Wei et al. show that Spata2-deficient cells show resistance to RIPK1-dependent apoptosis and necroptosis and are partially protected against RIPK1-independent apoptosis. Spata2 deficiency also promotes M1 ubiquitination of RIPK1 to inhibit RIPK1 kinase activity.
Keywords: CYLD, necroptosis, RIPK1, Spata2
Abstract
Stimulation of cells with TNFα leads to the formation of the TNF-R1 signaling complex (TNF-RSC) to mediate downstream cellular fate decision. Activation of the TNF-RSC is modulated by different types of ubiquitination and may lead to cell death, including apoptosis and necroptosis, in both RIPK1-dependent and RIPK1-independent manners. Spata2 (spermatogenesis-associated 2) is an adaptor protein recruited into the TNF-RSC to modulate the interaction between the linear ubiquitin chain assembly complex (LUBAC) and the deubiquitinase CYLD (cylindromatosis). However, the mechanism by which Spata2 regulates the activation of RIPK1 is unclear. Here, we report that Spata2-deficient cells show resistance to RIPK1-dependent apoptosis and necroptosis and are also partially protected against RIPK1-independent apoptosis. Spata2 deficiency promotes M1 ubiquitination of RIPK1 to inhibit RIPK1 kinase activity. Furthermore, we provide biochemical evidence for the USP domain of CYLD and the PUB domain of the SPATA2 complex preferentially deubiquitinating the M1 ubiquitin chain in vitro. Spata2 deficiency also promotes the activation of MKK4 and JNK and cytokine production independently of RIPK1 kinase activity. Spata2 deficiency sensitizes mice to systemic inflammatory response syndrome (SIRS) induced by TNFα, which can be suppressed by RIPK1 inhibitor Nec-1s. Thus, Spata2 can regulate inflammatory response and cell death in both RIPK1-dependent and RIPK1-independent manners.
TNFα is an important proinflammatory cytokine critically involved in mediating innate immunity response. TNFα-induced trimerization of TNFR1 leads to the formation of a transient intracellular multiprotein complex, called the TNF-R1 signaling complex (TNF-RSC, also called complex I). The TNF-RSC orchestrates a complex pattern of modifications on key mediators of signaling by different ubiquitination linkages, including M1, K48, and K63, in a spatiotemporal-specific manner that collectively decides whether a cell (and, ultimately, an organism) may live or die (Walczak 2011; Ofengeim and Yuan 2013; de Almagro et al. 2015). The intracellular DD (death domain) motif of the trimerized TNFR1 complex recruits TRADD, an adaptor protein, and RIPK1 via homotypic interactions with their respective DD motifs. TRADD interacts with FADD, a critical adaptor for the recruitment and activation of caspase-8, to mediate apoptosis. TRADD is also involved in the recruitment of TRAF2/TRAF5, which function as redundant adaptors to recruit cIAP1/2, two important E3 ubiquitin ligases that mediate the ubiquitination of RIPK1 by K63- and K11-linked chains (Dynek et al. 2010; de Almagro et al. 2015). The ubiquitination of RIPK1 in the TNF-RSC regulates the activation of its kinase activity, which in turn activates RIPK3 and MLKL to promote necroptosis, a regulated necrotic cell death pathway (Berger et al. 2014; de Almagro et al. 2017).
A major downstream event of the TNF-RSC mediated by the TRADD/TRAF2/cIAP1 complex is the recruitment of the linear ubiquitin chain assembly complex (LUBAC) to mediate M1 (also known as Met1-linked or linear ubiquitin chain)-linked ubiquitin modification of the TNF-RSC (Haas et al. 2009; Gerlach et al. 2011; Ikeda et al. 2011). The LUBAC includes heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1), shank-associated RH domain-interacting protein (SHARPIN), and the catalytic subunit HOIL-1-interacting protein (HOIP) (Kirisako et al. 2006; Ikeda et al. 2011). RIPK1 is known to be a substrate of the LUBAC in the TNF-RSC (Gerlach et al. 2011). M1-linked ubiquitination of the TNF-RSC has been shown to be important for blocking TNFα-mediated apoptosis; however, the consequences of M1 ubiquitination of RIPK1 are unknown.
CYLD (cylindromatosis), a deubiquitinating enzyme that targets both M1- and K63-linked ubiquitin chains (Komander et al. 2009), is recruited to the TNF-RSC to negatively modulate the ubiquitination levels of RIPK1, TNFR1, NEMO, and TRADD to inhibit the NF-κB pathway and promote both apoptosis and necroptosis (Hitomi et al. 2008; Draber et al. 2015). CYLD is recruited to the TNF-RSC by its interaction with the LUBAC in association with spermatogenesis-associated 2 (SPATA2) (Kupka et al. 2016; Schlicher et al. 2016; Wagner et al. 2016). Spata2 was identified as a gene involved in mediating necroptosis from a genome-wide siRNA screen (Hitomi et al. 2008). SPATA2 is rapidly recruited into the TNF-RSC upon stimulation by TNFα together with the deubiquitinating enzyme CYLD and the LUBAC. The putative PUB domain of SPATA2 binds to the catalytic USP domain of CYLD and activates CYLD-mediated hydrolysis of ubiquitin chains (Elliott et al. 2016). On the other hand, a conserved PUB-interacting motif in SPATA2 binds with the HOIP PUB domain. Spata2-deficient cells show resistance to RIPK1-dependent apoptosis (RDA) and necroptosis (Hitomi et al. 2008; Kupka et al. 2016; Schlicher et al. 2016; Wagner et al. 2016). However, it is still not clear how SPATA2 might regulate the activation of the RIPK1 kinase.
Here, we report that Spata2 regulates the activation of RIPK1 by modulating its M1 ubiquitination. Loss of Spata2 promotes the M1 ubiquitination of RIPK1, which leads to inhibition of RIPK1 kinase activity and RDA and necroptosis. We show that the USP–CYLD/PUB–SPATA2 complex preferentially deubiquitinates the M1 ubiquitin chain in vitro. On the other hand, Spata2 deficiency also promotes the activation of MKK4 and JNK and production of proinflammatory cytokines independently of RIPK1 kinase activity, leading to hypersensitivity of Spata2−/− mice to systemic inflammatory response syndrome (SIRS), which can be inhibited by RIPK1 inhibitor Nec-1s.
Results
Spata2 promotes necroptosis
We first confirmed that knockdown of Spata2 using siRNA and shRNA protected against necroptosis of L929 cells induced by zVAD.fmk or TNFα (Supplemental Fig. S1A). Furthermore, we found that the sensitivity to necroptosis was restored upon the complementation of Spata2 in Spata2 knockdown cells (Supplemental Fig. S1B).
We next examined the expression of Spata2 in different tissues in mice and found that the expression of Spata2 was detectable in multiple tissues such as the brain, heart, and spleen but most abundantly in the lungs, intestines, and testes (Supplemental Fig. S2A). To characterize the function of Spata2 in vivo, we derived Spata2−/− mice using Spata2+/− ES2 cells generated by the Knockout Mouse Project (KOMP) Repository that had a deletion in the entire coding region for Spata2 of amino acids 2–517 (Supplemental Fig. S2B). The chimeras were crossed to C57BL/6N mice, and their heterozygous Spata2+/− offspring were intercrossed to yield homozygous Spata2−/− mice at the expected Mendelian frequency. Spata2−/− mice were backcrossed with C57BL/6 mice for 10 generations. The deletion of Spata2 was determined by genotyping of tail DNA and Western blotting to confirm knockout of Spata2 expression in Spata2−/− mice, Spata2−/− mouse embryonic fibroblasts (MEFs), and Spata2−/− bone marrow-derived macrophages (BMDMs) (Supplemental Fig. S2C,D). Spata2−/− mice are fertile and healthy with no obvious abnormalities in major organs (data not shown). Thus, Spata2 is dispensable for normal development and adult life in a controlled environment.
Since Spata2 is expressed in both MEFs and BMDMs (Supplemental Fig. S2D), we next characterized the sensitivity of primary Spata2−/− BMDMs and MEFs to necroptosis. We found that the treatment of mouse TNFα plus IDN-6556, a pan-caspase inhibitor that has been tested in human clinical trials (Linton et al. 2005), was sufficient to induce the death of primary wild-type BMDMs, whereas Spata2−/− BMDMs were highly resistant (Fig. 1A,B). Spata2−/− MEFs also showed resistance to necroptosis induced by TNFα plus IDN-6556 or zVAD.fmk (Fig. 1C,D). Since the cell death induced by mouse TNFα plus zVAD.fmk was effectively inhibited by Nec-1s (Fig. 1D), we conclude that Spata2 is involved in regulating the RIPK1-dependent cell death pathway.
