In this study, Peter et al. present crystal structures of the 4EHP-binding regions of GIGYF1 and GIGYF2 in complex with 4EHP, which reveal the molecular basis for the selectivity of the GIGYF1/2 proteins for 4EHP. Their findings provide structural insights into the assembly of 4EHP–GIGYF1/2 repressor complexes and reveal that GIGYF1/2 proteins are required for repressive activity.
Keywords: eIF4E, translational regulation, translational repression
Abstract
The eIF4E homologous protein (4EHP) is thought to repress translation by competing with eIF4E for binding to the 5′ cap structure of specific mRNAs to which it is recruited through interactions with various proteins, including the GRB10-interacting GYF (glycine–tyrosine–phenylalanine domain) proteins 1 and 2 (GIGYF1/2). Despite its similarity to eIF4E, 4EHP does not interact with eIF4G and therefore fails to initiate translation. In contrast to eIF4G, GIGYF1/2 bind selectively to 4EHP but not eIF4E. Here, we present crystal structures of the 4EHP-binding regions of GIGYF1 and GIGYF2 in complex with 4EHP, which reveal the molecular basis for the selectivity of the GIGYF1/2 proteins for 4EHP. Complementation assays in a GIGYF1/2-null cell line using structure-based mutants indicate that 4EHP requires interactions with GIGYF1/2 to down-regulate target mRNA expression. Our studies provide structural insights into the assembly of 4EHP–GIGYF1/2 repressor complexes and reveal that rather than merely facilitating 4EHP recruitment to transcripts, GIGYF1/2 proteins are required for repressive activity.
The initiation of cap-dependent translation involves a series of sequential steps that start with the assembly of the eIF4F on the mRNA 5′ cap structure (Jackson et al. 2010). The eIF4F complex consists of the cap-binding protein eIF4E, the RNA helicase eIF4A, and the scaffold protein eIF4G, which bridges the interaction between the other two subunits in the complex. eIF4G also interacts with eIF3 and mediates the recruitment of the preinitiation complex (PIC; comprising a 40S ribosomal subunit and associated factors) to the mRNA to initiate translation (Jackson et al. 2010).
The assembly of the eIF4F complex is regulated by multiple mechanisms. One major mechanism involves a broad class of eIF4E-binding proteins (4E-BPs) that compete with eIF4G for binding to eIF4E, thereby inhibiting translation initiation (Mader et al. 1995; Marcotrigiano et al. 1999). eIF4G and the 4E-BPs share a conserved, canonical (C) 4E-binding motif with the sequence YX4LΦ (where Y, X, L, and Φ represent Tyr, any amino acid, Leu, and a hydrophobic residue, respectively), which binds to the dorsal surface of eIF4E opposite to the cap-binding pocket (Matsuo et al. 1997; Marcotrigiano et al. 1999; Gross et al. 2003). Both eIF4G and the 4E-BPs also contain variable noncanonical (NC) 4E-binding motifs that bind to an eIF4E hydrophobic lateral surface, increasing the affinity of the interaction (Kinkelin et al. 2012; Paku et al. 2012; Lukhele et al. 2013; Igreja et al. 2014; Peter et al. 2015a,b; Sekiyama et al. 2015; Grüner et al. 2016). Because eIF4G and 4E-BPs bind to the same surfaces on eIF4E, their binding is mutually exclusive, resulting in translation activation and inhibition, respectively.
An alternative mechanism that inhibits the recruitment of the eIF4F complex involves the recognition of the mRNA 5′ cap by another member of the eIF4E family, the 4E homologous protein (4EHP; also known as eIF4E2) (Rom et al. 1998; Joshi et al. 2004). Despite its sequence and structural similarity to eIF4E (Supplemental Fig. S1A; Rosettani et al. 2007), 4EHP does not interact with eIF4G and thus fails to initiate translation (Rom et al. 1998; Joshi et al. 2004; Hernandez et al. 2005).
4EHP is recruited to specific mRNAs by RNA-binding proteins and thus acts as a sequence-specific rather than a general translational repressor. For example, Drosophila melanogaster 4EHP is specifically recruited to and represses translation of caudal and hunchback mRNAs through interactions with the RNA-binding proteins Bicoid (Bcd) and Brain tumor (Brat), respectively (Cho et al. 2005, 2006). Mammalian 4EHP has been implicated in post-transcriptional mRNA regulation through its interaction with the nucleocytoplasmic shuttling protein 4E-T (eIF4E transporter), which is a component of P bodies (Kubacka et al. 2013). In mouse oocytes, the homeobox protein Prep1 recruits 4EHP to inhibit the translation of Hoxb4 mRNA (Villaescusa et al. 2009). 4EHP also forms a translational repressor complex with GIGYF2 (GRB10-interacting GYF [glycine–tyrosine–phenylalanine domain] protein 2 [GYF2]), a protein involved in the insulin signaling pathway (Giovannone et al. 2009; Morita et al. 2012). This repressor complex is recruited to specific mRNAs by the zinc finger protein ZNF598 (Morita et al. 2012). Alternatively, the 4EHP–GYF2 complex is recruited to mRNAs containing AU-rich elements (AREs) in their 3′ untranslated regions (UTRs) by tristetraprolin (TTP) (Tao and Gao 2015; Fu et al. 2016). Thus, through its association with diverse binding partners, 4EHP regulates the translation of mRNAs involved in a broad range of biological process, and disruption of its expression results in perinatal lethality in mice (Morita et al. 2012).
Current models suggest that 4EHP-binding proteins (4EHP-BPs) interact with 4EHP through a canonical 4EHP-binding motif with the sequence YXYX4LΦ that is present in GYF1/2 proteins (Fig. 1A; Supplemental Fig. S1B). Although this motif consists of a canonical 4E-binding motif extended by only two N-terminal residues (YX) (Cho et al. 2005; Morita et al. 2012), GYF2 does not bind to eIF4E in vivo (Morita et al. 2012). Conversely, eIF4G, which contains a canonical motif, binds to eIF4E but not 4EHP (Rom et al. 1998; Joshi et al. 2004; Hernandez et al. 2005). In contrast, some 4E-BPs, such as Homo sapiens 4E-BP1–3 and 4E-T, which lack the additional YX residues, interact with both eIF4E and 4EHP (Rom et al. 1998; Rosettani et al. 2007; Kubacka et al. 2013). This suggests that the canonical motif is unlikely to be the sole specificity determinant for 4EHP or eIF4E and that the structural basis for this molecular discrimination is unknown. Additionally, structural insights into 4EHP complexes are limited to a complex with the 4E-BP1 canonical motif, which binds to 4EHP in vitro but not in vivo (Rom et al. 1998; Rosettani et al. 2007).
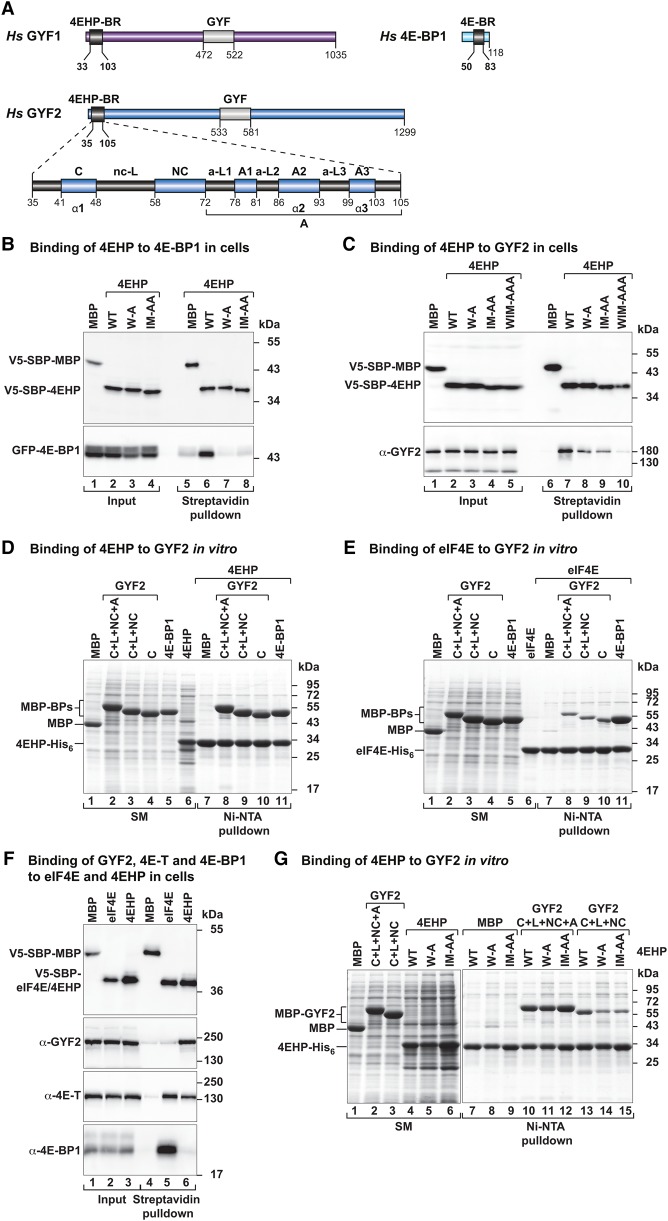
Figure 1.
