Abstract
Background
Langerhans cell (LC) networks play key roles in immunity and tolerance at body surfaces. LCs are established prenatally and can be replenished from blood monocytes. Unlike skin-resident dermal DCs (dDCs)/interstitial-type DCs and inflammatory dendritic epidermal cells appearing in dermatitis/eczema lesions, LCs lack key monocyte-affiliated markers. Inversely, LCs express various epithelial genes critical for their long-term peripheral tissue residency.
Objective
Dendritic cells (DCs) are functionally involved in inflammatory diseases; however, the mechanisms remained poorly understood.
Methods
In vitro differentiation models of human DCs, gene profiling, gene transduction, and immunohistology were used to identify molecules involved in DC subset specification.
Results
Here we identified the monocyte/macrophage lineage identity transcription factor Kruppel-like factor 4 (KLF4) to be inhibited during LC differentiation from human blood monocytes. Conversely, KLF4 is maintained or induced during dermal DC and monocyte-derived dendritic cell/inflammatory dendritic epidermal cell differentiation. We showed that in monocytic cells KLF4 has to be repressed to allow their differentiation into LCs. Moreover, respective KLF4 levels in DC subsets positively correlate with proinflammatory characteristics. We identified epithelial Notch signaling to repress KLF4 in monocytes undergoing LC commitment. Loss of KLF4 in monocytes transcriptionally derepresses Runt-related transcription factor 3 in response to TGF-β1, thereby allowing LC differentiation marked by a low cytokine expression profile.
Conclusion
Monocyte differentiation into LCs depends on activation of Notch signaling and the concomitant loss of KLF4.
Keywords: Kruppel-like factor 4, Runt-related transcription factor 3, TGF-β1 signaling, Notch, lineage decision, monocyte differentiation
Langerhans cells (LCs) are abundantly occurring dendritic cells (DCs) in environment-exposed stratified epithelia.1 They represent a well-defined DC subset marked by unique immunophenotypic ultrastructural and functional characteristics.2 Among various known human DC family members, LCs are identified as CD1ahiCD207+ cells that express several epithelial molecules (eg, E-cadherin, tumor-associated calcium signal transducer 2 [TACSTD2]/TROP2,3 epithelial cell adhesion molecule [EpCAM]/TROP1,4 and claudin-15), allowing them to undergo long-lasting adhesion to keratinocytes. LCs efficiently stimulate regulatory T cells and are involved in mediating tolerance to epithelial and environmental antigens.6 Consistently, even in the absence of microbial or danger signals, LCs continuously migrate to T-cell areas of skin-draining lymph nodes.1
LCs lack expression of monocyte/macrophage lineage-affiliated markers, including CD11b, CD36, lysozyme, CD209/DC-specific intercellular adhesion molecule–grabbing nonintegrin, and c-fms/monocyte colony-stimulating factor,7 as well as the early myeloid lineage marker myeloperoxidase.8 However, LC development is dependent on macrophage colony-stimulating factor receptor (CD115, c-fms), and blood monocytes can differentiate into LCs.9–12
Human monocytes can also differentiate into monocyte-derived dendritic cells (moDCs; induced by GM-CSF and IL-4), resembling inflammatory DCs in vivo (eg, inflammatory dendritic epidermal cells [IDECs]), or into macrophages. Unlike what is observed for LCs, monocyte lineage markers are maintained in these (sub)lineages.13 Moreover, moDCs possess a much higher capacity to synthesize proinflammatory cytokines.14,15 The transcriptional mechanism underlying monocyte/macrophage/moDC versus peripheral blood CD14+ monocyte-derived Langerhans cell (moLC) differentiation remains elusive.
LC differentiation from human hematopoietic progenitor cells is marked by upregulation of transcription factors.16–18 LCs express high levels of PU.1, and ectopic PU.1 promotes LC differentiation from myeloid progenitors.19 PU.1 transactivates Runt-related transcription factor 3 (RUNX3), which is essential for LC development.20 RUNX3 induction by PU.1 is context dependent because ectopic PU.1 in myeloid progenitors stimulates monocyte/macrophage and LC differentiation, depending on the presence or absence of cosignals.19 Therefore the critical molecular determinants of LC commitment are still undefined.
