In this study, Kim et al. show that flux of Cdc20 through kinetochores accelerates mitotic exit by promoting its dephosphorylation by kinetochore-localized protein phosphatase 1, which allows Cdc20 to activate the APC/C. The microtubule attachment status of kinetochores therefore optimizes mitotic duration by controlling the balance between opposing Cdc20 fates.
Keywords: cell division, kinetochore, spindle assembly checkpoint, APC/C, Cdc20, protein phosphatase 1
Abstract
Mitotic duration is determined by activation of the anaphase-promoting complex/cyclosome (APC/C) bound to its coactivator, Cdc20. Kinetochores, the microtubule-interacting machines on chromosomes, restrain mitotic exit when not attached to spindle microtubules by generating a Cdc20-containing complex that inhibits the APC/C. Here, we show that flux of Cdc20 through kinetochores also accelerates mitotic exit by promoting its dephosphorylation by kinetochore-localized protein phosphatase 1, which allows Cdc20 to activate the APC/C. Both APC/C activation and inhibition depend on Cdc20 fluxing through the same binding site at kinetochores. The microtubule attachment status of kinetochores therefore optimizes mitotic duration by controlling the balance between opposing Cdc20 fates.
Genome stability requires that cells spend sufficient time in mitosis to align and segregate their chromosomes with high accuracy. Mitotic duration is determined by the lag between activation of Cdk1–cyclin B and full activation of the anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase. The APC/C ubiquitinates and targets for degradation securin and cyclin B, thereby triggering anaphase onset and mitotic exit (Barford 2011). During mitosis, APC/C activity requires the essential cofactor Cdc20. Phosphorylation of APC/C subunits by Cdk1 promotes APC/C activation by enhancing the affinity of Cdc20 for the APC/C (Fujimitsu et al. 2016; Qiao et al. 2016; Zhang et al. 2016). However, Cdk phosphorylation of Cdc20 also negatively regulates its ability to bind to and activate the APC/C (Kramer et al. 2000; Yudkovsky et al. 2000; Labit et al. 2012; Hein and Nilsson 2016). Thus, Cdc20 dephosphorylation during mitosis has the potential to be a control point that regulates the timing of APC/C activation and therefore the duration of the mitotic phase of the cell cycle.
Cdc20 is also central to the spindle assembly checkpoint, a mechanism that promotes accurate chromosome segregation by delaying anaphase onset until all of the kinetochores, the microtubule-binding machines of mitotic chromosomes (Cheeseman 2014; Musacchio and Desai 2017), have established attachments to spindle microtubules. At unattached kinetochores, the spindle checkpoint pathway—comprised of the Bub1/Bub3 complex, the Mad1/Mad2 complex, Mad3 (known as BubR1 in vertebrates and Drosophila), and the kinases Mps1 and Plk1—generates a diffusible complex containing Cdc20 that inhibits the APC/C (Lara-Gonzalez et al. 2012; Musacchio 2015).
Cdc20 exhibits kinetochore localization that is conserved in fungi, Drosophila, and vertebrates (Kallio et al. 2002; Li et al. 2010; Yang et al. 2015). Recruitment of Cdc20 depends on the kinetochore-localized components Bub1 and BubR1 (Li et al. 2010; Di Fiore et al. 2015), which share a common Cdc20 interaction motif known as the ABBA motif or Phe box (Diaz-Martinez et al. 2015; Di Fiore et al. 2015). However, the contribution of Cdc20 kinetochore localization to its roles in activating and/or inhibiting the APC/C has remained unclear.
In prior work, it was found that the kinetochore-localized Bub1/Bub3 complex, which is central to spindle checkpoint signaling, paradoxically promotes anaphase onset via a mechanism that can be uncoupled from the checkpoint (Kim et al. 2015; Yang et al. 2015). Here, we show that Cdc20 rapidly fluxes through kinetochores in a Bub1 ABBA motif-dependent manner. This localized flux facilitates APC/C activation by promoting removal of inhibitory Cdk phosphorylation on Cdc20 by kinetochore-localized protein phosphatase 1 (PP1). Notably, flux of Cdc20 through the same binding site is also required for checkpoint-mediated APC/C inhibition when kinetochores are unattached. These results indicate that kinetochores optimize mitotic duration by channeling Cdc20 into opposing fates, with the balance between them controlled by microtubule attachment.