GYF1/2 proteins use canonical, noncanonical, and auxiliary sequences to bind to 4EHP. (A) GYF1/2 proteins contain a central GYF domain and an N-terminal 4EHP-binding region (4EHP-BR). The 4EHP-BR includes canonical, noncanonical, and auxiliary motifs (A1–3) connected by linker sequences (nc-L and auxiliary linkers 1–3 [a-L1–3]). The 4E-binding region (4E-BR) of 4E-BP1 contains canonical and noncanonical motifs. (B,C) Western blots showing the interaction between V5-SBP-4EHP (wild type or the indicated mutants) and GFP-4E-BP1 (full-length) or endogenous GYF2. The proteins were pulled down using streptavidin-coated beads. V5-SBP-MBP (maltose-binding protein) served as negative control. The inputs (1.5% for the V5-tagged proteins and 1% for the GFP-tagged proteins) and bound fractions (3%–5% for the V5-tagged proteins and 20% for GYF2 and GFP-4E-BP1) were analyzed by Western blotting using anti-V5, anti-GFP, and anti-GYF2 antibodies. (D,E) Ni-NTA pull-down assays showing the interactions between GYF2 fragments (C+L+NC+A, C+L+NC, and C) and 4EHP-His6 (M1–F234) (D) or eIF4E-His6 (E). 4E-BP1 and MBP served as positive and negative controls, respectively. The GYF2 and 4E-BP1 peptides contain an N-terminal MBP tag and a C-terminal GB1 tag. The starting material (SM; 1.3% for MBPs and 6% for 4EHP and purified eIF4E) and bound fractions (7%–10%) were analyzed by SDS-PAGE followed by Coomassie blue staining. (F) The interaction between V5-SBP-eIF4E or 4EHP proteins and endogenous GYF2, 4E-T, and 4E-BP1 was analyzed in HEK293T cell lysates using streptavidin pull-downs. The input (1% for 4E-BP1 and 4E-T and 1.5% for V5-SBP-tagged proteins and GYF2) and bound fractions (20% for 4E-BP1 and 4ET, 30% for GYF2, and 5% for the V5-SBP-tagged proteins) were analyzed by Western blotting using the indicated antibodies. (G) Ni-NTA pull-down assay showing the interaction between 4EHP (M1–F234, wild type, or the indicated mutants) and GYF2 fragments. MBP served as a negative control. Samples were analyzed as described in D. The starting material (2% for the MBP-tagged proteins and 4%–12% for the 4EHP proteins) and bound fractions (10%) were analyzed by SDS-PAGE followed by Coomassie blue staining.
To obtain molecular insights into the assembly of 4EHP repressor complexes, we determined the crystal structures of 4EHP in complex with the binding regions of human 4E-BP1, GYF1, and GYF2. The structures reveal that, in addition to the known canonical motifs that bind to the dorsal surface of 4EHP, 4E-BP1 and GYF1/2 also make contacts with the lateral surface of 4EHP using noncanonical motifs, indicating that lateral binding is a common feature observed in both 4EHP and eIF4E complexes. Remarkably, GYF1/2 proteins, but not 4E-BP1, contain C-terminal auxiliary sequences (A) that extend the interface, contacting 4EHP residues that are not conserved in eIF4E. Our studies reveal the molecular basis for the selectivity of GYF1/2 proteins for 4EHP over eIF4E and provide mechanistic insights into the regulation of cap-dependent translation initiation by 4EHP-repressive complexes.
Results
GYF1/2 proteins bind to the dorsal and lateral surfaces of 4EHP
Given that the canonical motif in 4E-BP1 binds to the dorsal surface of 4EHP in a way similar to how it binds to eIF4E in vitro (Tee et al. 2004; Rosettani et al. 2007), we initially investigated whether the noncanonical sequences in 4E-BP1 could also bind to the lateral surface of 4EHP, as observed in the eIF4E–4E-BP1 complex (Igreja et al. 2014; Peter et al. 2015a). We substituted residues I85 and M101 on the lateral surface of 4EHP with alanine (IM-AA mutant) (Supplemental Fig. S1A; Supplemental Table S1). These residues are structurally equivalent to eIF4E residues I63 and I79, which are required for the noncanonical motifs in 4E-BPs to bind to the lateral surface of eIF4E (Igreja et al. 2014; Peter et al. 2015a). As a control, a 4EHP mutant carrying the W95A substitution on the dorsal surface (mutant W-A [Supplemental Table S1], corresponding to the eIF4E W73A mutant) was designed to disrupt canonical motif binding.
The substitutions in either the dorsal or the lateral surface of V5-SBP-tagged 4EHP disrupted interactions with GFP-tagged 4E-BP1 in HEK293T cells (Fig. 1B, lanes 7,8). Conversely, substitutions in either the canonical (C* mutant) or noncanonical (NC* mutant) motif of 4E-BP1 abolished its interaction with 4EHP (Supplemental Fig. S2A). These results indicate that the interactions between the noncanonical 4E-BP1 sequences and the 4EHP lateral surface are also critical for complex stability.
The interaction between GYF2 and 4EHP requires a canonical 4EHP-binding motif at the N terminus of the protein (YXYX4LФ) (Fig. 1A; Supplemental Fig. S1B; Morita et al. 2012). Accordingly, alanine substitutions of the four conserved residues (Y, Y, L, and Ф) in this motif abolished full-length GYF1/2 binding to V5-SBP-4EHP in human cells (Supplemental Fig. S2B,C). However, it is not known whether GYF proteins contain noncanonical sequences. We therefore examined the effects of substitutions at the dorsal and lateral surfaces of 4EHP on interactions with GYF1/2 proteins. The substitutions at either surface reduced but did not abolish 4EHP binding to endogenous GYF2 in human cells (Fig. 1C, lanes 8,9 vs. 7). The interaction was abolished only when the substitutions on both surfaces were combined (Fig. 1C, lane 10). In contrast, the dorsal and lateral substitutions disrupted binding with endogenous GYF1 (Supplemental Fig. S2D). Thus, 4EHP uses its dorsal and lateral surfaces to interact with the GYF1/2 proteins, suggesting that these proteins also contain noncanonical motifs.
GYF1/2 proteins contain noncanonical and auxiliary 4EHP-binding sequences
The noncanonical motifs in 4E-BPs are typically located 12–30 residues C-terminal to the canonical motifs and contain hydrophobic residues (Igreja et al. 2014; Peter et al. 2015a,b). During a search for potential noncanonical motifs in GYF1/2 proteins, we identified a hydrophobic motif 12 residues C-terminal to the canonical motif containing a conserved Phe that we termed the noncanonical motif (Fig. 1A; Supplemental Fig. S1B). The following 30 residues (which we termed auxiliary sequences) are also well conserved and contain several short motifs that may potentially interact with 4EHP, as was observed previously in the D. melanogaster protein Mextli in complex with eIF4E (Peter et al. 2015b).
To more precisely define the GYF1/2 sequences that interact with 4EHP, we performed in vitro pull-down assays using recombinant proteins expressed in Escherichia coli. In particular, we tested the binding of 4EHP to GYF1/2 fragments comprising the canonical motif and the noncanonical sequences (i.e., noncanonical linker and motif; L+NC) with and without the auxiliary sequences (fragments C+L+NC and C+L+NC+A) (Supplemental Table S1) as well as a fragment comprising only the canonical motif. 4EHP expressed with a hexahistidine (His6) tag pulled down all three GYF1/2 fragments expressed with an N-terminal maltose-binding protein (MBP) tag (Fig. 1D; Supplemental Fig. S2E), indicating that the canonical motifs are sufficient for 4EHP binding in vitro. The eIF4E-binding region of human 4E-BP1 interacted with 4EHP to a similar extent (Fig. 1D; Supplemental Fig. S2E, lane 11). However, the GYF1/2 fragments bound to eIF4E much less efficiently than 4E-BP1 (Fig. 1E; Supplemental Fig. S2F), indicating that GYF1/2 proteins exhibit selectivity for 4EHP over eIF4E in vitro in the absence of cellular factors.
In contrast to the results obtained in vitro, endogenous GYF1/2 proteins interacted with 4EHP but not eIF4E in cell lysates, as reported previously (Fig. 1F; Supplemental Fig. S2G; Rom et al. 1998; Morita et al. 2012). Similar results were obtained with overexpressed GYF1/2 (Supplemental Fig. S2H,I), suggesting that although GYF1/2 proteins can bind to eIF4E in vitro, their binding affinity may be too low to compete with other 4E-BPs present in cell lysates for binding to eIF4E. As expected, the 4E-T protein interacted with both eIF4E and 4EHP (Fig. 1F; Kubacka et al. 2013). In contrast, although endogenous 4E-BP1 did not bind to 4EHP in cell lysates (Fig. 1F; Supplemental Fig. S2G; Rom et al. 1998), it did bind when it was overexpressed (Fig. 1B). Thus, in cell lysates, the selectivity of GYF1/2 proteins and 4E-BP1 for 4EHP and eIF4E, respectively, is likely to be determined by their affinities and concentrations relative to those of other competing proteins.
GYF1/2 auxiliary sequences increase affinity for 4EHP
Although the GYF1/2 fragments with and without auxiliary sequences associated with wild-type 4EHP in in vitro pull-down assays (Fig. 1G; Supplemental Fig. S2J, lanes 10,13), they were differentially affected by mutations on the 4EHP dorsal and lateral surfaces. The GYF1/2 fragments, including the auxiliary sequences, were insensitive to the mutations (Fig. 1G; Supplemental Fig. S2J, lanes 10–12). In contrast, the binding of the fragments lacking the auxiliary sequences was reduced or abolished by the mutations (Fig. 1G; Supplemental Fig. S2J, lanes 14,15). Thus, the GYF1/2 auxiliary sequences contribute to the stability of the complexes with 4EHP and compensate for the destabilizing effects of the mutations in the dorsal and lateral 4EHP surfaces.
To evaluate the thermodynamic contribution of the GYF1/2 canonical, noncanonical, and auxiliary sequences to the affinity for 4EHP, we performed isothermal titration calorimetry (ITC) experiments. GYF1/2 peptides containing only the canonical motif exhibited dissociation constants (KDs) for 4EHP in the high nanomolar range (360 nMGYF1 ± 120 nMGYF1 and 290 nMGYF2 ± 160 nMGYF2) (Supplemental Fig. S3A,B; Supplemental Table S2). Addition of the noncanonical linker and motif (C+L+NC peptides) increased the affinity for 4EHP by 20-fold to 30-fold (KDs of 12 nMGYF1 ± 2 nMGYF1 and 14 nMGYF2 ± 1 nMGYF2) (Supplemental Fig. S3C,D; Supplemental Table S2), confirming the importance of the noncanonical sequences for complex formation. Importantly, addition of the auxiliary sequences increased the affinity even further by 30-fold to 40-fold (C+L+NC+A peptides; KDs of 0.4 nMGYF1 ± 0.2 nMGYF1 and 0.3 nMGYF2 ± 0.1 nMGYF2) (Supplemental Fig. S3E,F; Supplemental Table S2) relative to that of the peptides lacking the auxiliary sequences.
Collectively, the affinity measurements indicate that the GYF1/2 auxiliary sequences contribute substantially to the affinity for 4EHP by further stabilizing the interactions mediated by the canonical motif and the noncanonical sequences.