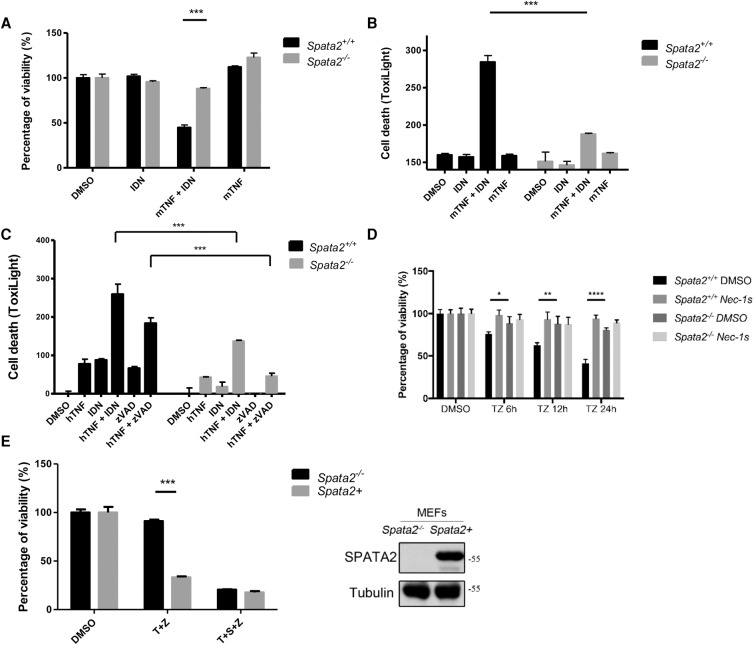
Figure 1.
SPATA2 is required for TNFα-induced necroptosis in BMDMs and MEFs. (A,B) Primary Spata2+/+ or Spata2−/− BMDMs were treated with DMSO, 1 µM IDN-6556, or 20 ng/mL mouse TNFα for 16 h. Cell viability and cell death were evaluated using CellTiterGlo and ToxiLight assay, respectively. (C) Primary Spata2+/+ or Spata2−/− MEFs were treated with DMSO, 1 µM IDN-6556, 20 µM zVAD.fmk, or 20 ng/mL human TNFα for 16 h. Cell death was measured by ToxiLight assay. (D) Immortalized Spata2+/+ or Spata2−/− MEFs were pretreated with Nec-1s for 30 min and treated with 20 µM zVAD.fmk and 20 ng/mL human TNFα for the indicated periods of time. Cell viability was measured by CellTiterGlo assay. (E) Immortalized Spata2−/− MEFs and Spata2+ MEFs were pretreated with or without 100 nM SM-164 for 30 min and then treated with 10 ng/mL human TNFα and 50 µM zVAD.fmk for 24 h. Cell viability was measured by CellTiterGlo assay. (Right) Western blotting was used to determine the levels of Spata2. Error bars represent standard error of the mean (SEM) from three technical replicates.
To address the possible clonal variation issue, we complemented the expression of Spata2 in Spata2−/− MEFs (referred to as Spata2+ MEFs below) (Supplemental Fig. S2E) and found that the expression of Spata2 in Spata2−/− MEFs restored the sensitivity to necroptosis induced by TNFα plus zVAD.fmk (Fig. 1E).
While Spata2−/− MEFs were resistant to necroptosis induced by TNFα plus zVAD.fmk, the resistance was lost upon the cotreatment of TNFα and small molecule IAP antagonist SM-164 (Fig. 1E; Lu et al. 2008). In order to confirm the effect of SM-164 on Spata2−/− MEFs, we used an xCELLigence RTCA biosensor to monitor cell viability in real time and found that the attenuation of TNFα/zVAD.fmk-induced necroptosis was abolished by SM-164 (Supplemental Fig. S3). Since SM-164 promotes the degradation of cIAP1/2 (Lu et al. 2008), this result suggests that cIAP1/2 are involved in mediating the effect of Spata2.
SPATA2 promotes the formation of complex IIb
We next characterized the changes in the biochemical hallmarks of necroptosis, including the phosphorylation of RIPK1 and MLKL. Phosphorylation of RIPK1 at S166 is a biomarker for the activation of RIPK1 (Degterev et al. 2008; Berger et al. 2014; Ofengeim et al. 2015). We developed an anti-phospho-S166 RIPK1 antibody that can be used to immunoprecipitate activated RIPK1 (Supplemental Fig. S4A,B). The ability of the p-S166 antibody to isolate complex IIb by immunoprecipitation was demonstrated using mass spectrometry analysis. From the immunocomplex in necroptotic cells pulled down by anti-p-S166 RIPK1, we identified RIPK1 as the dominant species along with other known components of complex IIb, including RIPK3, FADD, caspase-8, cFLIP, and TRADD (Supplemental Fig. S4C).
Using this anti-p-S166 RIPK1 antibody, we found that the levels of activated RIPK1 were significantly reduced in Spata2−/− MEFs and Spata2−/− immortalized BMDMs (iBMDMs) stimulated by TNFα plus zVAD.fmk (Fig. 2A,B). The binding of activated RIPK1 with caspase-8 and FADD in wild-type but not Spata2−/− MEFs was confirmed by mass spectrometry analysis (Supplemental Fig. S4D). Furthermore, the phosphorylation of MLKL at S345, a biomarker for necroptosis, was detected in Spata2+/+ MEFs but was significantly inhibited in Spata2−/− MEFs (Fig. 2C). Thus, Spata2 deficiency blocks the activation of RIPK1 as well as phosphorylation of MLKL, the downstream execution event of necroptosis. Consistent with the requirement of cIAP1/2 for the dependency of necroptosis on SPATA2, the treatment of SM-164 eliminated the involvement of SPATA2 for mediating RIPK1 activation in MEFs (Fig. 2D). Taken together, these results suggest that Spata2 is involved in TNFα-mediated necroptosis by promoting the activation of RIPK1 in a cIAP1/2-dependent manner.
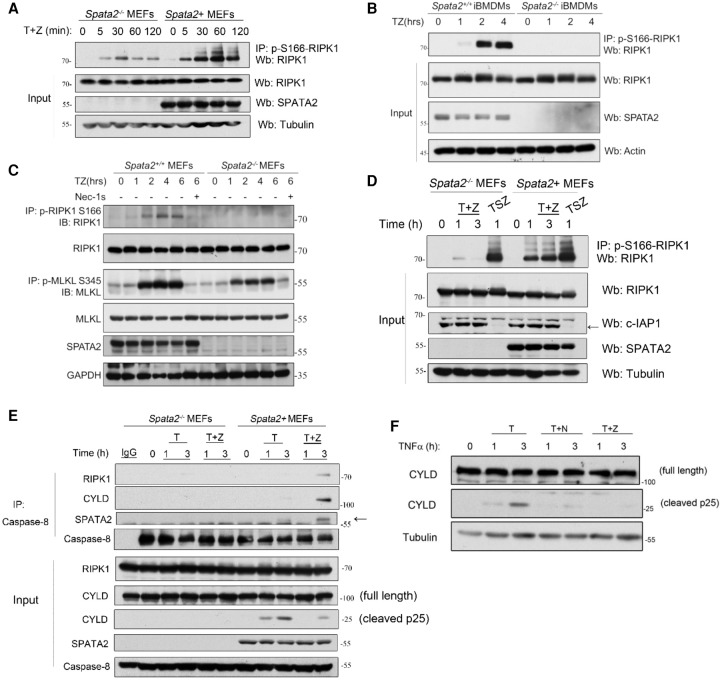
Figure 2.
SPATA2 promotes the activation of the RIPK1 kinase and the formation of complex IIb. (A) Immortalized Spata2−/− MEFs and Spata2+ MEFs were treated with 100 ng/mL human TNF and 50 µM zVAD.fmk for the indicated periods of time. The cell lysates were analyzed by immunoprecipitation using p-S166 RIPK1 antibody followed by Western blotting using the indicated antibodies. (B) Spata2+/+ iBMDMs and Spata2−/− iBMDMs were treated with 100 ng/mL human TNFα plus 50 µM zVAD.fmk for the indicated periods of time. The cell lysates were subjected to immunoprecipitation using anti-p-S166-RIPK1 antibody, and the immunocomplexes were analyzed by Western blotting. (C) Immortalized Spata2+/+ MEFs and Spata2−/− MEFs were pretreated with 20 µM Nec-1s for 30 min and then treated with 50 ng/mL human TNFα and 50 µM zVAD.fmk for the indicated periods of time. The cell lysates were analyzed by immunoprecipitation using anti-p-S166 RIPK1 or p-S345 MLKL antibody as indicated. The immunocomplexes or cell lysates were analyzed by Western blotting using the indicated antibodies. (D) Immortalized Spata2−/− MEFs and Spata2+ MEFs were pretreated with or without 100 nM SM-164 for 30 min and then treated with 100 ng/mL human TNFα for the indicated periods of time. The cell lysates were analyzed by immunoprecipitation using anti-p-S166 and analyzed by Western blotting. (E) Immortalized Spata2−/− MEFs and Spata2+ MEFs were treated with 100 ng/mL human TNFα and 20 µM zVAD.fmk for the indicated periods of time. The cell lysates were analyzed by immunoprecipitation using anti-caspase-8 followed by Western blotting using the indicated antibodies. (F) MEFs were pretreated with or without 20 µM Nec-1s for 10 min and then treated with 100 ng/mL human TNFα and 50 µM zVAD.fmk for the indicated periods of time. The cell lysates were analyzed by Western blotting using the indicated antibodies. GAPDH and Tubulin were used as loading controls.