Results and Discussion
To define the role of kinetochore-localized Cdc20 in controlling mitotic duration, we used the first mitotic division of the Caenorhabditis elegans embryo as a model system. As in other species, CDC-20 (also known as FZY-1) (Kitagawa et al. 2002) localized to the kinetochores of C. elegans holocentric chromosomes between prometaphase and early anaphase (Fig. 1A; Supplemental Movies S1, 2). In contrast, no kinetochore localization was observed for an in situ tagged essential APC/C subunit (Apc1MAT-2) (Supplemental Fig. S1A), indicating that kinetochore-localized CDC-20 is not bound to the APC/C. Depletion of the kinetochore component BUB-1 prevented CDC-20 kinetochore localization (Fig. 1B), consistent with prior work in mammalian cells (Di Fiore et al. 2015; Jia et al. 2016). Fluorescence recovery after photobleaching (FRAP) experiments revealed that the population of CDC-20 at kinetochores is highly dynamic, with a half-life of 1.2 sec (Fig. 1C); in contrast, neither BUB-1 nor the outer kinetochore scaffold protein KNL-1, to which the BUB-1/BUB-3 complex binds, exhibited rapid turnover (Fig. 1C; data not shown). Fast Cdc20 turnover at kinetochores is also observed in human cells (Kallio et al. 2002), indicating that rapid flux of Cdc20 through kinetochores is widely conserved.
Figure 1.
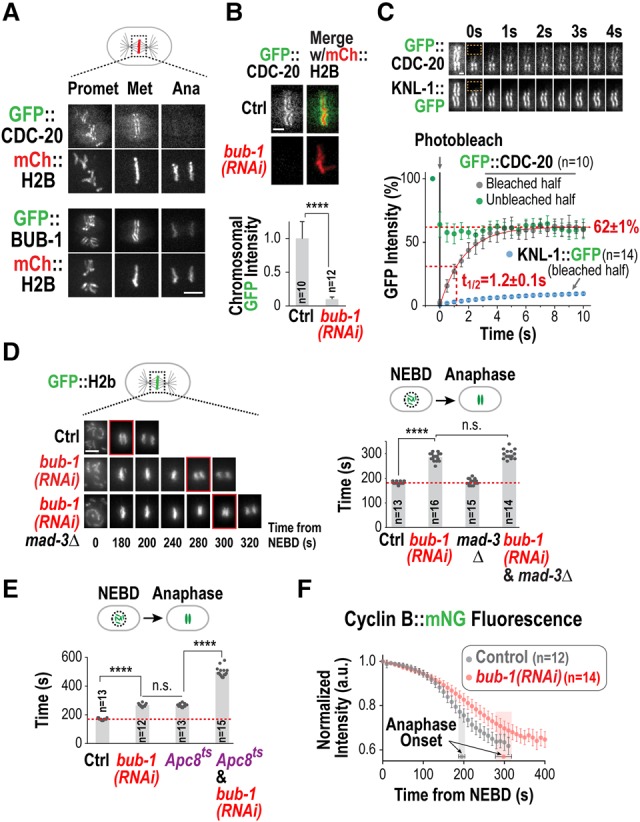
BUB-1 promotes APC/C activation and is required to recruit a dynamic CDC-20 pool to the kinetochore. (A) Images from a time-lapse sequence of the indicated GFP fusions in embryos also coexpressing mCherry(mCh)::H2b. Bar, 5 µm. (B) Metaphase images (top) and quantification (bottom) of GFP::CDC-20 chromosomal fluorescence for the indicated conditions. Error bars are the 95% confidence interval. (****) P < 0.0001 (Mann-Whitney test). Bar, 2 µm. (C) FRAP analysis. (Top) Metaphase images of GFP::CDC-20-expressing or KNL-1::GFP-expressing embryos. Time 0 indicates the first bleached frame. (Bottom) Quantification of GFP fluorescence. Error bars are the 95% confidence interval. Bar, 1 µm. (D) Images from time-lapse sequences (left) and quantification of the nuclear envelope breakdown (NEBD)–anaphase onset interval (right) for the indicated conditions. Time below panels is in seconds relative to NEBD. P-values are from Mann-Whitney test. Bar, 5 µm. (E) Quantification of the NEBD–anaphase onset interval for the indicated conditions. Imaging was performed at 22°C, at which the Apc8MAT-3 temperature-sensitive mutant is viable. (****) P < 0.0001 (Mann-Whitney test). (F) Plot of normalized Cyclin B::mNeongreen (mNG) fluorescence for the indicated conditions. Error bars are the 95% confidence interval; anaphase onset times are indicated. See also Supplemental Figure S1.
BUB-1 is required for flux of the APC/C activator CDC-20 through kinetochores, and prior work in C. elegans and Saccharomyces cerevisiae has shown that the kinetochore-localized Bub1/Bub3 complex promotes anaphase onset (Kim et al. 2015; Yang et al. 2015), suggesting a potential link between these two phenomena. In the one-cell C. elegans embryo, BUB-1 depletion increased the interval between nuclear envelope breakdown (NEBD) and anaphase onset by ∼50% (Fig. 1D); this increase was unaffected by removal of the checkpoint component MAD-3 (Fig. 1D) and was also observed in a bub-3 deletion (Kim et al. 2015). Two pieces of evidence indicate that BUB-1 depletion delayed anaphase onset by compromising APC/C activation. First, BUB-1 depletion exhibited a genetic interaction with a temperature-sensitive mutation in the essential APC/C subunit Apc8MAT-3 [mat-3(or344); referred to as Apc8ts] (Rappleye et al. 2002). BUB-1 depletion and the Apc8ts mutation each delayed anaphase onset by ∼50%; by comparison, when the two perturbations were combined, anaphase onset was delayed by ∼200% (Fig. 1E; Supplemental Movie S3). Second, BUB-1 depletion slowed degradation of the APC/C substrate Cyclin B (CYB-1) (Fig. 1F; Supplemental Fig. S1B,C), whereas depletion of the checkpoint component MAD-2 had no effect (Supplemental Fig. S1D), indicating that mitotic duration is not controlled by the spindle checkpoint during this division.