The overall architecture of the 4EHP–GYF1/2 and 4EHP–4E-BP1 complexes
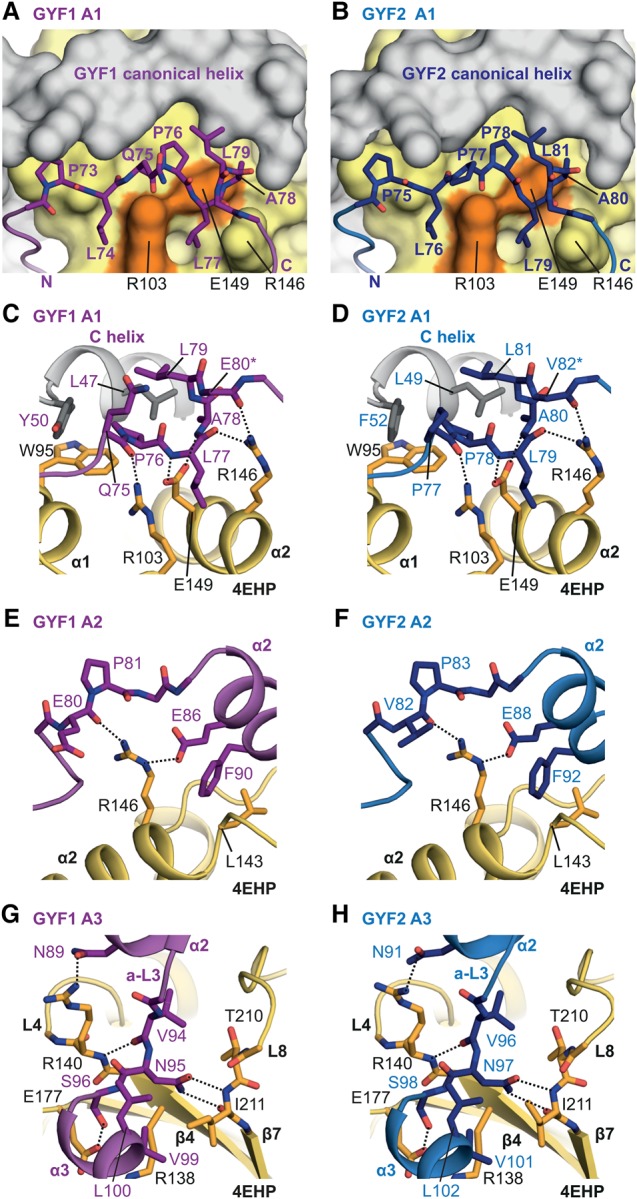
To understand the structural basis of complex formation and selectivity, we determined the crystal structures of human 4EHP bound to GYF1 and GYF2 fragments (C+L+NC+A peptides; residues 33–103GYF1 and 35–105GYF2) at 2.9 Å and 2.3 Å resolution, respectively (Fig. 2A–E; Table 1; Supplemental Fig. S4A,B). We also determined the crystal structure of 4EHP bound to a peptide comprising the 4E-BP1 canonical motif and noncanonical sequences (C+L+NC) and the corresponding peptide in GYF2 at 1.9 Å and 2.0 Å resolution, respectively (Fig. 2F–H; Table 1; Supplemental Fig. S4C,D).
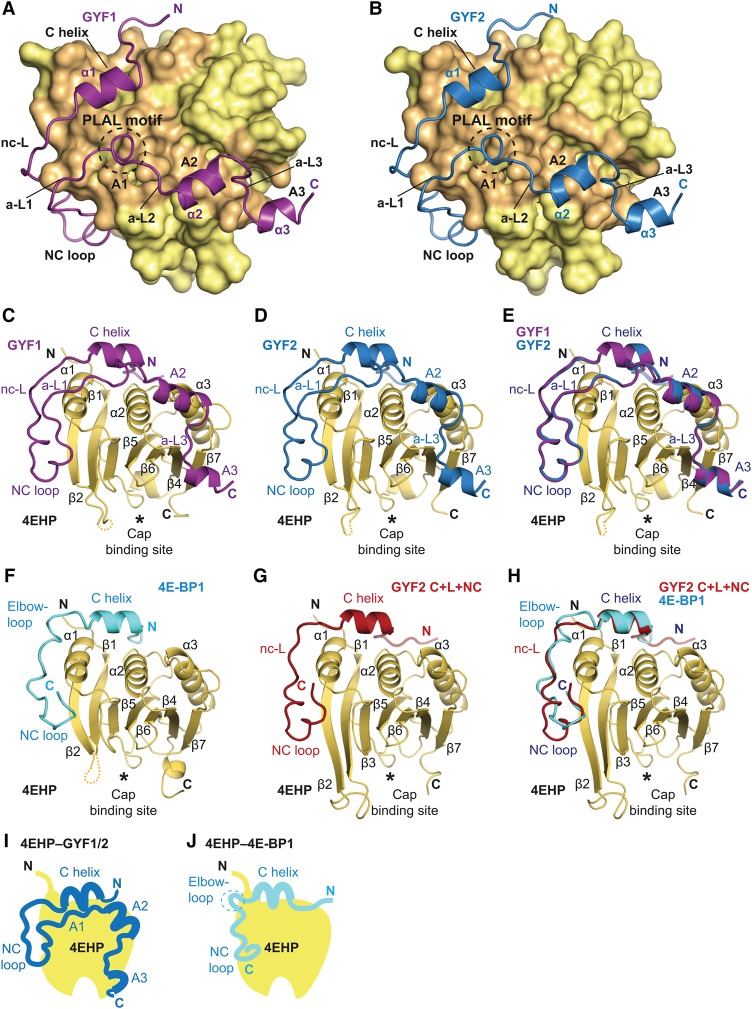
Figure 2.
Overall structures of GYF1, GYF2, and 4E-BP1 bound to 4EHP. (A,B) Overview of the structures of 4EHP bound to GYF1/2 (C+L+NC+A) fragments. The 4EHP surface is shown in yellow, and surface residues within a radius of 4 Å of the bound GYF1 or GYF2 peptides are colored in orange. The GYF1 and GYF2 peptides are colored in purple and blue, respectively. Selected secondary structure elements in the GYF1/2 peptides are indicated. The invariant PLAL motif of GYF1/2 is circled with a dashed line. (C,D) Cartoon representation of the structures of 4EHP bound to GYF1/2. Selected secondary structure elements are labeled in black for 4EHP and in color for GYF1/2. (E) Superposition of the structures of 4EHP bound to GYF1 and GYF2. For clarity, the 4EHP molecule from the 4EHP–GYF1 complex was omitted. The structures of the complexes are very similar, and overall root mean square deviations do not exceed 0.32 Å over 227 Cα atoms. (F) Structure of 4EHP bound to 4E-BP1. Selected secondary structure elements are labeled in black for 4EHP and in cyan for 4E-BP1. (G) Structure of 4EHP bound to the GYF2 C+L+NC fragment. Selected secondary structure elements are labeled in black and red for 4EHP and GYF2, respectively. (H) Superposition of the structures of 4EHP bound to the 4E-BP1 and GYF2 C+L+NC peptides. For clarity, the 4EHP molecule from the 4EHP–4E-BP1 complex was omitted. (I,J) Schematic representations of 4EHP bound to GYF1/2 and 4E-BP1.
Table 1.
Data collection and refinement statistics
The cap-binding protein 4EHP adopts a eIF4E-like fold (Fig. 2C–G; Rosettani et al. 2007), and its dorsal and lateral surfaces are very similar to those of eIF4E at the structural and sequence levels (Supplemental Figs. S1A, S4E,F). Upon 4E-BP1 or GYF1/2 binding, no major conformational changes are observed between the 4EHP structures presented in this study compared with a previously determined structure of 4EHP bound to the 4E-BP1 canonical motif (Rosettani et al. 2007).
The 4E-BP1 and GYF1/2 canonical motifs adopt a helical conformation on the dorsal surface of 4EHP, whereas the noncanonical sequences bind to the lateral surface of 4EHP using a binding mode similar to that described for 4E-BP1 in complex with eIF4E (Fig. 2C–J; Supplemental Fig. S4E,F; Peter et al. 2015a; Sekiyama et al. 2015). The binding mode and conformation for the GYF2 fragment comprising the canonical motif and the noncanonical sequences were not influenced by the auxiliary region, as the two GYF2 structures in complex with 4EHP are very similar across common elements irrespective of whether the auxiliary region was present (Supplemental Fig. S4G).
The distinguishing structural feature observed in the 4EHP–GYF1/2 complexes containing the auxiliary sequences is the unprecedented binding mode between these sequences and 4EHP (Fig. 2A–E). The auxiliary sequences extend the binding interface to 2190 Å2 compared with 1380 Å2 in their absence. The auxiliary sequences in GYF1/2 can be delineated into three short sequence motifs, termed auxiliary motifs 1–3 (A1–3); connect to the noncanonical motif by the auxiliary linker 1 (a-L1); and are interconnected by a-L2 and a-L3 (Figs. 1A, 2A,B; Supplemental Fig. S1B).
The GYF1/2 canonical helix stabilizes the interaction with 4EHP and the auxiliary sequences
The canonical helices in GYF1/2 and 4E-BP1 bind to the 4EHP dorsal surface through interactions analogous to those observed for the eIF4G and 4E-BP canonical motifs in complex with eIF4E (Fig. 3A,B; Supplemental Fig. S5A–D; Marcotrigiano et al. 1999; Gross et al. 2003; Kinkelin et al. 2012; Peter et al. 2015a,b; Grüner et al. 2016). The most conserved interactions are mediated by residues corresponding to LФ in the YXYX4LФ consensus sequence (M46 and L47GYF1, M48 and L49GYF2, and L59 and M60BP1) and the second Tyr side chain in the canonical 4EHP-binding motif (Y41GYF1 and Y43GYF2, corresponding to Y54BP1) (Fig. 3A,B; Supplemental Fig. S5A–D).