We further characterized the downstream events in Spata2-deficient cells mediated by TNFR1 signaling. The cleavage of CYLD induced by TNFα is mediated by caspase-8 (O'Donnell et al. 2011). A 25-kDa CYLD cleavage product was detected in Spata2+ MEFs but not in Spata2−/− MEFs stimulated by TNFα, suggesting a reduction in the activation of caspase-8 under Spata2-deficient conditions (Fig. 2E; Supplemental Fig. S5A). The CYLD cleavage induced by TNFα could be inhibited by both Nec-1s and zVAD.fmk or the RIPK1 D138N knock-in kinase-dead mutation (Polykratis et al. 2014), indicating a role of RIPK1 kinase activity in the activation of caspase-8 (Fig. 2F; Supplemental Fig. S5B). The interaction of RIPK1, CYLD, and SPATA2 with caspase-8 was detectable in Spata2+ MEFs stimulated by human TNFα plus zVAD.fmk but not in Spata2−/− MEFs (Fig. 2E), suggesting that the formation of complex IIb, indicated by the interaction of RIPK1 and caspase-8, was inhibited by Spata2 deficiency. Spata2 deficiency also blocked the interaction of RIPK1, FADD, and CYLD in complex IIb in Spata2−/− MEFs (Supplemental Fig. S5C). Taken together, Spata2 is required for the formation of complex IIb. Furthermore, SPATA2 interacts with CYLD in complex IIb and is required for the recruitment of CYLD into complex IIb.
SPATA2 regulates apoptosis
The kinase activity of RIPK1 has also been known to mediate TNFα-mediated apoptosis, termed RDA, when cells are deficient for TAK1, IKKs, or cIAP1/2 (Mihaly et al. 2014; Dondelinger et al. 2015). Spata2−/− MEFs showed a partial resistance against RDA induced by the TNFα and TAK1 inhibitor (5Z)-7-oxozeaenol (5Z-7) compared with Spata2+ MEFs (Fig. 3A). The addition of Nec-1s provided further protection in both Spata2−/− and Spata2+/+ MEFs (Fig. 3B). Spata2 deficiency also protected against RDA induced by the TNFα and IKK inhibitor TPCA-1 (Fig. 3C).
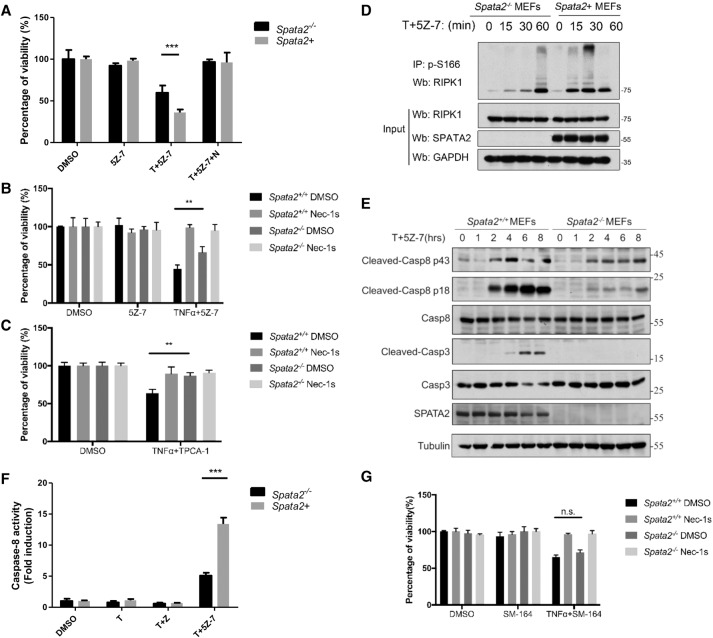
Figure 3.
SPATA2 is required for TNFα-induced apoptosis. (A,B) Immortalized Spata2+/+ MEFs and Spata2−/− MEFs were pretreated with 20 µM Nec-1s for 30 min and treated with 100 nM 5Z-7 and 50 ng/mL human TNFα for 12 h. Cell viability was measured by CellTiterGlo assay. (C) Immortalized Spata2+/+ MEFs and Spata2−/− MEFs were pretreated with 20 µM Nec-1s for 30 min and treated with 100 nM TPCA-1 and 50 ng/mL human TNFα for 12 h. Cell viability was measured by CellTiterGlo assay. (D) Immortalized Spata2−/− MEFs and Spata2+ MEFs were treated with 100 nM 5Z-7 and 100 ng/mL human TNFα for the indicated periods of time. The cell lysates were analyzed by immunoprecipitation using anti-p-S166 RIPK1. The immunocomplexes were analyzed by Western blotting. (E) Immortalized Spata2+/+ MEFs and Spata2−/− MEFs were treated with 100 nM 5Z-7 and 100 ng/mL human TNFα for the indicated periods of time. The cell lysates were analyzed by Western blotting using the indicated antibodies. (F) Immortalized Spata2−/− MEFs and Spata2+ MEFs were treated with 100 nM 5Z-7 and 100 ng/mL human TNFα for 12 h. Caspase-8 activity was measured according to the manufacturer's protocol. (G) Immortalized Spata2+/+ MEFs and Spata2−/− MEFs were pretreated with 20 µM Nec-1s and 25 nM SM-164 for 30 min and treated with 50 ng/mL human TNFα for 12 h. Cell viability was measured by CellTiterGlo assay. Error bars represent SEM from three technical replicates. Tubulin was used as a loading control.
Since the kinase activity of RIPK1 is important in RDA, we next checked the phosphorylation of RIPK1 in Spata2-deficient cells. We found that Spata2 deficiency reduced the activation of RIPK1 as indicated by the appearance of p-S166 RIPK1 (Fig. 3D). Consistent with the activation of apoptosis, the treatment of TNFα and 5Z-7 induced the cleavage and activation of caspase-8, which was partially reduced in Spata2−/− MEFs (Fig. 3E,F).
We next characterized the effect of Spata2 deficiency in RDA induced by TNFα plus SM-164. While Spata2 deficiency protected against RDA induced by TNFα plus TAK1 inhibitor or IKK1 inhibitor, Spata2−/− MEFs showed no resistance to RDA induced by TNFα plus SM-164 (Fig. 3G). Thus, cIAP1/2 are required for Spata2 to mediate both RDA and necroptosis.
We also tested the role of Spata2 in mediating apoptosis in a RIPK1-independent manner. The treatment of TNFα plus the protein synthesis inhibitor cycloheximide (CHX) induces apoptosis independently of RIPK1. We found that Spata2−/− MEFs were partially protected against TNFα plus CHX-induced apoptosis as well (Supplemental Fig. S6).
Taken together, these data suggest that Spata2 deficiency provides partial protection against RDA and RIPK1-independent apoptosis in a cIAP1/2-dependent manner.
SPATA2 regulates the TNF-RSC
As reported, SPATA2 is recruited with CYLD to the TNF-RSC upon TNFα stimulation (Supplemental Fig. S7A; Elliott et al. 2016; Schlicher et al. 2016; Wagner et al. 2016). We found that CYLD was not recruited to the TNF-RSC in Spata2−/− MEFs like it was in wild-type MEFs upon TNFα stimulation but was restored by the expression of a Spata2 expression vector (Spata2+ cells) (Fig. 4A; Supplemental Fig. S7A). While the recruitment of TRADD and HOIP, the catalytic subunit of the LUBAC, in the TNF-RSC was not altered by Spata2 deficiency, the ubiquitination modification of RIPK1 in the TNF-RSC was reduced, and the recruitment of A20, a key ubiquitin-editing enzyme for RIPK1, in the TNF-RSC was increased in Spata2−/− MEFs. The interaction of SPATA2 and CYLD was induced by TNFα in Spata2+ cells (Supplemental Fig. S7B).
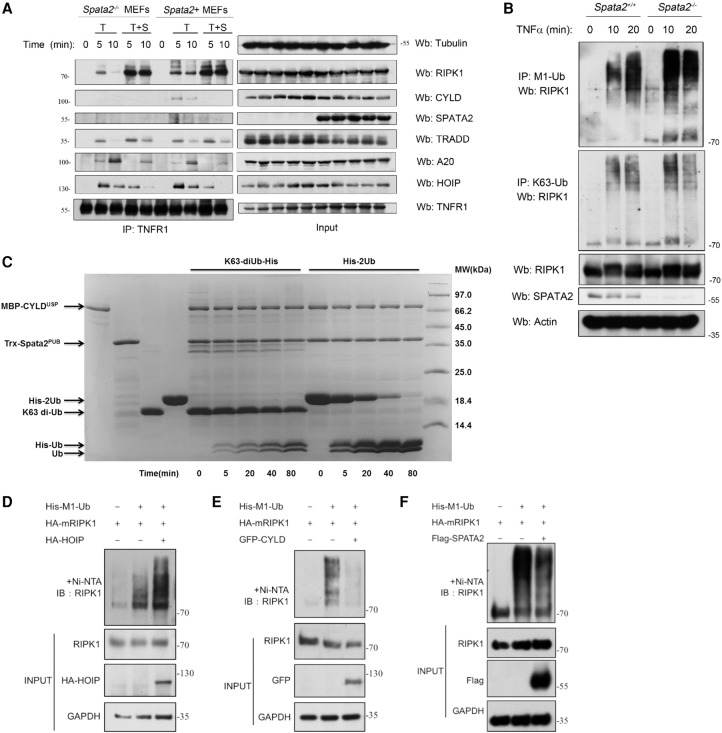
Figure 4.