To determine whether the role of BUB-1 in promoting APC/C activation was related to CDC-20 kinetochore recruitment, we specifically altered the ability of BUB-1 to bind CDC-20. C. elegans BUB-1 has a potential CDC-20-interacting ABBA motif (Fig. 2A; Supplemental Fig. S2A), and a yeast two-hybrid assay confirmed that this motif mediates interaction with CDC-20 (Fig. 2B). Using a targeted RNAi-resistant transgene integration system (Moyle et al. 2014), we replaced endogenous BUB-1 with either wild-type or ABBA mutant BUB-1 at endogenous expression levels (Fig. 2C). The BUB-1 ABBA mutant significantly reduced CDC-20 kinetochore localization and delayed anaphase onset to an extent similar to BUB-1 depletion (Fig. 2D,E; Supplemental Movie S4). Moreover, the delay in anaphase onset cause by the BUB-1 ABBA mutant was significantly enhanced in the presence of the Apc8ts mutant (Supplemental Fig. S2B; Supplemental Movie S5). These results indicate that CDC-20 recruitment to kinetochores by the BUB-1 ABBA motif promotes anaphase onset.
Figure 2.
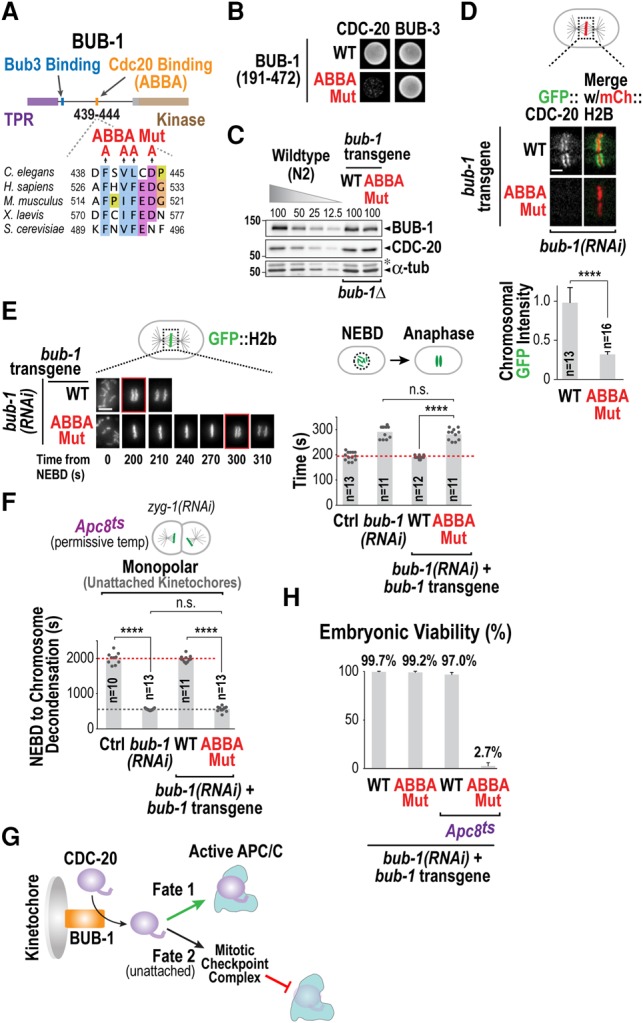
The ABBA motif of BUB-1 recruits CDC-20 to both promote APC/C activation and generate a checkpoint signal at unattached kinetochores. (A) Schematic of BUB-1 and its conserved ABBA motif. Alanine mutations introduced in the ABBA motif are indicated. (B) Yeast two-hybrid analysis of a BUB-1 fragment that interacts with both CDC-20 and BUB-3. (C) Immunoblots of transgene-encoded BUB-1 (wild-type [WT] or ABBA mutant [Mut]) and endogenous CDC-20 in the absence of endogenous BUB-1. A dilution curve of wild-type (N2) worms is on the same blot for comparison. α-Tubulin served as a loading control. The asterisk denotes residual CDC-20 signal after reblotting. (D) Metaphase images (top) and quantification (bottom) of GFP::CDC-20 chromosomal fluorescence for the indicated conditions. Error bars are the 95% confidence interval. (****) P < 0.0001 (Mann-Whitney test). Bar, 2 µm. (E) Images from time-lapse sequences (left) and quantification of the NEBD–anaphase onset interval (right) of embryos expressing GFP::H2b for the indicated conditions. Time below panels is seconds relative to NEBD. (****) P < 0.0001 (Mann-Whitney test). Bar, 5 µm. (F) Quantification of checkpoint signaling in two-cell embryos in the presence of unattached kinetochores for the indicated conditions. Mitotic duration is the NEBD–chromosome decondensation interval. (****) P < 0.0001 (Mann-Whitney test). (G) Schematic summarizing two opposing fates of CDC-20 that is fluxing through kinetochores via interaction with the ABBA motif of BUB-1. (H) Embryo viability analysis for the indicated conditions. At least 14 worms and 2330 embryos were scored per condition. Error bars represent the standard deviation. See also Supplemental Figure S2.