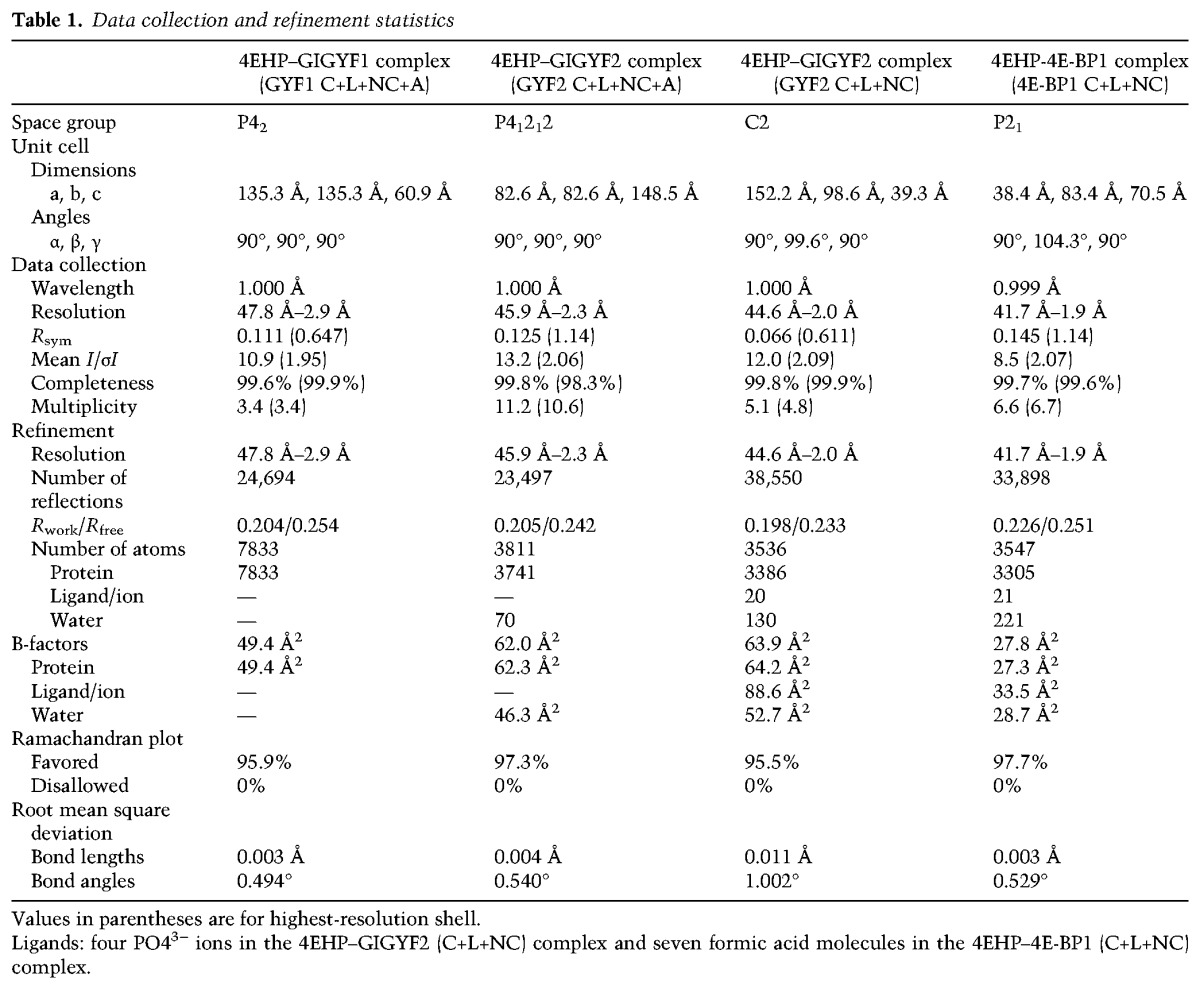
Figure 3.
The interactions between the canonical and noncanonical sequences of GYF2 and 4E-BP1 with 4EHP. (A,B) Close-up views of the interactions between the 4EHP dorsal surface and the GYF2 and 4E-BP1 canonical helices. 4E-BP1 residue R63 is colored in dark blue after its Cγ atom and is highlighted with a black dashed box. The corresponding residues in GYF2 (F52) are also highlighted by a black dashed box. (C,D) Close-up views of the interaction between the 4EHP lateral surface and the GYF2 and 4E-BP1 noncanonical linkers. (E,F) Close-up views of the interactions between the 4EHP lateral surface and the GYF2 and 4E-BP1 noncanonical loops. Selected interface residues are shown as sticks. For clarity, all residues labeled with an asterisk are shown without their side chain.
The first Tyr in the canonical 4EHP-binding motif (YXYX4LФ) was suggested to contribute to the binding specificity of 4EHP-BPs (Cho et al. 2005; Villaescusa et al. 2009). Our structural analysis does not support such a role for this Tyr (Y39GYF1 and Y41GYF2). Although its aromatic ring is in contact with P554EHP (Fig. 3A; Supplemental Fig. S5C,D), this interaction is not unique to 4EHP-BPs, as P554EHP forms a similar contact with I52BP1 at an equivalent position in the motif in the 4E-BP1 complex (IXYX4LФBP1) (Fig. 3B).
An important difference between the 4E-BP1 and GYF1/2 canonical motifs is that the latter do not possess an Arg/Lys/Gln residue at position 9 in the extended canonical motif [extended motif: YX(R/K)X2LФX2(R/K/Q)]. This residue typically contributes to the interaction with eIF4E (Marcotrigiano et al. 1999; Peter et al. 2015a,b). Instead, GYF1/2 proteins contain an aromatic residue at this position (Y50GYF1 and F52GYF2) (Fig. 3A; Supplemental Figs. S1B, S5C,D), which establishes hydrophobic contacts with W954EHP and stabilizes the GYF1/2 auxiliary motifs through intramolecular interactions with the invariant Pro residue in the PLAL motif (P76GYF1 and P78GYF2) (see below), thus rationalizing the conservation of both residues.
The noncanonical linkers contribute to structural stability in the 4EHP complexes
Following the canonical helix, the noncanonical linkers orient the peptide chains to engage with the 4EHP lateral surface (Fig. 2A–G). In the structures of 4E-BPs bound to eIF4E, these linkers adopt a specific “elbow loop” conformation (Peter et al. 2015a,b). An analogous structural feature is observed in the 4EHP–4E-BP1 complex, where P66BP1, P71BP1, and P72BP1 restrict the flexibility of the backbone conformation in the elbow loop (Fig. 3D; Supplemental Fig. S5E,F). Stabilizing hydrophobic contacts, such as between the invariant H1004EHP (corresponding to H784E) and P71BP1, ensure that the overall elbow conformation is almost identical in the eIF4E- and 4EHP-bound complexes (Fig. 3D; Supplemental Fig. S5E,F).
One important difference, however, is that the N774E side chain is in hydrogen-bonding distance to the R63BP1 guanidium group in position 9 of the extended canonical motif and to the T68BP1 carbonyl oxygen in the linker region in the eIF4E–4E-BP1 complex (Supplemental Fig. S5F; Peter et al. 2015a). In the 4EHP complex, these contacts cannot be maintained by S99, and, consequently, R63BP1 does not contribute to complex stability (Fig. 3D; Supplemental Fig. S5E,F).
Comparison of the GYF1/2 conformations in complex with 4EHP reveals that the linker region is also arranged in a single preferred conformation (Supplemental Fig. S4H) that is distinct from the elbow loop conformation observed in the complexes with 4E-BP1 (Fig. 3, C vs. D). The invariant H1004EHP plays a crucial role in anchoring the GYF1/2 linker to the 4EHP surface through van der Waals contacts with I58GYF2 (V56GYF1), while its imidazole ring is also in hydrogen-bonding distance to the D55GYF2 (E53GYF1) carbonyl oxygen (Fig. 3C; Supplemental Fig. S5G,H). The principal stabilizing hydrophobic interaction in the GYF1/2 linker region is between the invariant P59GYF2 (P57GYF1) and F974EHP, which is structurally equivalent to the interaction between P72BP1 and F974EHP (Fig. 3C,D; Supplemental Fig. S5E–H).
Noncanonical loops mediate conserved contacts at the 4EHP lateral surface
The noncanonical loops in 4E-BP1 and the GYF1/2 proteins engage a hydrophobic pocket on the lateral surface of 4EHP, which is lined by residues Y64, I85, and M101 (corresponding to eIF4E residues F47, I63, and I79, respectively) (Fig. 3E,F; Supplemental Fig. S5I–L). The conformation of the GYF1/2 noncanonical loops is stabilized by an extensive and conserved network of contacts across the 4EHP lateral surface. The 4E-BP1 and GYF1/2 noncanonical loops differ in conformation and align only at major contact points (Fig. 3, E vs. F).
Strikingly, a carbon–π interaction, through which the conserved aromatic residue Y644EHP (equivalent to F474E) makes contacts with I70GYF2 (V68GYF1) to anchor this loop at the lateral surface of 4EHP (Fig. 3E; Supplemental Fig. S5K,L), is conserved in the 4EHP–4E-BP1 complex (Y644EHP–V81BP1) as well as in all eIF4E–4E-BP and eIF4E–eIF4G complex structures (Peter et al. 2015a; Grüner et al. 2016), underscoring the role of this aromatic residue (Y644EHP, F474E) in positioning the noncanonical loops at the lateral surface of eIF4E proteins. The invariant F67GYF2 (F65GYF1) is critically positioned at the sharp turn of the peptide and stabilizes this conformation via hydrophobic contacts with Y644EHP, K834EHP, and I854EHP. The C-terminal residues in the GYF1/2 and 4E-BP1 noncanonical loops (V68 and Q70GYF1, I70 and Q72GYF2, and V81 and S83BP1) mediate similar backbone interactions with the 4EHP residues H100 and V102 (Fig. 3E,F; Supplemental Fig. S5I–L).
4EHP-specific interactions with GYF1/2 auxiliary motifs close to the cap-binding site
The a-L1 and the invariant PLAL motif at the start of the auxiliary sequences adapt to a composite surface formed between the GYF1/2 canonical helix and the 4EHP surface (Fig. 4A–D). The linker (a-L1) is fixed in position via H1004EHP, which can participate in polar contacts with the carbonyl oxygens of Q70GYF1 or D71GYF1 (Q72GYF2 or E73GYF2). The R1034EHP guanidinium group is in hydrogen-bonding distance to the Q75GYF1 (P77GYF2) carbonyl oxygen, which rationalizes the conservation of the R103 residue in 4EHP but not eIF4E. The invariant P76GYF1 (P78GYF2) in the PLAL motif coordinates an intramolecular carbon–π interaction with Y50GYF1 (F52GYF2). Importantly, key interactions of 4EHP with the a-L1 and PLAL motif are mediated by 4EHP-specific residues; e.g., E149, which fixes the orientation of the GYF1/2 chain via hydrogen bonds to L77GYF1 (L79GYF2) and A78GYF1 (A80GYF2), as well as R1464EHP, which contacts E80GYF1 (V82GYF2) and L77GYF1 (L79GYF2) (Fig. 4C,D). Therefore, the a-L1 and the PLAL motif interactions are highly specific and involve residues present only in 4EHP (R103 and E149) and thus would not be possible with eIF4E.