SPATA2 regulates the M1 ubiquitination of RIPK1. (A) Immortalized Spata2−/− MEFs and Spata2+ MEFs were pretreated with or without 100 nM SM-164 for 20 min and then treated with 100 ng/mL human TNFα for the indicated periods of time. The cell lysates were subjected to immunoprecipitation using anti-TNFR1. The immunocomplexes were analyzed by Western blotting using the indicated antibodies. (B) Immortalized Spata2+/+ MEFs and Spata2−/− MEFs were stimulated by 500 ng/mL human TNFα for the indicated periods of time and then lysed in 6 M urea. The cell lysates were analyzed by immunoprecipitation using M1 chain-specific or K63 chain-specific ubiquitin antibody. The immunocomplexes were then analyzed by Western blotting using the indicated antibodies. (C) An in vitro deubiquitination assay was set up using purified MBP-CYLDUSP, Trx-Spata2PUB, His-2Ub, and K63-diUb. The deubiquitination process was monitored by taking samples at different time intervals and was resolved by SDS-PAGE. (D–F) 293T cells were transfected with HA-mRIPK1 and His-M1-Ub with HA-HOIP, GFP-CYLD, or Flag-Spata2 for 24 h. The cells were lysed with 8 M urea buffer and analyzed by pull-down using Ni-NTA beads. The pull-down was further analyzed by Western blotting using the indicated antibodies. GAPDH and Tubulin were used as loading controls.
Consistent with the requirement of cIAP1/2, the treatment of SM-164 eliminated the recruitment of SPATA2 and CYLD into the TNF-RSC induced by TNFα (Fig. 4A). The treatment of TNFα with SM-164 reduced the recruitment of HOIP and A20 into the TNF-RSC and significantly increased the association of RIPK1 with TNFR1. Notably, the effect of Spata2 deficiency on TNF-RSC and RIPK1 activation was lost upon the treatment of SM-164. Thus, the regulation of TNF-RSC and RIPK1 activation by SPATA2 critically depends on cIAP1/2.
M1 ubiquitination antagonizes RIPK1 kinase activation
Since Spata2 deficiency blocks the recruitment of CYLD into the TNF-RSC without affecting the recruitment of the LUBAC, we predict that M1 ubiquitination of RIPK1 mediated by the LUBAC in TNFα-stimulated cells might occur without the deubiquitinating activity of CYLD and thus leads to an unbalanced ubiquitination pattern. Next, we explored the role of Spata2 in regulating the M1 ubiquitination of RIPK1 in TNFα stimulation using an anti-M1 ubiquitin-specific or anti-K63 ubiquitin-specific chain antibody as described (Newton et al. 2008). We found that, in cells stimulated by TNFα, the M1 ubiquitinated RIPK1 species were increased with a higher molecular weight in Spata2−/− MEFs compared with Spata2+ MEFs, while K63 ubiquitinated RIPK1 showed a weak difference (Fig. 4B). As the ubiquitinated RIPK1 associated with TNFR1 stimulated by TNFα was decreased in Spata2−/− MEFs (Supplemental Fig. S7A), the increasing M1 ubiquitinated RIPK1 species may exist in a cytosolic compartment, suggesting that Spata2 may regulate the M1 ubiquitination of RIPK1, which controls its recruitment to the TNF-RSC.
To explore the mechanistic basis of this selective effect on M1 ubiquitination in Spata2-deficient conditions, we expressed and purified the recombinant USP domain of CYLD (the catalytic domain) and the PUB domain of SPATA2 from Escherichia coli and tested their activity using an in vitro deubiquitinating assay (Fig. 4C). We found that the CYLDUSP/SPATA2PUB complex preferentially hydrolyzed 2-Ubi in M1 linkage to monoubiquitin in vitro compared with K63 di-Ubi, suggesting that the CYLD/Spata2 complex has selectivity toward the M1 ubiquitin chain.
As Spata2 regulates the RIPK1 ubiquitination pattern, we next characterized the regulation of RIPK1 M1 ubiquitination by HOIP, CYLD, and SPATA2 in 293T cells. We found that overexpression of HOIP in 293T cells increased the M1 ubiquitination of RIPK1, while overexpression of SPATA2 or CYLD limited the M1 ubiquitination of RIPK1 (Fig. 4D–F). These results demonstrate that RIPK1 is an important ubiquitination/deubiquitination substrate of the HOIP–SPATA2–CYLD complex.
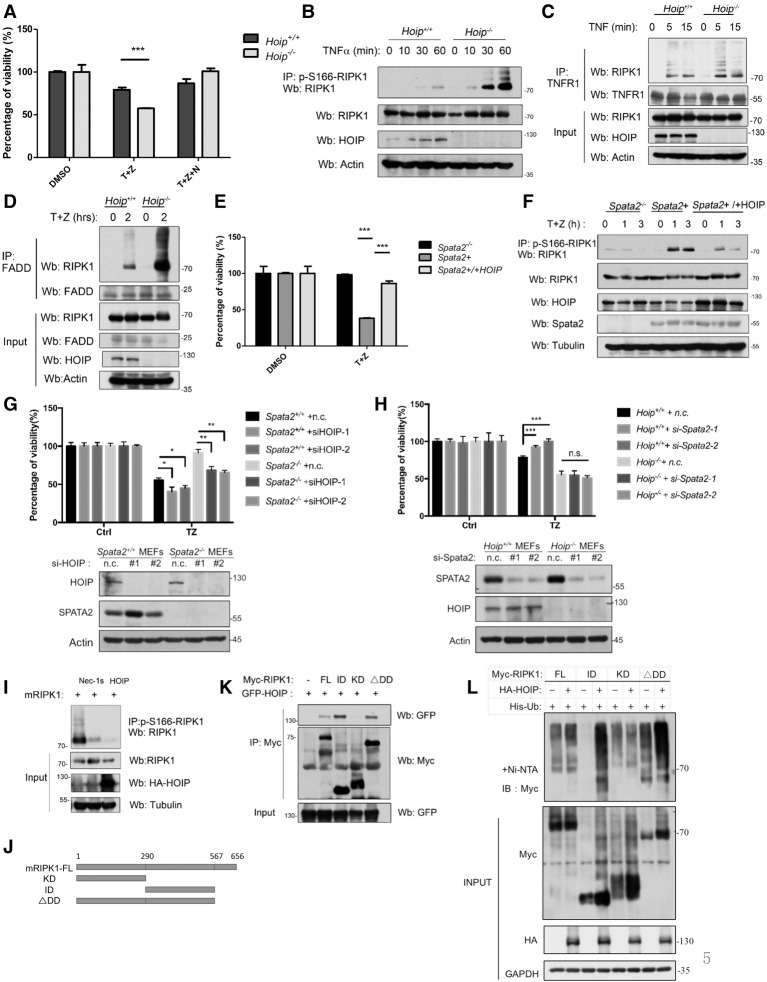
To characterize the significance of M1 ubiquitination of RIPK1 mediated by the LUBAC, we compared the sensitivity of Hoip+/+ and Hoip−/− MEFs to necroptosis. We found that Hoip−/− MEFs were significantly more sensitive to necroptosis induced by TNFα plus zVAD.fmk, and the levels of p-S166 RIPK1 were significantly higher than those of wild type (Fig. 5A,B). HOIP deficiency did not affect the recruitment of RIPK1 into the TNF-RSC but promoted the recruitment of RIPK1 into complex IIb, defined by the interaction of RIPK1 and FADD (Fig. 5C,D). Furthermore, the sensitivity of Spata2+ MEFs to TNFα/zVAD.fmk-induced necroptosis and increased activation of RIPK1 could be suppressed by the expression of HOIP (Fig. 5E,F).
Figure 5.
M1 ubiquitination antagonizes RIPK1 kinase activation. (A) Hoip+/+ MEFs and Hoip−/− MEFs were pretreated with or without 20 µM Nec-1s for 10 min and then treated with 10 ng/mL human TNFα and 20 µM zVAD.fmk for 24 h. Cell viability was measured by CellTiterGlo assay. (B) Hoip+/+ MEFs and Hoip−/− MEFs were treated with 100 ng/mL human TNFα and 50 µM zVAD.fmk for the indicated periods of time. The cell lysates were analyzed by immunoprecipitation with anti-p-S166 RIPK1. The immunocomplexes were analyzed by Western blotting using the indicated antibodies. (C) Hoip+/+ MEFS and Hoip−/− MEFs were treated with 100 ng/mL human TNFα for the indicated periods of time. The cell lysates were subjected to immunoprecipitation using anti-TNFR1 and then analyzed by Western blotting using the indicated antibodies. (D) Hoip+/+ MEFS and Hoip−/− MEFs were treated with 100 ng/mL human TNFα plus 50 µM zVAD.fmk for the indicated periods of time. The cell lysates were subjected to immunoprecipitation using anti-FADD and then analyzed by Western blotting using the indicated antibodies. (E) Spata2−/− MEFs, Spata2+ MEFs, and Spata2+ MEFs overexpressing HOIP were treated with 10 ng/mL human TNFα and 50 µM zVAD.fmk for 24 h. Cell viability was measured by CellTiterGlo assay. (F) Spata2−/− MEFs, Spata2+ MEFs, and Spata2+ MEFs overexpressing HOIP were treated with 10 ng/mL human TNFα and 50 µM zVAD.fmk for the indicated periods of time. The cell lysates were analyzed by immunoprecipitation using anti-p-S166 RIPK1. The immunocomplexes were then analyzed by Western blotting using the indicated antibodies. (G) Spata2+/+ MEFs and Spata2−/− MEFs were transfected with HOIP siRNA or nontargeting control (n.c.) for 24 h and treated with 50 ng/mL TNFα plus 50 µM zVAD.fmk for 16 h. Cell viability and cell death were evaluated using CellTiterGlo assay. (H) Hoip+/+ MEFs and Hoip−/− MEFs were transfected with Spata2 siRNA or nontargeting control (n.c.) for 24 h and treated with 50 ng/mL TNFα plus 50 µM zVAD.fmk for 16 h. Cell viability was evaluated using CellTiterGlo assay. (I) 293T cells were transfected for HA-mRIPK1 with or without HA-HOIP for 24 h and treated with or without 20 µM Nec-1s for the last 6 h. The cell lysates were analyzed by immunoprecipitation using antip-S166 RIPK1. The immunocomplexes were analyzed by Western blotting using the indicated antibodies. (J) Schematic representation of the Myc-tagged mouse RIPK1 truncations used. (K) 293T cells were transfected with expression vectors for different domains of Myc-mRIPK1 (as shown in J) with GFP-HOIP for 24 h. The cell lysates were analyzed by immunoprecipitation using anti-Myc antibody. The immunocomplexes were then analyzed by Western blotting using the indicated antibodies. (L) 293T cells were transfected with expression vectors for different domains of Myc-mRIPK1 with or without HA-HOIP for 24 h. The cell lysates were subjected to His pull-down with Ni-NTA beads and analyzed by Western blotting using the indicated antibodies. Error bars represent SEM from three technical replicates. GAPDH and Tubulin were used as loading controls.