Methods
In vitro culture of CD34+ cord blood cells
For CD34+ progenitor cell–derived Langerhans cell (p-LC), CD34+ progenitor cell–derived monocyte-derived dendritic cell (p-moDC), or monocyte generation, a previously described 2-step culture model21,22 was used with slight modifications. In brief, sorted CD34+ cells were cultured in CellGro DC (CellGenix, Freiburg, Germany) medium supplemented with 10% FCS, 100 ng/mL GM-CSF, 20 ng/mL stem cell factor (SCF), 50 ng/mL FMS-like tyrosine kinase 3 ligand (FLT3L), and 2.5 ng/mL TNF-α for 5 days before subculturing in RPMI (Sigma, St Louis, Mo; 110% FCS) under lineage-specific cytokine conditions (100 ng/mL monocyte colony-stimulating factor, 50 ng/mL FLT3L, 20 ng/mL SCF, 2.5 ng/mL TNF-α, and 2 ng/mL IL-6 for monocytes; 100 ng/mL GM-CSF, 2.5 ng/mL TNF-α, and 25 ng/mL IL-4 for p-moDCs; and 100 ng/mL GM-CSF, 2.5 ng/mL TNF-α, and 1 ng/mL TGF-β1 for p-LCs). Clusters were purified by means of 1 g of sedimentation, as previously described.23 For generating moDCs, purified CD14+ blood monocytes were cultured in RPMI medium supplemented with GM-CSF (100 ng/mL) and IL-4 (25 ng/mL) in the presence of 10% FCS for 7 days. For generating moLCs, purified CD14+ blood monocytes were cultured in 24-well plates low/2 int hi coated with Delta-1, as previously described,24 in the presence of GM-CSF (100 ng/mL) and TGF-β1 (10 ng/mL) for 5 to 6 days (RPMI medium and 10% FCS). All cultures were supplemented with GlutaMAX (2.5 mmol/L; Gibco/Invitrogen, Grand Island, NY) and penicillin/streptomycin (125 U/mL each; PAA, Pasching, Austria).
RNA isolation and quantitative PCR
Cells were harvested and total RNA was isolated with the RNeasy Micro Kit (Qiagen, Hilden, Germany). Purified RNA was reverse transcribed with oligo-dT-primers (Eurofins MWG GmbH, Ebersberg, Germany) and reverse transcriptase (M-MLV-RT-H-; Fermentas, Waltham, Mass), according to the manufacturer’s instructions. Quantitative PCR was performed in a Roche LightCycler (Roche, Mannheim, Germany) with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Carlsbad, Calif). Values were normalized to hypoxanthine phosphoribosyltransferase (HPRT). Primers are listed in Table E1 in this article’s Online Repository at www.jacionline.org.
Flow cytometry
Flow cytometric staining and analyses were performed, as previously described.25 Flow cytometric analysis was performed with an LSRII instrument (BD Biosciences, San Jose, Calif) and FlowJo software (TreeStar, Ashland, Ore). For FACS sorting, the BD FACSAria flow cytometer (BD Biosciences) was used. Used antibodies are listed in Table E2 in this article’s Online Repository at www.jacionline.org.
Western blot analysis
For Western blot analysis, lysates of 1 to 2 × 106 cells per lane were loaded, and resolved proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Temecula, Calif). Membranes were probed with antibodies against Kruppel-like factor 4 (KLF4; Santa Cruz Biotechnology, Dallas, Tex), RUNX3 (a kind gift of S. Sakaguchi, Vienna, Austria), or β-actin (Sigma-Aldrich, St Louis, Mo), followed by horseradish peroxidase–conjugated goat anti-rabbit IgG antibodies (Pierce Biotechnology, Rockford, Ill). Antibody binding was visualized with the chemiluminescent substrates SuperSignal West Pico or West Dura (Pierce Biotechnology).
Cytokine measurement
Cells were seeded (1 × 104 to 2 × 104/200 μL) in 96-well plates, and supernatants were collected 48 hours later, as previously described.26 Cytokine (IL-6, IL-10, IL-8, TNF-α, and IL-12p40) levels were quantified with the Luminex system (Luminex, Austin, Tex).
Statistical analysis
Statistical analysis was performed with the paired, 2-tailed Student t test or ANOVA. P values of less than .05 were considered significant.
Detailed descriptions of cytokines and reagents, sources for cells and skin tissue, isolation of immune cells for microarray studies, mRNA microarray and data analysis, retroviral vectors, confocal microscopy, and the chromatin immunoprecipitation assay are presented in the Methods section in this article’s Online Repository at www.jacionline.org.