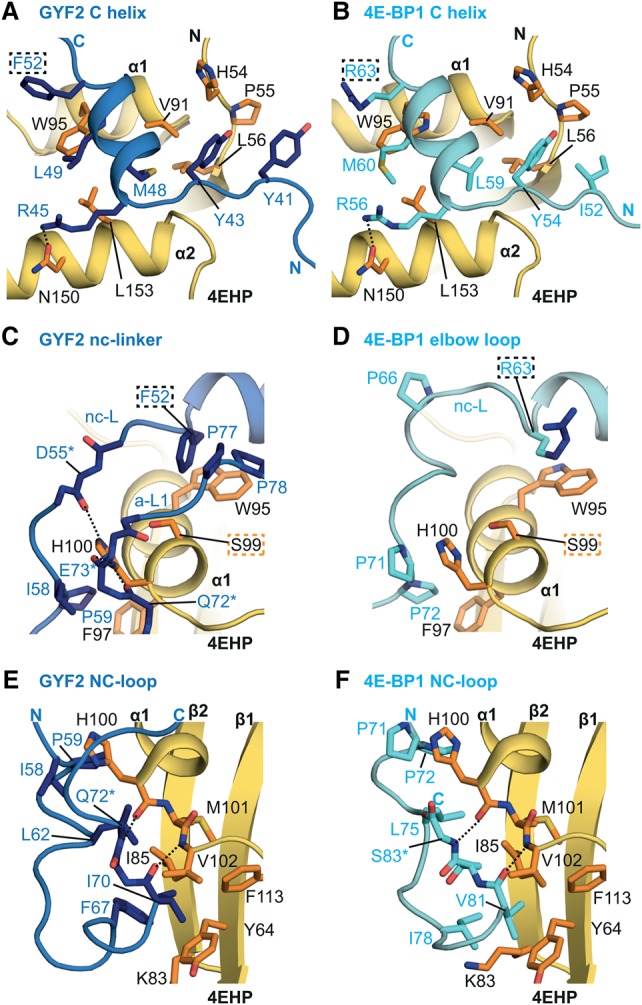
Figure 4.
Interaction between the GYF1/2 auxiliary sequences and 4EHP. (A,B) Close-up view of the arrangement of the linker a-L1 and the PLAL motif (A1) in GYF1/2 proteins at the 4EHP dorsal surface. The surface of 4EHP is shown in yellow, and the surfaces of the GYF1/2 canonical helices are shown in gray. The positions of the 4EHP unique residues R103 and E149 are highlighted in orange, and selected GYF1/2 residues are shown as either purple (GYF1) or blue (GYF2) sticks. (C–H) Close-up views of the interactions between 4EHP and the GYF1/2 auxiliary sequences (A1, A2, and A3). Selected GYF1/2 residues and 4EHP interface residues are shown as sticks. The GYF1/2 canonical helices are colored in gray.
The auxiliary sequences A2 and A3 arrange into two helical elements (auxiliary helices α2 and α3, respectively) (Fig. 2A–E), which are connected by a conserved VNS linker (linker 3 [a-L3]). Helix α2 (A2) shows some conformational heterogeneity across all complex structures (Supplemental Fig. S4H), most likely due to weak contacts at the interface (Fig. 4E–H). In contrast, a-L3 aligns well between the six complex structures. The linker VNS sequence enters a surface groove on 4EHP and interacts closely with 4EHP [e.g., through invariant S96GYF1 (S98GYF2)], which maintains a backbone hydrogen bond to R1384EHP, while its hydroxyl group is in polar contact with the 4EHP-specific residue E177 (Fig. 4G,H). As a consequence of these interactions, helix α3 (A3) is positioned in close proximity to the 4EHP cap-binding pocket (Fig. 2C,D) and is stabilized in this orientation through hydrophobic contacts between V99GYF1 (V101GYF2) and L100GYF1 (L102GYF2) and the aliphatic side chain of R138 and I211 in 4EHP (Fig. 4G,H). However, the GYF1/2 peptides containing all of the 4EHP-binding elements did not contribute to 4EHP's affinity for the m7GpppG cap analog as observed by ITC (Supplemental Table S2; Supplemental Fig. S3G,H), suggesting that additional GYF1/2 sequences may contribute to enhance 4EHP binding to capped mRNAs.
The auxiliary sequences contribute to complex stability in vivo
To assess the biological significance of the interactions mediated by the auxiliary sequences, we substituted 4EHP residues R103 and E149, which interact with the GYF1/2 PLAL motif, with leucine residues, as is observed in eIF4E (4EHP RE-LL mutant). The RE-LL substitutions strongly reduced binding to endogenous GYF2 compared with wild-type 4EHP or the 4EHP dorsal and lateral mutants in human cells (Fig. 5A, lanes 7–10). All of the mutations disrupted binding to endogenous GYF1 (Supplemental Fig. S6A, lanes 7–10). As a control, binding of 4E-BP1 was not affected by the RE-LL substitutions (Fig. 5A, lane 10), indicating that the mutations do not disrupt the 4EHP fold.
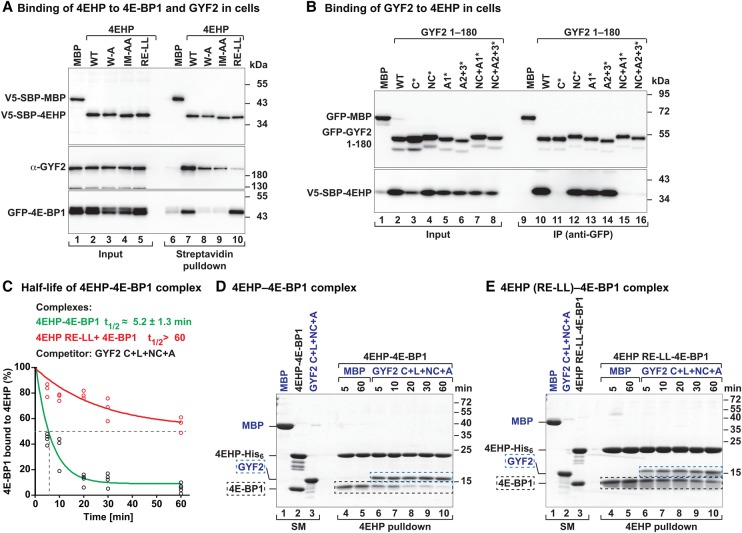
Figure 5.
The auxiliary interactions are crucial for the formation of the 4EHP–GYF complex. (A) Western blot showing the interaction of endogenous GYF2 or GFP-4E-BP1 with V5-SBP-4EHP (wild type or the indicated mutants). The proteins were pulled down using streptavidin-coated beads. The inputs (1.5% for the V5-tagged proteins and 1% for GYF2 and GFP-4E-BP1) and bound fractions (3% for the V5-tagged proteins and 20% for GYF2 and GFP-4E-BP1) were analyzed by Western blot using the indicated antibodies. (B) Interaction of V5-SBP-4EHP with GFP-GYF2 (residues 1–180; either wild type or the indicated mutants). The proteins were immunoprecipitated using anti-GFP antibodies. GFP-MBP served as negative control. The inputs (1.5% for the GFP-tagged proteins and 0.5% for V5-SBP-4EHP) and immunoprecipitates (7.5% for the GFP-tagged proteins and 30% for V5-SBP-4EHP) were analyzed by Western blot using anti-GFP and anti-V5 antibodies. (C–E) Purified 4EHP–4E-BP1 complexes containing 4EHP-His6 (wild type or the RE-LL mutant) were incubated in the presence of equimolar amounts of the GYF2 C+L+NC+A peptide C-terminally tagged with GB1 or MBP as a negative control. The proteins bound to 4EHP were pulled down using Ni-NTA beads at the indicated time points and analyzed by SDS-PAGE and Coomassie blue staining. C shows the quantification of the amount of 4E-BP1 still associated with 4EHP. n = 3. The half-life of the 4EHP–4E-BP1 complex (t1/2) in the presence of the competitor protein is represented as the mean ± SD. D and E show representative SDS-PAGE gels. The positions of the GYF2 and 4E-BP1 peptides are marked by blue and black dashed boxes, respectively. The lanes labeled SM (starting material) show the purified complexes and peptides used in the assay.
We also analyzed the impact of amino acid substitutions in the GYF1/2 proteins on complex formation. Substitutions in the GYF1/2 noncanonical (NC*) or auxiliary (A1* and A2+3*) motifs did not affect binding to overexpressed 4EHP in human cells. This is consistent with structural data showing that R103 and E149 in 4EHP interact via their side chains with the backbone atoms of the GYF1/2 auxiliary sequences (Fig. 5B; Supplemental Fig. S6B, cf. lanes 12–14 and 10). However, binding was disrupted when the mutations in the noncanonical and auxiliary motifs were combined (Fig. 5B; Supplemental S6B, lanes 15, 16 vs. 10). Together with the data showing that the mutations in the canonical motif prevent GYF1/2 binding to 4EHP (Fig. 5B; Supplemental Fig. S6B), this indicates that the canonical motif is necessary but not sufficient for 4EHP-binding in vivo.
We further assessed the relevance of R103 and E149 toward complex stability in competition assays using preassembled 4EHP–4E-BP1 complexes containing either the wild type or the RE–LL 4EHP mutant. These complexes were challenged with an equimolar amount of the GYF2 fragment comprising all 4EHP-interacting elements. The amount of 4E-BP1 that remained bound to 4EHP was determined over time (Fig. 5C–E). The GYF2 fragment displaced 50% of 4E-BP1 from the preassembled 4EHP–4E-BP1 complexes in 5 min ± 1.2 min. Under the same conditions, the GYF2 fragment displaced only 40% of 4E-BP1 bound to the 4EHP RE-LL mutant after 60 min of incubation (Fig. 5C–E). Collectively, the competition experiments together with the observation that the GYF1/2 proteins do not associate with the 4EHP RE-LL mutant in cell lysates (Fig. 5A; Supplemental Fig. S6A) indicate that the auxiliary sequences afford GYF2 a competitive advantage over 4E-BP1 for binding to 4EHP.