Next, we characterized the effect of directly blocking M1 ubiquitination of RIPK1 by knocking down HOIP in Spata2−/− MEFs and the effect of knocking down Spata2 in Hoip−/− cells. We found that knocking down HOIP could significantly promote cell death in both Spata2+/+ MEFs and Spata2−/− MEFs induced by TNFα/zVAD.fmk (Fig. 5G). On the other hand, knocking down Spata2 failed to protect against cell death induced by TNFα/zVAD.fmk in Hoip−/− MEFs (Fig. 5H). These results indicate that the effect of Spata2 is dependent on the M1 ubiquitination of RIPK1.
Finally, the activation of RIPK1 in 293T cells upon overexpression of RIPK1 was also suppressed by the coexpression of HOIP (Fig. 5I). Taken together, these results suggest that the activation of RIPK1 is negatively regulated by its M1 ubiquitination.
To further characterize the M1 ubiquitination of RIPK1 mediated by HOIP, we investigated the domain of RIPK1 that interacted with HOIP. We expressed HOIP with different domains of RIPK1 in 293T cells and found that full-length, ΔDD (C-terminal DD deletion), and ID (intermediate domain only), but not KD (kinase domain only), RIPK1 was able to interact with HOIP (Fig. 5J,K). Thus, the ID of RIPK1 is important for interacting with HOIP. In addition, we characterized the ability of HOIP to mediate the ubiquitination of RIPK1 and found that the ubiquitination of the RIPK1 ID was most significantly increased (compared with that of full-length, ΔDD, and KD) when coexpressed with HOIP (Fig. 5L). These results suggest that RIPK1 may have multiple M1 ubiquitination sites, and the ID of RIPK1 might be the major target for the M1 ubiquitination by HOIP. Since the ID contains the RHIM motif critical for RIPK1 to interact with RIPK3 to form complex IIb (Li et al. 2012), this result suggests that elevating M1 ubiquitination of the ID of RIPK1 by HOIP might directly interfere with the binding of RIPK3.
Spata2 deficiency increases the activation of MKK4 and JNK
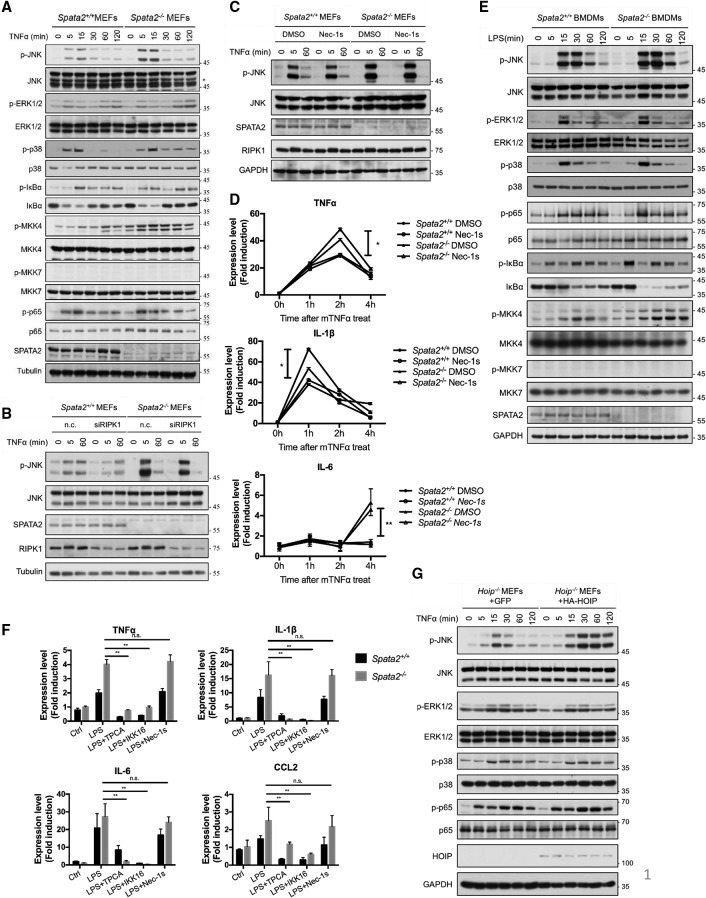
Since the ubiquitination of RIPK1 controls multiple aspects of TNFα signaling (O'Donnell et al. 2007) and since Spata2 deficiency leads to an increase of RIPK1 M1 ubiquitination, we next examined whether loss of Spata2 affects the other signaling events downstream from TNFR1. Spata2 deficiency was reported to increase JNK activation in A549 cells (Kupka et al. 2016). We found that Spata2 had no significant effect on the NF-κB activation in MEFs cells when treated with TNFα, including IκBα phosphorylation and degradation as well as the phosphorylation of p65, while JNK activation was considerably increased after stimulation by TNFα for 15 min under Spata2-deficient conditions (Fig. 6A). Furthermore, we explored the involvement of MKK4 or MKK7, two known activators of JNK (Yan et al. 1994; Tournier et al. 1999; Mazzitelli et al. 2011). Upon TNFα stimulation, the phosphorylation of MKK4 was significantly increased in Spata2-deficient cells, while p-MKK7 was undetectable (Fig. 6A).
Figure 6.
Loss of SPATA2 increases the activation of MKK4 and JNK kinases and inflammatory cytokine production. (A) Spata2+/+ MEFs and Spata2−/− MEFs were treated with 50 ng/mL human TNFα for the indicated periods of time, and the cell lysates were analyzed by Western blotting using the indicated antibodies. (B) Spata2+/+ MEFs and Spata2−/− MEFs were transfected with the indicated nontargeting control (n.c.) or RIPK1 siRNA for 48 h and then treated with human TNFα for the indicated periods of times. The cell lysates were analyzed by Western blotting using the indicated antibodies. (C) Spata2+/+ MEFs and Spata2−/− MEFs were pretreated with Nec-1s for 30 min and then treated with human TNFα for the indicated periods of time. The cell lysates were analyzed by Western blotting using indicated antibodies. (D) Primary Spata2+/+ and Spata2−/− BMDMs were treated with murine TNFα for the indicated periods of time. The mRNA levels of indicated genes were measured by quantitative RT–PCR. (E) Spata2+/+ and Spata2−/− iBMDMs were treated with LPS for the indicated periods of time and then analyzed by Western blotting using the indicated antibodies. (F) Spata2+/+ and Spata2−/− iBMDMs were treated with LPS with or without 20 µM Nec-1s, 100 nM TPCA-1, and 100 nM IKK-16 for 4 h. The mRNA levels of the indicated genes were measured by quantitative RT–PCR. The fold of induction was calculated based on the levels in untreated cells. Error bars represent SEM from three technical replicates. (G) Hoip−/− MEFs expressing GFP or HA-HOIP were treated with human TNFα for the indicated periods of time and analyzed by Western blotting using the indicated antibodies. GAPDH and Tubulin were used as loading controls.
While Nec-1s treatment had no inhibitory effect on the JNK activation, RIPK1 knockdown reduced the JNK activation, suggesting that the activation of JNK is independent of RIPK1 kinase activity but may be partially dependent on the scaffold function of RIPK1 (Fig. 6B,C). Similarly, after treatment of primary BMDM cells with mouse TNFα up to 4 h, the IL-6, TNFα, and IL-1β mRNA levels were greatly increased in Spata2−/− MEFs, which were not inhibited by the RIPK1 kinase inhibitor Nec-1s (Fig. 6D), indicating that Spata2 deficiency might promote the activation of TNFα-mediated transcription independently of RIPK1 kinase activity.
To further explore the role of Spata2 in cytokine production, we next examined the involvement of the TLR pathway in iBMDM cells. JNK activation was also increased under Spata2-deficient conditions in iBMDM cells treated with LPS for 15–30 min. (Fig. 6E). In addition, we found that the M1 ubiquination of RIPK1 with LPS stimulation was also increased in Spata2−/− BMDMs (Supplemental Fig. S8). Consistently, the phosphorylation of MKK4, but not MKK7, was also increased in Spata2-deficient cells stimulated by LPS. Similarly, the transcription of downstream cytokine genes—including TNFα, IL-6, IL-1β, and CCL2—following LPS treatment for 4 h was also enhanced upon the loss of Spata2 (Fig. 6F). The increased cytokine production induced by LPS in both wild-type and Spata2−/− cells was not blocked by Nec-1s but was effectively inhibited by the IKK inhibitors TPCA-1 and IKK-16 (Fig. 6F). These results suggest that the increased cytokine production in Spata2-deficient cells stimulated by LPS is independent of RIPK1 kinase activity but may be dependent on IKK activity.