Results
KLF4 is inversely regulated during LC versus monocyte differentiation of myeloid progenitor cells
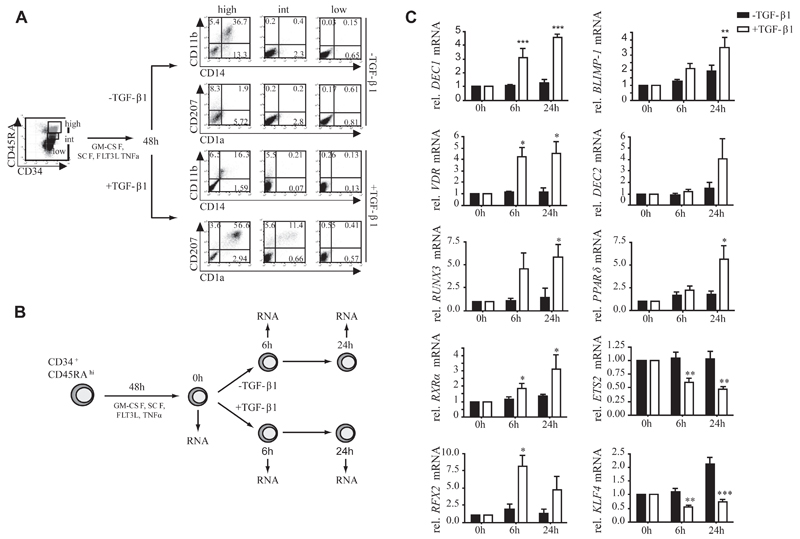
TGF-β1 induces the generation of CD1a+CD207+ p-LCs at the expense of monocytes/macrophages when added to serum-free, cytokine-supplemented cultures (GM-CSF, SCF, FLT3L, and TNF-α) of CD34+ hematopoietic progenitor cells.8 We screened for TGF-β1–regulated transcription factors during p-LC commitment. CD34+CD19− cells were subfractionated into CD45RAlow/−, CD45RAint, and CD45RAhi subsets (Fig 1, A, initial myelopoiesis).27–30 These subsets were prestimulated with GM-CSF, SCF, FLT3L, and TNF-α for 48 hours to bias their differentiation to monocytes. Thereafter, TGF-β1 was added to induce p-LC differentiation; parallel cultures were maintained without TGF-β1 addition to further promote monocyte development (Fig 1, A).8 Only CD45RAhi cells, representing the most differentiated myeloid progenitor subset, exhibited capacity to differentiate into CD1a+CD207+ p-LCs or CD14+CD11b+ monocytes (Fig 1, A). To enrich for functional LC precursors, we used these CD45RAhiCD34+ cells for microarray profiling (Fig 1, B). Conversely, all the subsequently described cell-culture experiments (see below) were done with total CD34+ cells.
Fig 1. Identification of transcription factors regulated during LC commitment.
A, Sorted CD34+CD45RAlow, CD34+CD45RAint, and CD34+CD45RAhigh progenitor subsets cultured for 7 days under monocyte-promoting (–TGF-β1) or LC-promoting (+TGF-β1) conditions. B, Schematic representation of microarray set-up. C, Quantitative PCR validation of selected transcription factors (n = 4-8, ± SEM). *P < .05, **P < .01, and ***P < .001, t test.
From mRNA analysis (Fig 1, B), we identified several “proof-of-concept” molecules being induced in TGF-β1–supplemented cultures, such as claudin-1, E-cadherin, CD207, or CCR6 (see Table E3 in this article’s Online Repository at www.jacionline.org); vice versa, monocyte/moDC-associated markers (CD36, CLEC10, MRC1, or FXIIIA)were repressed under LC conditions (see Table E4 in this article’s Online Repository at www.jacionline.org). Three groups of differentially regulated transcription factors were identified: (1) factors rapidly induced in TGF-β1–containing cultures, including DEC1, VDR, RUNX3, RXRα, and RFX2; (2) factors induced late at 24 hours in the presence of TGF-β1 (DEC2 and peroxisome proliferator-activated receptor [PPAR] γ) or slightly induced in absence of TGF-β1 (BLIMP-1); and (3) factors rapidly repressed in TGF-β1–containing cultures, including ETS-2 and KLF4. Among these, KLF4 was the only factor significantly inversely regulated under p-LC versus monocyte differentiation conditions (ie, repressed during p-LC [+TGF-β1] but induced during monocyte [−TGF-β1] differentiation; Fig 1, C). KLF4 induces monocyte differentiation,31,32 and therefore it might be involved in regulating alternate lineage fate options of shared monocyte/LC progenitor cells. KLF4 is a member of the family of Kruppel-like factors carrying conserved DNA binding zinc fingers but lacking homology outside DNA-binding regions.33 Apart from KLF10 (TIEG1), an immediate early TGF-β1–inducible factor in various cell types,34 and KLF6, none of the other KLF family members underwent significant regulation in our screen (see Table E5 in this article’s Online Repository at www.jacionline.org).