We next determined whether perturbing CDC-20 kinetochore localization affected spindle checkpoint activation at unattached kinetochores. In C. elegans embryos, spindle checkpoint activity is analyzed by triggering the assembly of monopolar spindles with unattached kinetochores at the two-cell stage, which increases mitotic duration by ∼60% (Essex et al. 2009). Notably, at the permissive temperature in the Apc8ts mutant, the same perturbation increases mitotic duration by ∼500%, significantly improving the sensitivity of checkpoint signaling analysis (Supplemental Fig. S2C–F; Supplemental Movie S6). Using this sensitive assay, we found that the BUB-1 ABBA mutant failed to activate the checkpoint in the presence of unattached kinetochores (Fig. 2F; Supplemental Movie S7) even though MAD-1 enrichment at unattached kinetochores was unaffected (Supplemental Fig. S2G). A requirement for the BUB-1 ABBA motif in spindle checkpoint signaling has also been reported in human cells (Di Fiore et al. 2015). Thus, CDC-20 recruited by the ABBA motif of BUB-1 to kinetochores has the ability to both promote anaphase onset and, when kinetochores are persistently unattached, delay anaphase onset (Fig. 2G).
While the BUB-1 ABBA mutant rescued the lethality of a bub-1 deletion (Supplemental Fig. S2H), it was synthetic-lethal in combination with the mild reduction in APC/C function caused by Apc8ts. At 22°C, Apc8ts and the BUB-1 ABBA mutant on their own supported viability of >97% of embryos; in contrast, only 3% of embryos generated by the double mutant were viable (Fig. 2H). Notably, the synthetic lethality observed in the double mutant was not associated with loss of checkpoint signaling in the BUB-1 ABBA mutant, as it was not observed when the mad-3 deletion, which is checkpoint signaling-defective, was combined with Apc8ts (Supplemental Fig. S2I). Thus, recruitment of CDC-20 by the ABBA motif is required for kinetochore-localized BUB-1 to accelerate APC/C activation, and this acceleration becomes essential for embryo viability when APC/C activity is mildly compromised.
Next, we focused on addressing how CDC-20 fluxing through the kinetochore is altered to accelerate APC/C activation. It was shown previously in vitro that the ability of the APC/C coactivators Cdc20 and Cdh1 to bind to and activate the APC/C is inhibited by Cdk1/2-dependent phosphorylation (Kramer et al. 2000; Yudkovsky et al. 2000; Labit et al. 2012). Cdk targeted phosphorylation sites in the Cdc20 N terminus are in the vicinity of the conserved C-box that directly contacts the APC/C subunit Apc8 (Chang et al. 2015; Zhang et al. 2016). Phosphorylation of human Cdc20 by Plk1/Bub1 on Ser92 was also shown to inhibit the recruitment of E2 ubiquitin-conjugating enzymes that promote ubiquitin chain elongation on APC/C substrates (Craney et al. 2016; Jia et al. 2016). However, the function of Cdc20 phosphorylation in a cellular context is currently unclear.
C. elegans CDC-20 possesses three putative Cdk sites in its N terminus, which are evolutionarily conserved (note that Ser92 is not conserved outside of vertebrates) (Fig. 3A). To assess whether phosphorylation of these sites controls APC/C activation in vivo, we developed a targeted RNAi-resistant transgene insertion system (Supplemental Fig. S3A–D) to replace endogenous CDC-20 with either wild-type CDC-20 or a triple-alanine (3A) mutant. Strikingly, mutation of the three Cdk sites resulted in embryos racing through mitosis and entering anaphase prior to full chromosome congression (Fig. 3B; Supplemental Fig. S3E; Supplemental Movie S8). The accelerated mitosis phenotype of CDC-20 3A was due to premature APC/C activation, as Cyclin B degradation started concomitantly with NEBD (Fig. 3C). Conversely, a phosphomimetic triple-aspartate (3D) mutant (Supplemental Fig. S3F) resulted in an embryo permeability defect, osmotic sensitivity, and lethality (Supplemental Fig. S3G,H)—phenotypes associated with compromised APC/C function (Rappleye et al. 2002). We confirmed that one or more of the mutated sites are phosphorylated in vivo through phosphorylation-dependent gel mobility shifts and phos tag gels (Fig. 3D–F). Mutation of individual sites indicated that Thr32, which is closest to the C-box and is likely phosphorylated in vivo based on phos tag gel analysis, is the major site whose mutation to alanine is responsible for accelerated APC/C activation (Supplemental Fig. S4A–E). This result is consistent with phosphorylation reducing the affinity of the C-box interaction with the APC/C. A prior study in human cells suggested that Cdc20 N-terminal phosphorylation is mediated by Cyclin A–Cdk2 and promotes mitotic entry (Hein and Nilsson 2016); however, the CDC-20 3A mutant did not alter interphase duration, and depletion of the single C. elegans Cyclin A (CYA-1) did not accelerate mitosis (Supplemental Fig. S4F,G).