We next explored how Spata2 might regulate the downstream signal transduction. Since Spata2 deficiency promotes M1 ubiquitination, we expressed a lentiviral expression vector for HOIP (the catalytic subunit of the LUBAC that mediates M1 ubiquitination) in Hoip−/− MEFs and stimulated the cells with TNFα. We found that increased expression of HOIP could also promote the phosphorylation of JNK, while p65, ERK, or p38 activation showed no difference (Fig. 6G). Next we overexpressed HOIP in BV2 cells and treated the cells with LPS. We found that overexpression of HOIP could also promote the activation of JNK and the mRNA levels of multiple cytokines, such as TNFα, IL-6, IL-1β, and CCL2, upon stimulation by LPS (Supplemental Fig. S9A,B).
Taken together, these results suggest that increased M1 ubiquitination in Spata2-deficient cells promotes the activation of MKK4 and JNK in a RIPK1 kinase activity-independent manner.
Spata2−/− mice are hypersensitized to SIRS
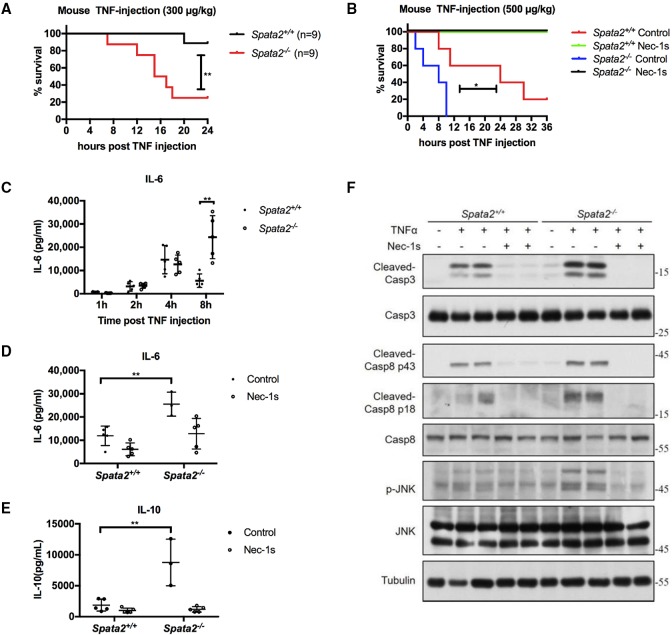
We next investigated the role of Spata2 in necroptosis and inflammation in vivo using a mouse model of SIRS. Administration of TNFα to mice mimics the pathological changes associated with the acute SIRS in vivo, which is RIPK1–RIPK3 kinase-dependent (Duprez et al. 2011). We challenged Spata2+/+ mice and Spata2−/− mice with TNFα in vivo. Surprisingly, with a sublethal dose injection of TNFα when Spata2+/+ mice appeared normal, Spata2−/− mice were already moribund. Thus, Spata2 deficiency sensitized mice to the lethal effect of TNFα instead of giving protection. Interestingly, the lethality of Spata2−/− mice was still completely rescued by the RIPK1 inhibitor Nec-1s (Fig. 7A,B).
Figure 7.
Loss of Spata2 sensitizes mice to TNFα-induced SIRS in vivo. (A) Survival curves of Spata2+/+ and Spata2−/− mice injected with 300 µg/kg mouse TNFα intravenously. (**) P < 0.01. n = 9. Spata2+/+ is marked in black, and Spata2−/− is marked in red. (B) Survival curve of Spata2+/+ and Spata2−/− mice given 40 mg/kg Nec-1s or vehicle control intragastrically 30 min before injection of 500 µg/kg mouse TNFα intravenously. (*) P < 0.05. n = 5. Spata2+/+ with vehicle control is marked in red, Spata2+/+ with Nec-1s is marked in green, Spata2−/− with vehicle control is marked in blue, and Spata2−/− with Nec-1s is marked in black. (C–E) Sera from injected mice were collected at the indicated time points, and IL-6 or IL-10 levels were demonstrated using ELISA. Error bars represent the SEM from three technical replicates. (F) Tissues from the intestines of mice with the indicated treatment for TNFα injection after 3 h were analyzed by Western blotting using the indicated antibodies. Tubulin was used as a loading control.
Next, we characterized the levels of inflammatory cytokines in mice injected with TNFα. We found higher levels of IL-6 and IL-10 in the sera of Spata2-deficient mice, which could be reduced by the RIPK1 kinase inhibitor Nec-1s (Fig. 7C–E). Since TNFα injection of mice can induce apoptosis and cause damage in the gut (Linde et al. 2011), we next investigated the caspase cleavage and MAPK kinase activation in TNFα-injected mice. We found that Spata2 deficiency did not inhibit the cleavage and activation of caspase-8 and caspase-3 in the gut, while the activation of JNK was increased in the guts of Spata2−/− mice after TNFα injection (Fig. 7F).
Spata2−/− mice were also more sensitive than wild type when challenged with a high dose of LPS in vivo (Supplemental Fig. S10A–C). Spata2−/− mice showed an earlier drop in body temperature than wild type (data not shown), died within 14–18 h, and had higher levels of TNFα and IL-6 secretion in serum, while wild-type mice survived the LPS challenge and remained healthy beyond 24 h.
Taken together, these results suggest that Spata2 deficiency promotes the production of proinflammatory cytokine levels, resulting in the “cytokine storm,” which might play a key role in the lethal effect of TNFα and LPS in vivo.
Discussion
Here we demonstrate the role and mechanism by which Spata2 regulates TNFα-mediated cell death and inflammatory response. We show that Spata2 can control the kinase activity of RIPK1 and cell death signaling by modulating linear ubiquitination of RIPK1. Spata2 not only mediates the interaction between HOIP and CYLD at basal condition prior to stimulation but also affects the recruitment of CYLD to the TNF-RSC and complex II upon TNFα stimulation and thus might control the activation of RIPK1 by modulating its M1 ubiquitination under different conditions. On the other hand, the recruitment of A20 into the TNF-RSC, which requires M1 ubiquitination by the LUBAC, is also increased in the absence of Spata2, and, since A20 is a key ubiquitin-editing enzyme of RIPK1, the increased A20 recruitment might also affect the ubiquitination state of RIPK1 at the TNF-RSC. Thus, Spata2 might modulate the activity of RIPK1 in the TNF-RSC directly by controlling its M1 ubiquitination as well as indirectly through increased recruitment of A20.
The activation of RIPK1 kinase in the TNF-RSC is subject to dynamic regulation of ubiquitination and deubiquitination. While CYLD alone is known to hydrolyze M1- and K63-linked ubiquitin in vitro (Sato et al. 2015), we show here that, in the presence of the PUB domain from SPATA2, the USP domain of CYLD is selective toward the M1 ubiquitin chain. The preference of SPATA2/CYLD toward the M1 ubiquitin chain provides a mechanistic explanation for the preferential increases in the M1 ubiquitination levels of RIPK1 under Spata2-deficient conditions. The activation of RIPK1 kinase activity, which promotes its autophosphorylation, may drive the conformational changes required for RIPK1 to interact with other complex components to facilitate the transition from complex I into complex II. Since Spata2 deficiency inhibits the activation of RIPK1 kinase at both the TNF-RSC and complex II, we speculate that increased M1 ubiquitination of RIPK1 may antagonize its kinase activation as well as affect its interaction with other proteins. As the ubiquitination pattern on RIPK1 may present a “ubiquitin code” to decide whether a cell may live or die (Ofengeim and Yuan 2013), our results suggest that the elevated levels of M1 ubiquitination on RIPK1 may work as an inhibitory code to block the activation of its kinase activity.
On the other hand, the elevated levels of M1 ubiquitination in Spata2-deficient cells or upon overexpression of HOIP may also regulate the activation of JNK independently of RIPK1 kinase activity but may be dependent on the scaffold function of RIPK1 and mediated by its upstream regulator, MKK4. JNK activation is known to be critical for mediating the hepatocyte toxicity (Das et al. 2009). Mice that are deficient in both JNK1/2 in the hematopoietic compartment are strongly protected against Con-A-induced hepatitis with markedly reduced expression of TNFα. We speculate that increased M1 ubiquitination of unknown targets in Spata2-deficient cells upon TNFα and LPS stimulation may promote the activation of MKK4 and JNK, which in turn mediate the increased production of proinflammatory cytokines and hypersensitivity of Spata2−/− mice to TNFα and LPS injection. Interestingly, the lethality of Spata2−/− mice was still inhibited by Nec-1s, suggesting that cell death induced by the increased production of proinflammatory cytokines under Spata2-deficient conditions might still be RIPK1-dependent. Consistently, inhibition of RIPK1 pharmacologically and genetically also blocked the hepatitis induced by Con-A injection (Filliol et al. 2016).
