chap. 7, pp. 98–122. Oxford University Press, New York. [Google Scholar]
- Seeley WW, Carlin DA, Allman JM, Macedo MN, Bush C, Miller BL, Dearmond SJ. 2006. Early frontotemporal dementia targets neurons unique to apes and humans. Ann Neurol 60: 660–667. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford R, Rascovsky K, Kramer JH, Weiner M, Miller BL, Gorno-Tempini ML. 2008. Frontal paralimbic network atrophy in very mild behavioral variant frontotemporal dementia. Arch Neurol 65: 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. 2009. Neurodegenerative diseases target large-scale human brain networks. Neuron 62: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Zhou J, Kim EJ. 2011. Frontotemporal dementia: What can the behavioral variant teach us about human brain organization? Neuroscientist 18: 373–385. [DOI] [PubMed] [Google Scholar]
- Sha SJ, Takada LT, Rankin KP, Yokoyama JS, Rutherford NJ, Fong JC, Khan B, Karydas A, Baker MC, DeJesus-Hernandez M, et al. 2012. Frontotemporal dementia due to C9ORF72 mutations: Clinical and imaging features. Neurology 79: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden JS, Stopford CL, Julien CL, Thompson JC, Davidson Y, Gibbons L, Pritchard A, Lendon CL, Richardson AM, Varma A, et al. 2007. Cognitive phenotypes in Alzheimer's disease and genetic risk. Cortex 43: 835–845. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R. 2005. The human connectome: A structural description of the human brain. PLoS Comput Biol 1: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kotter R. 2007. Identification and classification of hubs in brain networks. PLoS One 2: e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele JC, Richardson JC, Olszewski J. 1964. Progressive supranuclear palsy. Arch Neurol 10: 333–360. [DOI] [PubMed] [Google Scholar]
- Thal DR, Attems J, Ewers M. 2014. Spreading of amyloid, tau, and microvascular pathology in Alzheimer's disease: Findings from neuropathological and neuroimaging studies. J Alzheimers Dis 42: S421–S429. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. 2011. Rich-club organization of the human connectome. J Neurosci 31: 15775–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon D, Loewenstein DA, Potter E, Greig MT, Agron J, Shen Q, Zhao W, Celeste Ramirez M, Santos I, Barker W, et al. 2011. Minimal atrophy of the entorhinal cortex and hippocampus: Progression of cognitive impairment. Dement Geriatr Cogn Disord 31: 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LC, Levine H 3rd, Mattson MP, Jucker M. 2006. Inducible proteopathies. Trends Neurosci 29: 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub S, Mesulam MM. 1996. From neuronal networks to dementia: Four clinical profiles. La demence: pourquoi, pp. 75–97.
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR Jr. 2007. 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer's disease. Brain 130: 1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR Jr, Boeve BF, Senjem ML, Baker M, Ivnik RJ, Knopman DS, Wszolek ZK, Petersen RC, Rademakers R, et al. 2009a. Atrophy patterns in IVS10 + 16, IVS10 + 3, N279K, S305N, P301L, and V337M MAPT mutations. Neurology 73: 1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR Jr, Boeve BF, Senjem ML, Baker M, Rademakers R, Ivnik RJ, Knopman DS, Wszolek ZK, Petersen RC, et al. 2009b. Voxel-based morphometry patterns of atrophy in FTLD with mutations in MAPT or PGRN. Neurology 72: 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106: 1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Wang SJ, Liu B, Ma ZL, Yang M, Zhang ZJ, Teng GJ. 2010. Resting brain connectivity: Changes during the progress of Alzheimer disease. Radiology 256: 598–606. [DOI] [PubMed] [Google Scholar]
- Zhou J, Greicius MD, Gennatas ED, Growdon ME, Jang JY, Rabinovici GD, Kramer JH, Weiner M, Miller BL, Seeley WW. 2010. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer's disease. Brain 133: 1352–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. 2012. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 73: 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham MP. 2012. Network centrality in the human functional connectome. Cereb Cortex 22: 1862–1875. [DOI] [PubMed] [Google Scholar]