The canonical and auxiliary regions promote complex self-association in solution
In the asymmetric unit in the 4EHP–GYF1/2 crystals, the GYF1/2 canonical and auxiliary motifs are part of a large interface (1008 Å2) connecting two neighboring complexes (Supplemental Fig. S7A–C), suggesting dimerization. We analyzed the solution properties of these complexes by small-angle X-ray scattering (SAXS). The SAXS parameters for the 4EHP–GYF2 complex were indeed consistent with those of a dimer (Supplemental Fig. S7A,D,E). We also observed that the self-association of the complexes was concentration-dependent (Supplemental Table S3). However, the SAXS measurements of complexes lacking the GYF2 auxiliary sequences were consistent with a monomeric state (Supplemental Fig. S7F,G; Supplemental Table S3).
The putative dimer interface is stabilized by residues that are 4EHP- and GYF-specific, including R202, M161, and Q159 in 4EHP and E46 and E47 in GYF2 (Supplemental Figs. S1A,B, S7A,B). We designed mutations in GYF2 and 4EHP to disrupt dimerization (dimerization mutant, D*) (Supplemental Table S1). These mutations did not affect complex assembly, as the mutated proteins still copurified as a complex (Supplemental Fig. S7H) and retained the same affinity for their partner as the wild-type proteins (Supplemental Fig. S3I,J; Supplemental Table S2). Importantly, however, the SAXS profile of the mutated complex demonstrated the best fit to a monomeric state (Supplemental Fig. S7I; Supplemental Table S3), indicating that the mutations effectively disrupt dimerization and validate the observed interface.
A 4EHP-specific residue reduces 4E-BP1 binding
To probe the molecular basis for the binding preference of 4E-BP1 for eIF4E over 4EHP observed in cell lysates, we measured the binding affinity of the 4E-BP1 peptide for 4EHP and eIF4E using ITC. The affinity of the 4E-BP1 peptide for 4EHP was 10-fold lower compared with eIF4E (KD = 55 nM ± 14 nM and KD = 5 nM ± 2 nM, respectively) (Supplemental Fig. S3K,L; Supplemental Table S2) and 100-fold lower compared with the GYF1/2 peptides for 4EHP (Supplemental Table S2).
A possible explanation for the lower affinity of 4E-BP1 for 4EHP compared with eIF4E is that R63BP1 directly interacts with N77 in the eIF4E complex, but this residue is replaced by a Ser (S99) in 4EHP, which breaks this critical contact (Supplemental Fig. S5E,F). Interestingly, a 4EHP mutant in which Ser99 was substituted with Asn (4EHP S99N) showed a 10-fold gain in affinity for 4E-BP1 to a level comparable with eIF4E (4 nM ± 1 nM and 5 nM ± 2 nM, respectively) (Supplemental Fig. S3M; Supplemental Table S2). The S99N 4EHP mutant also bound to endogenous 4E-BP1 (Supplemental Fig. S7J, lane 8 vs. 7), although not to the same level as observed for eIF4E. Binding of GYF2 was not affected by the S99N mutation because GYF proteins have an aromatic residue at the equivalent R63BP1 position (Y50GYF1 and F52GYF2), which mediates hydrophobic contacts with 4EHP.
The affinity measurements indicate that endogenous 4E-BP1 is unlikely to effectively compete with GYF1/2 proteins for 4EHP binding under equilibrium conditions in cell lysates. This is consistent with the in vivo data that show that 4E-BP1 bound to 4EHP only when overexpressed (Fig. 1B). Furthermore, a single amino acid substitution is responsible for the different affinities of eIF4E and 4EHP for 4E-BP1.
4EHP requires interaction with GYF1/2 proteins to down-regulate mRNA expression
To assess the functional relevance of the 4EHP–GYF1/2 complex in repressing mRNA targets, we tethered λN-HA-tagged 4EHP to an R-Luc reporter containing five binding sites for the λN tag (BoxB hairpins) in the 3′ UTR. To uncouple the effects on translation from the effects on mRNA stability, the reporter contained an internal polyadenosine stretch of 95 residues followed by the 3′ end of the noncoding RNA MALAT1, which is generated through endonucleolytic cleavage by RNase P and is thus not polyadenylated (Wilusz et al. 2012). An F-Luc-GFP reporter served as a transfection control. The λN-HA-4EHP protein repressed the expression of the R-Luc-5BoxB-A95-MALAT1 reporter relative to the λN-HA peptide (Fig. 6A) without causing corresponding changes in mRNA levels (Supplemental Fig. S8A,B). The levels of a reporter lacking the BoxB hairpins were not affected (Supplemental Fig. S8C–E), indicating that 4EHP recruitment is prerequisite for repression.
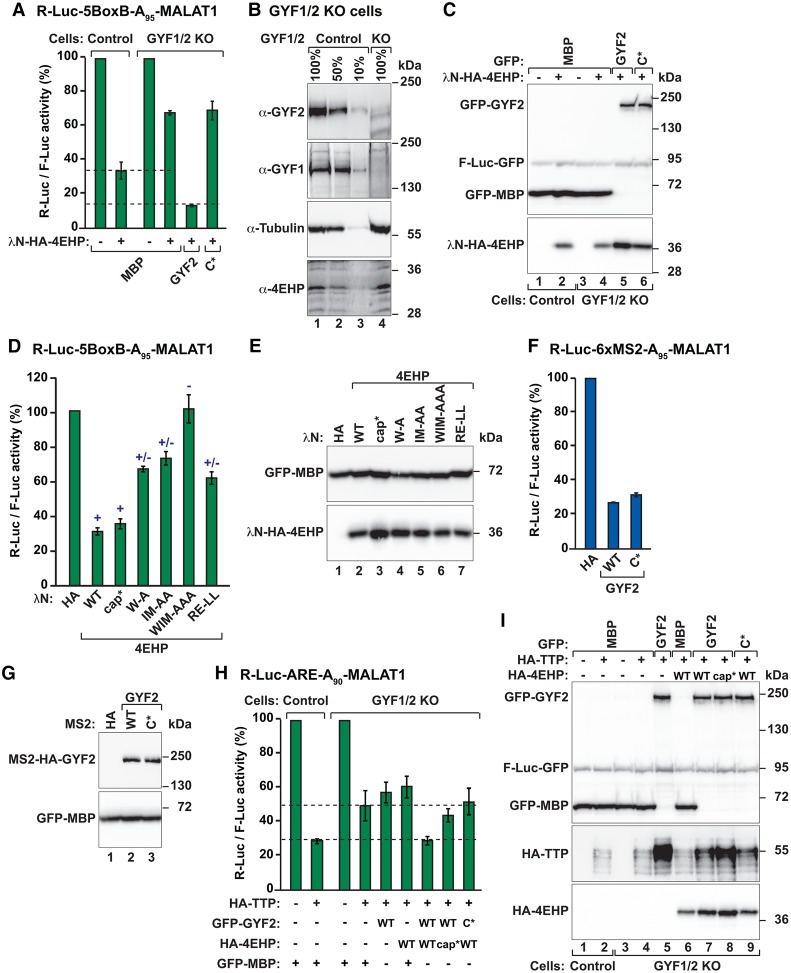
Figure 6.
4EHP requires interaction with GYF1/2 proteins to repress translation. (A) A complementation assay using the R-Luc-5BoxB-A95-MALAT1 reporter and λN-HA-4EHP (either wild type or the indicated mutants) was performed in control and GYF1/2-null HEK293T cells expressing GFP-MBP or GFP-GYF2 (wild type or canonical mutant). A plasmid expressing F-Luc-GFP served as the transfection control. R-Luc activity was normalized to that of the F-Luc transfection control and set to 100% in cells expressing the λN-HA peptide. Bars represent the mean values, and error bars represent standard deviations from three independent experiments. (B) Western blot analysis showing that full-length GYF1/2 levels were strongly reduced relative to control levels in the GYF1/2-null cell line. (C) Western blot analysis showing the expression of the λN-HA-4EHP and GFP-GYF2 proteins used in the assay shown in A. (D) Tethering assay using the R-Luc-5BoxB-A95-MALAT1 reporter and λN-HA-4EHP (wild type or mutants) in HEK293T cells. Samples were analyzed as described in A. (+) Binding to the GYF1/2 proteins; (+/−) reduced binding to the GYF1/2 proteins; (−) no binding to the GYF1/2 proteins. (E) Western blot showing the equivalent expression of the λN-HA-4EHP proteins used in the assay shown in D. (F) Tethering assay using the R-Luc-6xMS2-A95-MALAT1 reporter and MS2-HA-GYF2 (wild type or canonical mutant) in HEK293T cells. The cells were also cotransfected with GFP-MBP and F-Luc-GFP as transfection controls. R-Luc activity was normalized to that of the F-Luc transfection control and set to 100% in cells expressing MS2-HA. Samples were analyzed as described in A. (G) Western blot analysis showing the equivalent expression of the MS2-HA-GYF2 proteins. (H) Control HEK293T cells or cells depleted of GYF1/2 (KO) were transfected with the R-Luc-ARE-A90-MALAT1 reporter and plasmids expressing the indicated proteins. The F-Luc-GFP reporter served as a transfection control. R-Luc activity was normalized to that of the F-Luc transfection control and set to 100% in the absence of TTP for each cell line. (I) Western blot showing the expression of the proteins in the experiment shown in H. Note that TTP is stabilized in GYF1/2-null cells expressing GYF2. However, repression did not correlate with TTP levels but with the coexpression of wild-type GYF2 and 4EHP.
We used CRISPR–Cas9 gene editing to generate a GYF1/2-null HEK293T cell line in which the GYF1/2 levels were reduced below 10% of their control levels, whereas the expression of endogenous 4EHP was not affected (Fig. 6B, lane 4 vs. 1). In this cell line, the repression of the R-Luc mRNA reporter by tethered 4EHP was impaired (Fig. 6A) even though 4EHP was expressed at levels comparable with those observed in control cells (Fig. 6C, lane 2 vs. 4). The 4EHP-mediated repression was restored by transient expression of wild-type GFP-tagged GYF2 but not by the GYF2 canonical mutant (C*) that does not interact with 4EHP (Fig. 6A) despite comparable expression levels (Fig. 6C, lanes 5,6). Thus, 4EHP requires interactions with GYF1/2 proteins for full repressive activity.