The function of Spata2 requires the presence of cIAP1/2, as the treatment of SM-164 eliminated the protection of necroptosis by Spata2 deficiency. Other than their involvement in the TNFR1 signaling complex, c-IAP1 is also recruited to mediate the signaling of TNFR2 to ubiquitinate and degrade c-IAP2 and the adaptor protein TRAF2 to regulate the activation of MAP kinases (Li et al. 2002). The requirement of cIAP1/2 for Spata2 might suggest the involvement of TNFR2. Thus, Spata2 might also contribute to TNFR2 signaling. It might be interesting to investigate the role of the Spata2/CYLD complex in regulating MAP kinase activation and TNFR2 signaling in the future.
Materials and methods
Animals
Spata2+/− embryonic stem cells in the C57BL/6N background were obtained from the KOMP Repository. Germline-transmitting chimeras were generated from those cells, and knockout mice were obtained from breeding. Spata2−/− mice were backcrossed with C57BL/6 mice for 10 generations. Mice were housed and cared for in a specific pathogen-free environment, and all animal procedures were performed according to the protocols approved by the Standing Animal Care Committee at Interdisciplinary Research Center of Biology and Chemistry.
Injections, monitoring, and sampling
Mouse TNFα was diluted in endotoxin-free PBS and injected in a volume <0.2 mL. Mouse TNFα was injected intravenously, whereas 40 mg/kg Nec-1s was given intragastrically 30 min before mouse TNFα injection. Sera samples and tissue samples were collected at designated times after injection. Blood was obtained from the tail vein.
Cell culture
Primary MEFs were generated from embryonic day 12.5 (E12.5) embryos. Immortalized MEFs were generated by infecting primary MEFs with retrovirus made from pLenti CMV/TO SV40 small + large T vector (Addgene, plasmid no. 22298). Primary BMDMs were differentiated from BM cells with 40 ng/mL M-CSF for 6–7 d. iBMDMs were generated by infecting primary BMDMs with virus produced from the Cre-J2 cell line (Blasi et al. 1985). HEK293T cells were from Thermo Scientific. MEFs, BMDMs, and L929 and 293T cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin.
Reagents and antibodies
pMIG vector was a gift from Dr. William Hahn. The following commercial antibodies and reagents were used: TNFR1 (R&D Systems, AF-425-PB; and Abcam, ab19139), RIPK1 (Cell Signaling Technology, 3493), TRADD (Santa Cruz Biotechnology, sc-7868), FADD (Santa Cruz Biotechnology, sc-6036), A20 (Cell Signaling Technology, 5630), CYLD (Cell Signaling Technology, 8462; and Santa Cruz Biotechnology, sc-74435), cIAP-1 (Enzo Life Sciences, ALX-803-335), Caspase-8 (Santa Cruz Biotechnology, sc-7890; and Cell Signaling Technology, 4927), cleaved Caspase-8 (Cell Signaling Technology, 8592), Caspase-3 (Cell Signaling Technology, 9662), cleaved Caspase-3 (Cell Signaling Technology, 9661), HOIL-1 (Santa Cruz Biotechnology, sc-365523), SHARPIN (Proteintech, 14626-1-AP), JNK (Cell Signaling Technology, 9252), p-JNK (Cell Signaling Technology, 9255), ERK1/2 (Cell Signaling Technology, 4695), p-ERK1/2 (Cell Signaling Technology, 4370), p38 (Cell Signaling Technology, 9212), p-p38 (Cell Signaling Technology, 9216), IκBα (Cell Signaling Technology, 4812), p-IκBα (Cell Signaling Technology, 2859), p65 (Cell Signaling Technology, 8242), p-p65(Cell Signaling Technology 3033), MKK4 (Cell Signaling Technology, 9152), p-MKK4 (Cell Signaling Technology, 9156), MKK7 (Cell Signaling Technology, 4172), p-MKK7 (Cell Signaling Technology, 4171), linear ubiquitin (Merck, MABS199), Tubulin, Actin, and GAPDH (Sigma-Aldrich). K63 chain-specific and M1 chain-specific ubiquitin antibodies were gifts from Dr. Vishva Dixit of Genentech. Anti-SPATA2, HOIP, MLKL, and RIPK1 monoclonal antibodies were generated using standard methods after immunizing mice with purified recombinant proteins. CellTiter-Glo luminescent cell viability kit was from Promega. 7-Cl-O-Nec-1 (Nec-1s), SM-164, and zVAD.fmk were made by custom chemical synthesis. The source of LPS-EK O111:B4 was Sigma-Aldrich, and TNFα was from Cell Sciences.
Generation of anti-p-RIPK1 S166 and anti-p-MLKL S345 antibodies
The phospho-peptides CGVASFKTW-pS-KLTK for mouse RIPK1 and CFELSKTQN-pS-ISRTAK for mouse MLKL were synthesized and coupled to KLH carrier protein via Cys at the N terminus. Polyclonal anti-p-S166 RIPK1 and anti-p-S345 MLKL antibodies were produced in rabbits by Aibo Biotechnology.
Protein expression and RNAi in cell lines
Protein expression was introduced by retrovirus, which was produced by infecting HEK293T cells with pMIG vector carrying ORFs of genes and packaging vector VSVG and GAG. siRNA expression was transfected with HiPerFect (Qiagen) to L929 cells. The Spata2 targeting and RIPK1 targeting siRNAs were a pool of four of the siTARGET set (Dharmacon). The HOIP targeting siRNAs were 5′-CCUCGAAACUACCUCAACATT-3′ and 5′-CCUGAGCUCUAGUUUACCUTT-3′. shRNA was introduced by retrovirus. The sequence targeted by Spata2 shRNA was 5′-GCATGAATCTTGTACCCTGAA-3′. The knockdown efficiency was measured by RT–PCR with Spata2 primers 5′-GGGGAATTCAGTTCAATGGATACGAAGTAC-3′ and 5′-CCATGGTACCCTAGAGGGACTCCACAAGCT-3′ and actin primers 5′-CACACTGTGCCCATCTACG-3′ and 5′-CCAGACAGCACTGTGTTGG-3′.
Cell viability assays and caspase-8 activity assay
General cell survival was measured by ATP luminescence assay CellTiterGlo (Promega). The percentage of viability was normalized to readouts of untreated cells of each genotype. Release of adenylate kinase to cell culture supernatant, an indication of plasma membrane and necrotic cell death, was measured by ToxiLight assay (Lonza). The xCELLigence real-time cell analyzer (RTCA, Roche) measured focal adhesion of living cells in real time. Reduction in the “normalized cell index” was an indicator for cell death. Caspase-8 activity was measured by Caspase-Glo 8 assay (Promega) following the manufacturer's instructions.
Immunoprecipitation
Cells were lysed with NP-40 buffer (120 mM NaCl, 10 mM Tris-HCl at pH 7.4, 1 mM EDTA, 0.2% NP-40, 10% glycerol) supplemented with 1 mM PMSF, 1× protease inhibitor cocktail (Roche), 10 mM β-glycerophosphate, 5 mM NaF, and 1 mM Na3VO4. Debris was precipitated, and the lysate was incubated with an antibody overnight at 4°C. The immunocomplex was captured by protein A/G agarose (Life Technologies) with the appropriate antibodies for 1–2 h at 4°C. Beads were washed four times, and the immunocomplex was eluted from beads by loading buffer. K63 and linear ubiquitin immunoprecipitation followed the method described previously (Matsumoto et al. 2012).
His-ubiquitin pull-down assay
Cells were lysed with urea buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl at pH 8.0, 500 mM NaCl, 10% glycerol, 0.1% Triton X-100, 10 mM β-mercaptoethanol, 10 mM imidazole) and homogenized with a syringe. Ni-NTA agarose beads (Qiagen) were added and incubated for 4 h at room temperature with rotation. The mixture was then washed three times with lysis buffer + 20 mM imidazole and eluted using elution buffer (0.15 M Tris-HCl at pH 6.7, 5% SDS, 30% glycerol, 200 mM imidazole, 0.72 M β-mercaptoethanol).
Real-time RT–PCR analysis
Total RNA was isolated from 1 × 106 cells using the Trizol reagent (Invitrogen) according to the manufacturer's protocol. Subsequently, cDNA was generated from total RNA using the reverse transcriptase M-MLV (TaKaRa). The real-time RT–PCR analysis was performed using SYBR real-time PCR premixture (BioTeke) according to the manufacturer's instructions. The RNA levels of the genes of interest were measured using the following primers and probes: TNFα (5′-ATGGCCTCCCTCTCATCAGT-3′ and 5′-TGGTTTGCTACGACGTGGG-3′), IL-1β (5′-TGCCACCTTTTGACAGTGATG-3′ and 5′-AAGGTCCACGGGAAAGACAC-3′), IL-6 (5′-CTCTGCAAGAGACTTCCATCCA-3′ and 5′-GACAGGTCTGTTGGGAGTGG-3′), CCL2 (5′-CTGTAGTTTTTGTCACCAAGCTCA-3′ and 5′-GTGCTGAAGACCTTAGGGCA-3), and Actin (5′-AGATCAAGATCATTGCTCCTCCT-3′ and 5′-ACGCAGCTCAGTAACAGTCC-3′).
ELISA
Cell culture medium or blood obtained from mouse tail veins was centrifuged to remove cell debris. The levels of TNFα or IL-6 were determined using ELISA kits according to the manufacturer's instructions (R&D Systems).