Figure 3.
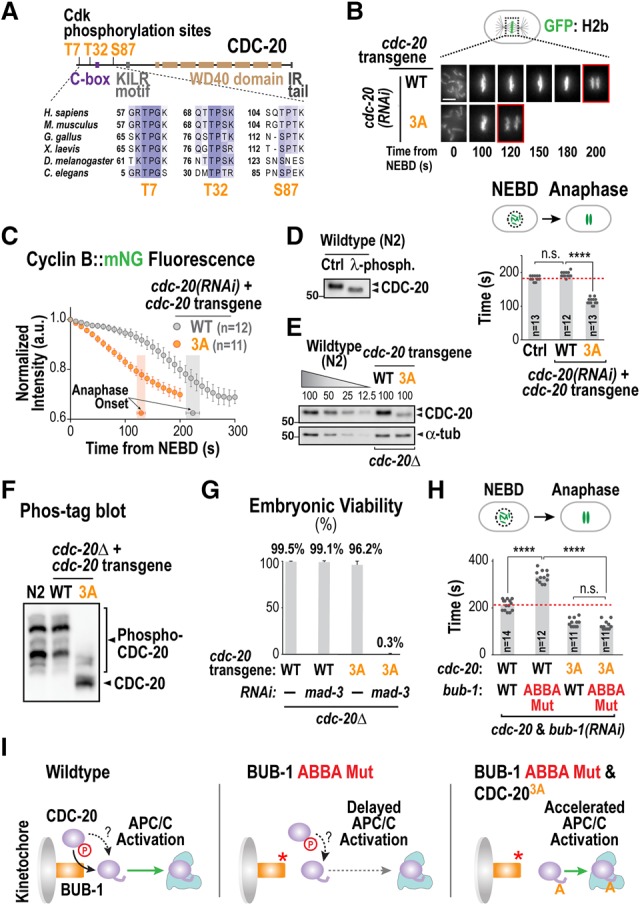
Cdk phosphorylation of CDC-20 restrains APC/C activation. (A) Schematic of CDC-20 structure and alignment highlighting conserved Cdk sites in the N terminus. (B) Images from time-lapse sequences (top) and quantification of the NEBD–anaphase onset interval (bottom) of embryos expressing GFP::H2b for the indicated conditions. Time below panels is in seconds relative to NEBD. (****) P < 0.0001 (Mann-Whitney test). Bar, 5 µm. (C) Plot of normalized Cyclin B::mNeongreen (mNG) fluorescence as in Figure 1C. (D) Immunoblot of endogenous CDC-20 with and without phosphatase treatment. (E) Immunoblot analysis of transgene-encoded CDC-20 (wild type [WT] or 3A) in the absence of endogenous CDC-20. α-Tubulin served as a loading control. Note that the 3A mutant is expressed at a lower level than wild-type CDC-20 but nonetheless accelerates APC/C activation and anaphase onset. (F) Analysis of CDC-20 phospho-isoforms by immunoblotting of a manganese phos tag gel. (G) Embryo viability analysis for the indicated conditions. At least 14 worms and 1300 embryos were scored per condition. (H) Quantification of the NEBD–anaphase onset interval of embryos expressing GFP::H2b for the indicated conditions. (****) P < 0.0001 (Mann-Whitney test). (I) Schematic summarizing the effect on APC/C activation of disrupting kinetochore localization or N-terminal Cdk phosphorylation of CDC-20. See also Supplemental Figures S3 and S4.
The CDC-20 3A and T32A mutants, while accelerating normal mitosis, retained the ability to significantly delay mitosis in the presence of unattached kinetochores (Supplemental Fig. S4H). This result suggests that the CDC-20 3A and T32A mutants can bind to the BUB-1 ABBA motif and are incorporated into the inhibitory complex generated by the spindle checkpoint at unattached kinetochores. In addition, while CDC-20 3A and T32A supported embryonic viability, they were synthetic-lethal/sick with depletion of the checkpoint component MAD-3 (Fig. 3G; data not shown), suggesting that, in the absence of inhibitory Cdk phosphorylation on CDC-20, mitotic duration is too short for establishment of correct kinetochore–microtubule attachments and requires time provided by the checkpoint for organismal survival.