In agreement with this conclusion, 4EHP activity in tethering assays correlated with GYF1/2 binding and was independent of cap binding. Indeed, 4EHP mutants with impaired GYF1/2 binding (+/−) exhibited reduced repressive activity, and repression was abolished by combined mutations that disrupt binding to GYF1/2 (Fig. 6D,E; Supplemental Fig. S8F,G). Unexpectedly, however, a 4EHP mutant that does not bind to the cap (cap*) (Supplemental Fig. S8H) but still binds to GYF1/2 (Supplemental Fig. S8I) repressed the expression of the reporter mRNA in a GYF1/2-dependent manner (Fig. 6D; Supplemental Fig. S9A,B). The 4EHP mutants did not repress the expression of a control mRNA lacking the BoxB-binding sites (Supplemental Fig. S9C–E).
Our results suggest that the GYF1/2 proteins confer repressive activity to the 4EHP–GYF1/2 complexes. Accordingly, GYF2 repressed mRNA reporter expression in tethering assays. GYF2 activity was independent of 4EHP because the GYF2 canonical mutant still repressed the expression of the R-Luc-6xMS2-A95-MALAT1 reporter as efficiently as wild-type GYF2 (Fig. 6F,G). GYF2 did not repress a reporter lacking the MS2-binding sites (Supplemental Fig. S9F,G).
Finally, to investigate 4EHP activity without artificial tethering, we used an R-Luc reporter that included two copies of the ARE present in the 3′ UTR of the TNF-α mRNA (R-Luc-ARE-A90-MALAT1). This reporter was repressed in control cells expressing TTP (Fig. 6H). TTP-induced repression was relieved in GYF1/2-null cells (Fig. 6H), although TTP was expressed at levels comparable with those observed in control cells (Fig. 6I, lane 4 vs. 2). In the GYF1/2-null cell line, repression was restored only when GYF2 and 4EHP were coexpressed but not when each protein was expressed individually (Fig. 6H,I). No restoration was observed when 4EHP was coexpressed with a GYF2 canonical mutant (C*). In contrast to the observations in tethering assays, a 4EHP mutant that does not bind the cap (cap*) was impaired in restoring TTP-mediated repression, although it was expressed at levels comparable with wild type (Fig. 6H,I). Collectively, these results indicate that the assembly of the 4EHP–GYF2 complex is required for full repression of target mRNA expression.
Discussion
GYF1/2 proteins are able to discriminate between 4EHP and eIF4E, but the molecular basis for this discrimination remains unknown (Morita et al. 2012). Here, we show that the 4EHP-binding region of GYF1/2 proteins comprises canonical and noncanonical motifs connected by a linker, which recognize the dorsal and lateral surfaces of 4EHP, respectively, in a manner similar to that observed for the diverse 4E-BPs and eIF4G bound to eIF4E (Kinkelin et al. 2012; Peter et al. 2015a,b; Sekiyama et al. 2015; Grüner et al. 2016). Thus, dorsal and lateral binding is conserved and widespread among eIF4E family proteins. Given this common binding interface, GYF1/2 proteins achieve their remarkable selectivity for 4EHP by virtue of unique auxiliary sequences (C-terminal to the noncanonical motif) that contact a surface on 4EHP, which is more divergent among eIF4E paralogs. In particular, the 4EHP-specific residues R103, R140, and E149 interact with GYF1/2 auxiliary sequences and are important for complex formation in vivo. These interactions stabilize the complex assembly by increasing the affinity of the interaction and may have evolved to ensure that in vivo GYF1/2 proteins efficiently compete for 4EHP binding with other potential binding partners such as 4E-BP1, which is more abundant than GYF1/2 but has lower affinity for 4EHP (Hein et al. 2015; this study).
Intriguingly, as a consequence of the 4EHP-specific interactions by the auxiliary sequences, the helix α3 (A3) of GYF1/2 is oriented in close structural proximity to the cap-binding pocket of 4EHP. However, the GYF2 peptide containing all of the 4EHP-binding elements did not increase the affinity of 4EHP for an m7GpppG cap analog (Supplemental Table S2). In the GYF1/2 proteins, there is a long stretch of Gly/Arg-rich sequence immediately following the auxiliary motifs. Given that such low-complexity Gly/Arg-rich regions often confer nonspecific RNA-binding properties to the proteins that contain them (Thandapani et al. 2013), it is tempting to speculate that the GYF1/2 proteins may play a role in stabilizing 4EHP bound to capped transcripts. Furthermore, because 4EHP has a reduced affinity for the cap structure compared with eIF4E (Rom et al. 1998; Zuberek et al. 2007), it is possible that the auxiliary sequence-mediated dimerization observed in this study may have some as yet undefined functions (e.g., increasing local concentration of repressor complexes on the mRNA), but this hypothesis needs to be tested in future studies. The affinity of 4EHP for capped mRNAs may also be stimulated by post-translational modifications such as ISG15 modification (Okumura et al. 2007) and monoubiquitinylation/diubiquitinylation (von Stechow et al. 2015), but whether these are synergistic with the GYF1/2 proteins is currently not known.
The mechanism of repressor complex assembly is likely to be divergent among the 4EHP interactors
Our study provides mechanistic insights into the assembly of 4EHP repressor complexes and raises the question of whether the binding mode is conserved among other 4EHP-BPs. However, it is important to note that although the D. melanogaster Brat protein sequence contains a canonical 4EHP-binding motif, the motif is buried within the hydrophobic core of a folded domain (the NHL domain) and therefore is unlikely to participate in interaction with 4EHP (Cho et al. 2006). Furthermore, the canonical motifs in D. melanogaster Bcd and the mammalian Prep1 proteins contain internal proline residues and are unlikely to adopt helical conformations, which are crucial for stable binding to 4EHP (Cho et al. 2006; Villaescusa et al. 2009). Thus, either the interaction with Bcd and Prep1 is indirect or the mode of binding has diverged. Only 4E-T features canonical and noncanonical motifs that bind directly to eIF4E and are likely to bind 4EHP in a similar manner (Kubacka et al. 2013; Peter et al. 2015a). Although the 4E-T orthologs do not contain motifs with similarity to the GYF1/2 auxiliary motifs, our structural data indicate that the precise sequence composition of these motifs may not be critical for interactions because the 4EHP-specific residues principally stabilize the complex via contacts with the auxiliary sequence's backbone. However, it remains to be seen whether 4E-T may indeed contain auxiliary sequences and what their mode of binding to 4EHP is.
4EHP–GYF1/2 complex assembly is required for post-transcriptional mRNA regulation
Because 4EHP has a reduced affinity for the cap structure compared with eIF4E (Rom et al. 1998; Zuberek et al. 2007), it has been proposed that it is recruited to specific mRNAs through interactions with proteins that are bound (directly or indirectly) to the mRNA, thus increasing its local concentration and competing with eIF4E in cis for binding to the 5′ cap (Cho et al. 2005, 2006; Villaescusa et al. 2009; Morita et al. 2012). According to this model, 4EHP should repress translation independently of GYF1/2 proteins when directly tethered to the 3′ UTR of an mRNA reporter. Unexpectedly, however, we observed not only that 4EHP loses its repressive activity in GYF1/2-null cells but also that its interaction with GYF1/2 proteins is in fact required for full repression. Thus, rather than merely facilitating 4EHP recruitment to an mRNA (e.g., by bridging the interaction between 4EHP and the zinc finger proteins ZNF598 and TTP), the GYF1/2 proteins act directly in the repression. In agreement with this conclusion, GYF1/2 repressed target transcripts in tethering assays independently of 4EHP binding. However, it is also evident that regulation of endogenous transcripts is dependent on the 4EHP–GYF1/2 complex assembly. Indeed, the TTP-mediated repression of an ARE-containing reporter in the GYF1/2-null cell line was restored only when 4EHP was coexpressed with GYF2 that was competent for binding to the 4EHP. Intriguingly, the cap binding by 4EHP was necessary for full repression in this context.
In summary, our studies reveal the structural basis for the assembly of a translational repressor complex consisting of 4EHP and its specific binding partners, the GYF1/2 proteins. We show that the GYF1/2 proteins directly contribute to the repressive activity of 4EHP, thus uncovering an unexpected facet of a mechanism that regulates mRNA expression.
Materials and methods
DNA constructs
The DNA constructs used in this study are described in the Supplemental Material and are listed in Supplemental Table S1. All of the constructs and mutations were confirmed by sequencing.
Protein expression and purification
All of the recombinant proteins were expressed in E. coli BL21 Star (DE3) cells (Thermo Fisher Scientific) grown in LB medium overnight at 20°C. The cells were lysed by sonication in lysis buffer containing 50 mM HEPES (pH 7.2), 200 mM (4EHP–4E-BP1) or 300 mM (4EHP–GYF1/2) NaCl, and 2 mM DTT supplemented with 5 µg/mL DNase I, 1 mg/mL lysozyme, and protease inhibitor cocktail (Roche). To purify the complexes containing 4EHP bound to GYF1, GYF2, or 4E-BP1 for crystallization and SAXS, His6-tagged 4EHP (residues A52–F234) was coexpressed with MBP-tagged GYF1 (residues K33–M103), GYF2 (residues A35–T105), or 4E-BP1 (residues T50–S83). The complexes were purified from cleared cell lysates using an amylose resin (New England Biolabs) followed by cleavage of the MBP and His6 tags with HRV3C protease overnight at 4°C. After cleavage of the tags, the complexes were separated from the MBP and His6 tags using a heparin column (5 mL of HiTrap Heparin HP, GE Healthcare) and further purified on a Superdex 75 column (GE Healthcare) in a buffer consisting of 10 mM HEPES (pH 7.2), 200 mM NaCl, and 2 mM DTT. The complexes were stored at −80°C or used directly for crystallization and SAXS. The 4EHP complexes used in the competition assays were expressed and purified as described above with the difference that the C-terminal His6 tag was not removed from the 4EHP and that the copurified 4E-BP1 peptide contained a C-terminal GB1 tag. The complexes were stored in a buffer containing 20 mM Na-phosphate (pH 7.0), 200 mM NaCl, and 5% (w/v) glycerol.
For the pull-down assays, eIF4E (full length) was expressed with a C-terminal His6 tag, purified from cleared cell lysates using a nickel column (5 mL of HisTrap HP, GE Healthcare), and further purified on a heparin column (5 mL of HiTrap Heparin HP, GE Healthcare) followed by size exclusion chromatography (Superdex 75 column, GE Healthcare) without removing the C-terminal His6 tag. The purified eIF4E-His6 was stored at −80°C in a buffer consisting of 20 mM Na-phosphate (pH 7.0) and 200 mM NaCl.