Mass spectrometric analysis
The binding partners of p-S166 RIPK1 were identified by mass spectrometry. The proteins obtained by immunoprecipitation against p-S166 RIPK1 in cells were trypsin-digested on beads. The resulting peptides were analyzed on a Thermo Scientific Orbitrap Fusion Tribrid mass spectrometer. Protein identification and quantification were done by MaxQuant. The tandem mass spectra were searched against the UniProt mouse protein database and a set of commonly observed contaminants. The precursor mass tolerance was set as 20 ppm, and the fragment mass tolerance was set as 0.5 Da. The cysteine carbamidomethylation was set as a static modification, and the methionine oxidation was set as a variable modification. The false discovery rate at the peptide spectrum match level and protein level was controlled below 1%. The quantification of proteins was done by the module of label-free quantification in MaxQuant. The unique peptides plus razor peptides were included for quantification.
Protein purification and ubiquitin chain digestion
The human CYLD USP domain (583–956) or Spata2 PUB domain (1–219) was expressed as N-terminal MBP-His6 or Trx-His6 fusion proteins in E. coli BL21 (DE3) and purified by Ni2+ affinity resin (GE Healthcare). M1-linked diubiquitin (His-2Ub) was expressed as an N-terminal His6 tag fusion protein in E. coli BL21 (DE3). K63-diUb protein was prepared as described (Pickart and Raasi 2005). The deubiquitination assay was set up by mixing 1 µM MBP-CYLDUSP, 2 µM Trx-Spata2PUB, and 40 µM K63-diUb or His-2Ub in a buffer containing 20 mM Tris (pH 7.5), 100 mM NaCl, and 1 mM DTT. The mixture was incubated at 37°C, and samples were taken at the indicated time intervals and resolved by SDS-PAGE.
Statistics and bioinformatics
Statistical analysis was performed using an unpaired two-tailed t-test. Statistical comparison of survival curves was performed using the log rank (Mantel-Cox) test. Differences were considered statistically significant if P < 0.05 (*), P < 0.01 (**), P < 0.001(***), or P < 0.0001(****). Data are expressed as the mean ± standard error of the mean.
Supplementary Material
Acknowledgements
We thank Dr. Vishva Dixit of Genentech for M1 and K63 antibodies, Dr. Henning Walczak for Hoip+/+ and Hoip−/− MEFs, and Dr. William Hahn of DFCI for pMIG vector. This work was supported in part by grants from the Chinese Academy of Sciences, the National Key R&D Program of China, the China Ministry of Science and Technology Program (2014ZX09102001-002), and the China National Natural Science Foundation (31530041); the National Institute of Neurological Disorders and Stroke (1R01NS082257), the National Institute on Aging (1R01AG047231), and the National Key R&D Program of China (2016YFA0501900) to J.Y.; the China National Natural Science Foundation (31401178) to H.P.; the Natural Science Foundation of Shanghai (16ZR1443900) to B.S.; and the Science and Technology Commission of Shanghai Municipality (15ZR1449100) and the China National Natural Science Foundation (31500597) to J.L.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.299776.117.
References
- Berger SB, Kasparcova V, Hoffman S, Swift B, Dare L, Schaeffer M, Capriotti C, Cook M, Finger J, Hughes-Earle A, et al. 2014. Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol 192: 5476–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, Rapp UR. 1985. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature 318: 667–670. [DOI] [PubMed] [Google Scholar]
- Das M, Sabio G, Jiang F, Rincon M, Flavell RA, Davis RJ. 2009. Induction of hepatitis by JNK-mediated expression of TNF-α. Cell 136: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almagro MC, Goncharov T, Newton K, Vucic D. 2015. Cellular IAP proteins and LUBAC differentially regulate necrosome-associated RIP1 ubiquitination. Cell Death Dis 6: e1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almagro MC, Goncharov T, Izrael-Tomasevic A, Duttler S, Kist M, Varfolomeev E, Wu X, Lee WP, Murray J, Webster JD, et al. 2017. Coordinated ubiquitination and phosphorylation of RIP1 regulates necroptotic cell death. Cell Death Differ 24: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. 2008. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 4: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y, Jouan-Lanhouet S, Divert T, Theatre E, Bertin J, Gough PJ, Giansanti P, Heck AJ, Dejardin E, Vandenabeele P, et al. 2015. NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol Cell 60: 63–76. [DOI] [PubMed] [Google Scholar]
- Draber P, Kupka S, Reichert M, Draberova H, Lafont E, de Miguel D, Spilgies L, Surinova S, Taraborrelli L, Hartwig T, et al. 2015. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep 13: 2258–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T, Declercq W, Libert C, Cauwels A, Vandenabeele P. 2011. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 35: 908–918. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, et al. 2010. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 29: 4198–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott PR, Leske D, Hrdinka M, Bagola K, Fiil BK, McLaughlin SH, Wagstaff J, Volkmar N, Christianson JC, Kessler BM, et al. 2016. SPATA2 links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol Cell 63: 990–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filliol A, Piquet-Pellorce C, Le Seyec J, Farooq M, Genet V, Lucas-Clerc C, Bertin J, Gough PJ, Dimanche-Boitrel MT, Vandenabeele P, et al. 2016. RIPK1 protects from TNF-α-mediated liver damage during hepatitis. Cell Death Dis 7: e2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, et al. 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471: 591–596. [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, et al. 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF–R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36: 831–844. [DOI] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. 2008. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471: 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. 2009. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10: 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupka S, De Miguel D, Draber P, Martino L, Surinova S, Rittinger K, Walczak H. 2016. SPATA2-mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep 16: 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang Y, Ashwell JD. 2002. TNF–RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416: 345–347. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. 2012. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde D, Nozomi T, Filip Van H, Benjamin V, Vera G, Tom Vanden B, Wim D, Claude L, Anje C, Peter V. 2011. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 35: 908–918. [DOI] [PubMed] [Google Scholar]
- Linton SD, Aja T, Armstrong RA, Bai X, Chen LS, Chen N, Ching B, Contreras P, Diaz JL, Fisher CD, et al. 2005. First-in-class pan caspase inhibitor developed for the treatment of liver disease. J Med Chem 48: 6779–6782. [DOI] [PubMed] [Google Scholar]
- Lu J, Bai L, Sun H, Nikolovska-Coleska Z, McEachern D, Qiu S, Miller RS, Yi H, Shangary S, Sun Y, et al. 2008. SM-164: a novel, bivalent Smac mimetic that induces apoptosis and tumor regression by concurrent removal of the blockade of cIAP-1/2 and XIAP. Cancer Res 68: 9384–9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Dong KC, Yu C, Phu L, Gao X, Hannoush RN, Hymowitz SG, Kirkpatrick DS, Dixit VM, Kelley RF. 2012. Engineering and structural characterization of a linear polyubiquitin-specific antibody. J Mol Biol 418: 134–144. [DOI] [PubMed] [Google Scholar]
- Mazzitelli S, Xu P, Ferrer I, Davis RJ, Tournier C. 2011. The loss of c-Jun N-terminal protein kinase activity prevents the amyloidogenic cleavage of amyloid precursor protein and the formation of amyloid plaques in vivo. J Neurosci 31: 16969–16976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly SR, Ninomiya-Tsuji J, Morioka S. 2014. TAK1 control of cell death. Cell Death Differ 21: 1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134: 668–678. [DOI] [PubMed] [Google Scholar]
- O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. 2007. Ubiquitination of RIP1 regulates an NF-kB-independent cell-death switch in TNF signaling. Curr Biol 17: 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, Green DR, Ting AT. 2011. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol 13: 1437–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengeim D, Yuan J. 2013. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol 14: 727–736. [DOI] [PubMed] [Google Scholar]
- Ofengeim D, Ito Y, Najafov A, Zhang Y, Shan B, DeWitt JP, Ye J, Zhang X, Chang A, Vakifahmetoglu-Norberg H, et al. 2015. Activation of necroptosis in multiple sclerosis. Cell Rep 10: 1836–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Raasi S. 2005. Controlled synthesis of polyubiquitin chains. Methods Enzymol 399: 21–36. [DOI] [PubMed] [Google Scholar]
- Polykratis A, Hermance N, Zelic M, Roderick J, Kim C, Van TM, Lee TH, Chan FK, Pasparakis M, Kelliher MA. 2014. Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J Immunol 193: 1539–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Goto E, Shibata Y, Kubota Y, Yamagata A, Goto-Ito S, Kubota K, Inoue J, Takekawa M, Tokunaga F, et al. 2015. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat Struct Mol Biol 22: 222–229. [DOI] [PubMed] [Google Scholar]
- Schlicher L, Wissler M, Preiss F, Brauns-Schubert P, Jakob C, Dumit V, Borner C, Dengjel J, Maurer U. 2016. SPATA2 promotes CYLD activity and regulates TNF-induced NF-κB signaling and cell death. EMBO Rep 17: 1485–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. 1999. The MKK7 gene encodes a group of c-Jun NH2-terminal kinase kinases. Mol Cell Biol 19: 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SA, Satpathy S, Beli P, Choudhary C. 2016. SPATA2 links CYLD to the TNF-α receptor signaling complex and modulates the receptor signaling outcomes. EMBO J 35: 1868–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak H. 2011. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunol Rev 244: 9–28. [DOI] [PubMed] [Google Scholar]
- Yan M, Dai T, Deak JC, Kyriakis JM, Zon LI, Woodgett JR, Templeton DJ. 1994. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature 372: 798–800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.