The above observations suggest that CDC-20 is inhibited by Cdk phosphorylation and is dynamically concentrated at kinetochores by the BUB-1 ABBA motif to promote its dephosphorylation (Fig. 3I, left panel). A prediction of this model is that preventing CDC-20 phosphorylation should bypass the role of its kinetochore localization in promoting APC/C activation. In agreement with this prediction, CDC-20 3A or T32A eliminated the delay caused by BUB-1 depletion/the ABBA mutant and made the NEBD–anaphase onset interval equivalent to that observed when only CDC-20 3A or T32A are present (Fig. 3H,I; Supplemental Fig. S4I,J).
We next focused on identifying the phosphatase that reverses inhibitory CDC-20 phosphorylation at the kinetochore. In human cells, PP1 and the B56 isoform of PP2A localize to kinetochores (Trinkle-Mulcahy et al. 2003; Foley et al. 2011). To visualize these phosphatases, we engineered in situ tagged GFP fusions of the two essential C. elegans PP1 catalytic (PP1c) subunits (GSP-1 and GSP2) and the two functionally redundant B56 orthologs (PPTR-1 and PPTR-2). The two PP1c orthologs localized to kinetochores with distinct temporal dynamics (Supplemental Fig. S5A). In contrast, no significant kinetochore signal was observed for the functional tagged B56 orthologs (Supplemental Fig. S5B,C).
To test whether PP1c acts on CDC-20 at kinetochores to reverse inhibitory Cdk phosphorylation, we used a PP1c-binding mutant of KNL-1 (Fig. 4A; Liu et al. 2010; Espeut et al. 2012), which significantly impaired kinetochore recruitment of both PP1c catalytic subunits (Supplemental Fig. S5D). The PP1c-binding site on KNL-1 is adjacent to a region with so-called “MELT” (Met–Glu–Leu–Thr) repeats that constitute phospho-dependent docking sites for BUB-1/BUB-3 (Fig. 4A; Primorac et al. 2013). Consistent with the idea that PP1c restricts KNL-1 “MELT” phosphorylation to limit BUB-1/BUB-3 kinetochore recruitment (London et al. 2012), significantly higher levels of BUB-1 and CDC-20 were observed at kinetochores in the KNL-1 PP1c-binding mutant (Fig. 4A). Notably, even though BUB-1 and CDC-20 were hyperrecruited to kinetochores, perturbing PP1c kinetochore localization delayed anaphase onset to an extent similar to preventing CDC-20 kinetochore localization by mutating the ABBA motif in BUB-1 (Fig. 4B); this delay was largely checkpoint-independent (Fig. 4B; Espeut et al. 2012). This result suggests that KNL-1-associated PP1c dephosphorylates CDC-20 as it is fluxing through kinetochores. Consistent with this, the CDC-20 3A and T32A phosphorylation mutants abolished the anaphase onset delay caused by the KNL-1 PP1c-binding mutant (Fig. 4C; Supplemental Fig. S5F). In addition, simultaneously perturbing PP1c and CDC-20 kinetochore localization using the KNL-1 PP1c-binding mutant and the BUB-1 ABBA mutant, respectively, (Fig. 4D), or generating a mutant form of KNL-1 that cannot recruit either PP1c or BUB-1/BUB-3 to kinetochores (Supplemental Fig. S5E) did not result in additive delays. Mitotic duration in the double CDC-20/KNL-1 mutants was slightly longer than in the CDC-20 3A or T32A mutants alone (Fig. 4C; Supplemental Fig. S5F) potentially because hyperrecruitment of CDC-20 to kinetochores due to perturbation of PP1c kinetochore localization may reduce its availability for APC/C activation. Taken together, these results suggest that PP1c bound to KNL-1 dephosphorylates CDC-20 dynamically recruited to kinetochores by the BUB-1 ABBA motif, thereby accelerating APC/C activation.
Figure 4.