Pull-downs, competition assays, coimmunoprecipitation, and Western blotting
The in vitro pull-down and competition assays were performed as described previously (Igreja et al. 2014; Peter et al. 2015a,b). All coimmunoprecipitation and pull-down assays in HEK293T cell lysates were performed in the presence of RNase A as described previously (Peter et al. 2015a). All of the Western blots were developed using the ECL Western blotting detection system (GE Healthcare). The antibodies used in this study are listed in Supplemental Table S4. A detailed description of these assays is included in the Supplemental Material.
ITC measurements and SAXS
The ITC measurements and SAXS experiments are described in the Supplemental Material.
Crystallization, data collection, and structure determination
A detailed description of the crystallization conditions and the structure determination process is included in the Supplemental Material. All diffraction data sets were recorded on a Pilatus 6M detector at the PXII beamline of the Swiss Light Source at a temperature of 100 K. The diffraction data and refinement statistics are summarized in Table 1.
Tethering and complementation assays
A detailed description of the procedure to generate the GYF1/2-null cell line is included in the Supplemental Material. For the complementation assays, HEK293T cells (wild-type or GYF1/2-null cells) were seeded in six-well plates (0.6 × 106 cells per well) and transfected using Lipofectamine 2000 (Invitrogen). The tethering reporters have been described previously (Kuzuoğlu-Öztürk et al. 2016). The transfection mixtures contained 0.25 µg of pEGFP-N3-F-Luc transfection control reporter, 0.5 µg of pCIneo-R-Luc-5BoxB-A95-MALAT1 (or pCIneo-R-Luc-A95-MALAT1 without BoxB), and 0.3 and 0.7 µg of the plasmids expressing the λN-HA and λN-HA-tagged 4EHP proteins, respectively. Cells were also cotransfected with plasmids expressing GFP-tagged proteins (0.25 μg of GFP-MBP, 1.8 μg of GFP-GYF2 wild type, and 1.2 μg of GFP-GYF2 C* mutant).
For the 4EHP tethering assays in HEK293T cells, wild-type or mutant λN-HA-4EHP proteins (0.3 µg for wild type and the cap* mutant, 0.8 µg for W-A, 1 µg for IM-AA, 1.2 µg for WIM-AAA, and 0.4 µg for RE-LL proteins) were cotransfected with the same amounts of reporter plasmids as described for the complementation assay. In the tethering assay with the GYF2 protein, the transfection mixture contained 0.25 µg of pEGFP-N3-F-Luc transfection control reporter; 0.5 µg of pCIneo-R-Luc-6xMS2-A95-MALAT1 or pCIneo-R-Luc-A95-MALAT1; 0.3 µg and 1 µg of the plasmids expressing the MS2-HA and MS2-HA-tagged GYF2 proteins, respectively; and 0.25 µg of GFP-MBP.
For the assay with the pCIneo-R-Luc-ARE-A90-MALAT1 reporter, wild-type and GYF1/2-null HEK293T cells were transfected with 1 µg of the ARE reporter and 0.25 µg of the pEGFP-N3-F-Luc transfection control reporter in the presence or absence of plasmids expressing 50 ng of λN-HA-TTPΔNIM, 0.2 µg of GFP-MBP, 1 µg of GFP-GYF2 (wild type or canonical mutant [C*]), and 0.5 µg of λN-HA-4EHP.
Firefly and Renilla luciferase activities were measured 2 d after transfection using the dual-luciferase reporter assay system (Promega).
Accession numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5NVK (4EHP–GYF1 C+L+NC+A), 5NVL (4EHP–GYF2 C+L+NC+A), 5NVM (4EHP–GYF2 C+L+NC), and 5NVN (4EHP–4E-BP1 C+L+NC).
Supplementary Material
Acknowledgments
We are grateful to M.Y. Chung and C. Weiler for excellent technical assistance, and H. Budde for help with cloning. We thank L. Langer for help with cloning, purification of some of the 4EHP complexes, and the initial crystallization trials. We thank R. Büttner and T. Raisch for the setup of crystallization screens, and the staff at the PX beamline of the Swiss Light Source, B21 beamline at the Diamond Light Source (Didcot, U.K.), and the SWING beamline at the SOLEIL synchrotron (Saint-Aubin, France) for assistance with data collection. We also thank R. Rambo for helpful comments on the SAXS data analysis, and O. Weichenrieder for insightful comments on the manuscript. This work was supported by Biostruct-X (ID 9926) and the Max Planck Society. D.P. purified, crystallized, collected, and solved the structures of all 4EHP complexes presented in this study together with F.S. ITC measurements were performed by D.P. and F.S. S.H. performed pull-downs and competition assays. R.W. generated the GYF1/2-null cell line; performed tethering, complementation, and pull-down assays; purified peptides/proteins; and generated many of the constructs used in pull-down assays. L.W. generated several constructs for expression in human cells and conducted immunoprecipitation experiments in HEK293T cells. D.P., F.S., and E.V. performed the SAXS experiments and the structural analysis. P.B. performed immunoprecipitations and generated the ARE reporter and the TTP expression vector. C.I. coordinated the project. E.I. was the principal investigator who supervised the project. D.P., C.I., E.V., and E.I. wrote the manuscript. All authors corrected the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.299420.117.
Freely available online through the Genes & Development Open Access option.
References
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. 2005. A new paradigm for translational control: inhibition via 5′–3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121: 411–423. [DOI] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. 2006. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol 16: 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R, Olsen MT, Webb K, Bennett EJ, Lykke-Andersen J. 2016. Recruitment of the 4EHP–GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA 22: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannone B, Tsiaras WG, de la Monte S, Klysik J, Lautier C, Karashchuk G, Goldwurm S, Smith RJ. 2009. GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum Mol Genet 18: 4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750. [DOI] [PubMed] [Google Scholar]
- Gruner S, Peter D, Weber R, Wohlbold L, Chung MY, Weichenrieder O, Valkov E, Igreja C, Izaurralde E. 2016. The structures of eIF4E–eIF4G complexes reveal an extended interface to regulate translation initiation. Mol Cell 64: 467–479. [DOI] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. 2015. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163: 712–723. [DOI] [PubMed] [Google Scholar]
- Hernandez G, Altmann M, Sierra JM, Urlaub H, Diez del Corral R, Schwartz P, Rivera-Pomar R. 2005. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech Dev 122: 529–543. [DOI] [PubMed] [Google Scholar]
- Igreja C, Peter D, Weiler C, Izaurralde E. 2014. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat Commun 5: 4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Cameron A, Jagus R. 2004. Characterization of mammalian eIF4E-family members. Eur J Biochem 271: 2189–2203. [DOI] [PubMed] [Google Scholar]
- Kinkelin K, Veith K, Grunwald M, Bono F. 2012. Crystal structure of a minimal eIF4E–Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA 18: 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubacka D, Kamenska A, Broomhead H, Minshall N, Darzynkiewicz E, Standart N. 2013. Investigating the consequences of eIF4E2 (4EHP) interaction with 4E-transporter on its cellular distribution in HeLa cells. PLoS One 8: e72761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuoglu-Ozturk D, Bhandari D, Huntzinger E, Fauser M, Helms S, Izaurralde E. 2016. miRISC and the CCR4–NOT complex silence mRNA targets independently of 43S ribosomal scanning. EMBO J 35: 1186–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhele S, Bah A, Lin H, Sonenberg N, Forman-Kay JD. 2013. Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure 21: 2186–2196. [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol 15: 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol 4: 717–724. [DOI] [PubMed] [Google Scholar]
- Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC, Alain T, Fonseca BD, Karashchuk G, Bennett CF, et al. 2012. A novel 4EHP–GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol 32: 3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Zou W, Zhang DE. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev 21: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paku KS, Umenaga Y, Usui T, Fukuyo A, Mizuno A, In Y, Ishida T, Tomoo K. 2012. A conserved motif within the flexible C-terminus of the translational regulator 4E-BP is required for tight binding to the mRNA cap-binding protein eIF4E. Biochem J 441: 237–245. [DOI] [PubMed] [Google Scholar]
- Peter D, Igreja C, Weber R, Wohlbold L, Weiler C, Ebertsch L, Weichenrieder O, Izaurralde E. 2015a. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol Cell 64: 467–479. [DOI] [PubMed] [Google Scholar]
- Peter D, Weber R, Kone C, Chung MY, Ebertsch L, Truffault V, Weichenrieder O, Igreja C, Izaurralde E. 2015b. Mextli proteins use both canonical bipartite and novel tripartite binding modes to form eIF4E complexes that display differential sensitivity to 4E-BP regulation. Genes Dev 29: 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom E, Kim HC, Gingras AC, Marcotrigiano J, Favre D, Olsen H, Burley SK, Sonenberg N. 1998. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem 273: 13104–13109. [DOI] [PubMed] [Google Scholar]
- Rosettani P, Knapp S, Vismara MG, Rusconi L, Cameron AD. 2007. Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J Mol Biol 368: 691–705. [DOI] [PubMed] [Google Scholar]
- Sekiyama N, Arthanari H, Papadopoulos E, Rodriguez-Mias RA, Wagner G, Leger-Abraham M. 2015. Molecular mechanism of the dual activity of 4EGI-1: dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc Natl Acad Sci 112: E4036–E4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Gao G. 2015. Tristetraprolin recruits eukaryotic initiation factor 4E2 to repress translation of AU-rich element-containing mRNAs. Mol Cell Biol 35: 3921–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Tee JA, Blenis J. 2004. Characterizing the interaction of the mammalian eIF4E-related protein 4EHP with 4E-BP1. FEBS Lett 564: 58–62. [DOI] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S. 2013. Defining the RGG/RG motif. Mol Cell 50: 613–623. [DOI] [PubMed] [Google Scholar]
- Villaescusa JC, Buratti C, Penkov D, Mathiasen L, Planaguma J, Ferretti E, Blasi F. 2009. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS One 4: e5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stechow L, Typas D, Carreras Puigvert J, Oort L, Siddappa R, Pines A, Vrieling H, van de Water B, Mullenders LH, Danen EH. 2015. The E3 ubiquitin ligase ARIH1 protects against genotoxic stress by initiating a 4EHP-mediated mRNA translation arrest. Mol Cell Biol 35: 1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. 2012. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26: 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberek J, Kubacka D, Jablonowska A, Jemielity J, Stepinski J, Sonenberg N, Darzynkiewicz E. 2007. Weak binding affinity of human 4EHP for mRNA cap analogs. RNA 13: 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.