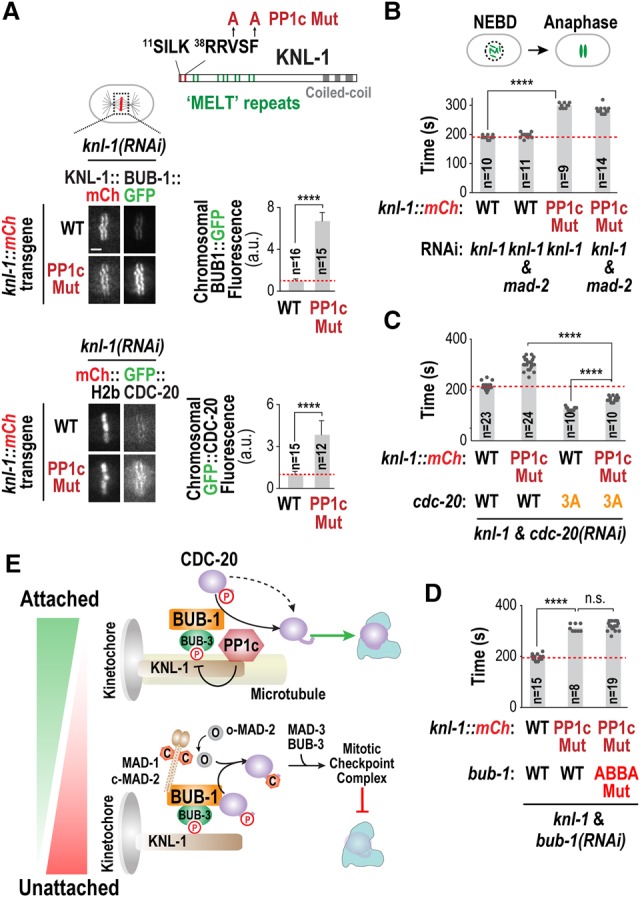
Kinetochore-localized PP1c is the likely enzyme dephosphorylating CDC-20 fluxing through the kinetochore. (A, top) Schematic of KNL-1 primary structure; the PP1-binding site (SILK–RRVSF), the “MELT” repeats that recruit the BUB-1/BUB-3 complex when phosphorylated, and the mutations engineered in KNL-1 to disrupt PP1c binding (PP1c Mut) are depicted. (Bottom) Images (left) and quantification (right) of chromosomal fluorescence for BUB-1::GFP and GFP::CDC-20 in the indicated conditions. (****) P < 0.0001 (Mann-Whitney test). Bar, 2 µm. (B–D) Quantification of the NEBD–anaphase onset interval of embryos expressing GFP::H2b for the indicated conditions. (****) P < 0.0001 (Mann-Whitney test). (E) Model summarizing the two opposing fates of CDC-20 fluxing through the kinetochore. Microtubule attachment shifts the balance by removing the MAD-1/MAD-2 checkpoint complex and potentially also by promoting PP1c kinetochore recruitment. See also Supplemental Figure S5.
The findings described here highlight a duality in the fate of CDC-20 that is fluxing through a specific binding site on the BUB-1/BUB-3 complex at kinetochores (Fig. 4E). When kinetochores are unattached, checkpoint kinases recruit the MAD-1/MAD-2 complex to kinetochores and catalyze conformational conversion of soluble MAD-2 into a form that binds CDC-20 to become part of the checkpoint complex that inhibits the APC/C (Fig. 4E). The ABBA motif of BUB-1 is required to deliver CDC-20 in this checkpoint signaling reaction, as its mutation abolishes checkpoint signaling without affecting MAD-1/MAD-2 recruitment. Unexpectedly, the pool of CDC-20 dynamically fluxing through the BUB-1 ABBA motif can also be dephosphorylated by kinetochore-localized PP1c to promote APC/C activation. We note that activation of CDC-20 by kinetochores accelerates but is not essential for APC/C activation either because CDC-20 is dephosphorylated in the cytoplasm or because phosphorylation kinetically slows but does not prevent APC/C activation. Thus, kinetochores act as kinetic accelerators that reverse the brake placed on APC/C activation by Cdk phosphorylation of CDC-20. As the checkpoint signaling reaction is sensitive to microtubule attachment, the choice between the two fates of CDC-20 will be attachment-regulated (Fig. 4E). Microtubule attachments remove MAD-1/MAD-2 but leave behind a substantial pool of BUB-1/BUB-3 (Jablonski et al. 1998; Kim et al. 2015); consequently, the CDC-20 cycling through the ABBA motif of BUB-1 at attached kinetochores is more likely to be dephosphorylated and promote APC/C activation. The localization profiles of kinetochore-localized phosphatases are also complex and potentially also microtubule attachment-regulated (Liu et al. 2010), which may further alter the balance of the two fates (Fig. 4E). By anchoring two opposing fates for CDC-20 at the same binding site on kinetochores and making the balance between them attachment-regulated, we speculate that chromosomes optimize mitotic duration in a “goldilocks” zone that ensures their accurate segregation while minimizing total time spent in a vulnerable cell cycle state when transcription, translation, and secretion are globally attenuated.
Materials and methods
C. elegans strains
C. elegans strains used in this study (Supplemental Tables S1, S2) were maintained at 20°C. A transposon-based strategy (Frokjaer-Jensen et al. 2008) was used to insert transgenes at a single locus in the genome. The details on strain construction are described in the Supplemental Material.
RNAi
For RNAi, dsRNAs (Supplemental Table S3) were injected into L4 worms and incubated for 40–48 h at 20°C before imaging. For double RNAi, dsRNAs were mixed in equal ratios, with a final concentration of >0.7 mg/mL for each dsRNA.
Imaging
Time-lapse imaging was performed on either a deconvolution microscope (DeltaVision Elite, Applied Precision) equipped with a CCD camera (pco.edge 5.5 sCMOS, PCO) and a 60× 1.42 NA plan apo N objective (Olympus) or an Andor Revolution XD confocal system (Andor Technology) with a spinning-disk confocal scanner unit (CSU-10, Yokogawa) mounted on an inverted microscope (Nikon, TE2000-E). Specific imaging conditions are detailed in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Matilde Galli and David Morgan for the Apc1MAT-2::GFP strain, the Caenorhabditis Genetics Center for providing mutant strains, and members of the Desai and Oegema laboratories for helpful discussions. This work was supported by a National Institutes of Health grant (GM074215) to A.D. P.L.-G. was supported by a Pew Latin American fellowship. F.M. was supported by the Deutsche Forschungsgemeinschaft (ME 4713/1-1). A.D. and K.O. received salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.302067.117.
References
- Barford D. 2011. Structure, function and mechanism of the anaphase promoting complex (APC/C). Q Rev Biophys 44: 153–190. [DOI] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. 2015. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM. 2014. The kinetochore. Cold Spring Harb Perspect Biol 6: a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craney A, Kelly A, Jia L, Fedrigo I, Yu H, Rape M. 2016. Control of APC/C-dependent ubiquitin chain elongation by reversible phosphorylation. Proc Natl Acad Sci 113: 1540–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Martinez LA, Tian W, Li B, Warrington R, Jia L, Brautigam CA, Luo X, Yu H. 2015. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J Biol Chem 290: 2431–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Davey NE, Hagting A, Izawa D, Mansfeld J, Gibson TJ, Pines J. 2015. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell 32: 358–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol 196: 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A, Dammermann A, Lewellyn L, Oegema K, Desai A. 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol Biol Cell 20: 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Maldonado M, Kapoor TM. 2011. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol 13: 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K, Grimaldi M, Yamano H. 2016. Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science 352: 1121–1124. [DOI] [PubMed] [Google Scholar]
- Hein JB, Nilsson J. 2016. Interphase APC/C-Cdc20 inhibition by cyclin A2–Cdk2 ensures efficient mitotic entry. Nat Commun 7: 10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. 1998. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107: 386–396. [DOI] [PubMed] [Google Scholar]
- Jia L, Li B, Yu H. 2016. The Bub1-Plk1 kinase complex promotes spindle checkpoint signalling through Cdc20 phosphorylation. Nat Commun 7: 10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. 2002. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol 158: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Moyle MW, Lara-Gonzalez P, De Groot C, Oegema K, Desai A. 2015. Kinetochore-localized BUB-1/BUB-3 complex promotes anaphase onset in C. elegans. J Cell Biol 209: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Law E, Tang L, Rose AM. 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr Biol 12: 2118–2123. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell 11: 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H, Fujimitsu K, Bayin NS, Takaki T, Gannon J, Yamano H. 2012. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J 31: 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. 2012. The spindle assembly checkpoint. Curr Biol 22: R966–R980. [DOI] [PubMed] [Google Scholar]
- Li D, Morley G, Whitaker M, Huang JY. 2010. Recruitment of Cdc20 to the kinetochore requires BubR1 but not Mad2 in Drosophila melanogaster. Mol Cell Biol 30: 3384–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol 188: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, Biggins S. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol 22: 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. 2014. A Bub1-Mad1 interaction targets the Mad1–Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol 204: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol 25: R1002–R1018. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Desai A. 2017. A molecular view of kinetochore assembly and function. Biology (Basel) 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, Musacchio A. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife 2: e01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao R, Weissmann F, Yamaguchi M, Brown NG, VanderLinden R, Imre R, Jarvis MA, Brunner MR, Davidson IF, Litos G, et al. 2016. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci 113: E2570–E2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Tagawa A, Lyczak R, Bowerman B, Aroian RV. 2002. The anaphase-promoting complex and separin are required for embryonic anterior-posterior axis formation. Dev Cell 2: 195–206. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andrews PD, Wickramasinghe S, Sleeman J, Prescott A, Lam YW, Lyon C, Swedlow JR, Lamond AI. 2003. Time-lapse imaging reveals dynamic relocalization of PP1γ throughout the mammalian cell cycle. Mol Biol Cell 14: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Tsuchiya D, Lacefield S. 2015. Bub3 promotes Cdc20-dependent activation of the APC/C in S. cerevisiae. J Cell Biol 209: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky Y, Shteinberg M, Listovsky T, Brandeis M, Hershko A. 2000. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem Biophys Res Commun 271: 299–304. [DOI] [PubMed] [Google Scholar]
- Zhang S, Chang L, Alfieri C, Zhang Z, Yang J, Maslen S, Skehel M, Barford D. 2016. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 533: 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