KLF4 protein is inversely regulated during moLC or moDC generation from CD14+ monocytes
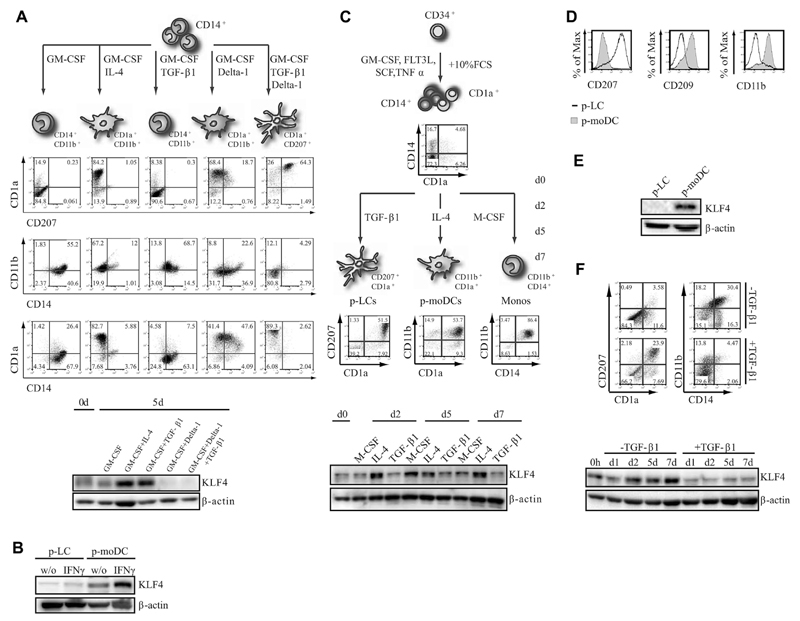
Murine inflammatory monocytes are KLF4+ and require KLF4 for differentiation.32 These cells can give rise to LCs under inflammatory conditions.9 Congruent with previous observations,12 the Notch ligand Delta-1 cooperates with TGF-β1 to induce LCs from CD14+ blood monocytes (moLCs; CD1a+CD207+ cells). KLF4 was repressed to virtually undetectable levels during moLC differentiation (Fig 2, A, right panel). Conversely, addition of TGF-β1 alone (ie, without Delta-1) to GM-CSF–supplemented cultures resulted in the generation of macrophages (CD14+CD11b+CD1a−) exhibiting marked upregulation of KLF4 protein (Fig 2, A). In fact, KLF4 expression levels were increased in GM-CSF plus TGF-β1–supplemented cultures compared with those in cells stimulated with GM-CSF only. Stimulation of monocytes with GM-CSF only in the presence of Delta-1 caused repression of KLF4 (Fig 2, A). KLF4 was also abundantly expressed in CD1a+ moDCs generated in the presence of GM-CSF plus IL-4 (Fig 2, A).
Fig 2. KLF4 is downregulated during LC differentiation.
A, KLF4 expression of CD14+ blood monocytes stimulated with indicated cytokines. B, KLF4 expression of FACS-purified p-LCs or p-moDCs (± IFN-γ). C, FACS and KLF4 expression analysis of day 7 p-moDCs, p-LCs, or progenitor-derived monocytes (p-monocytes). D and E, Lineage marker profile (Fig 2, D) and KLF4 expression (Fig 2, E) of CD1a+ cells of day 7 p-moDC or p-LC cultures. F, FACS and Western blot analysis of day 5 progenitors cultured with or without TGF-β1 for 7 days (n = 3).
IFN-γ was previously shown to enhance KLF4 expression in monocytes.35 Consequently, we analyzed whether KLF4 is induced in CD34+-derived p-LCs or p-moDCs in response to IFN-γ stimulation for 2 days. Although IFN-γ did not induce KLF4 in p-LCs, p-moDCs showed marked upregulation of KLF4 on treatment (Fig 2, B).
Time kinetic analyses of monocyte/LC precursors generated from CD34+ progenitors22 revealed weak KLF4 expression at day 0 (Fig 2, C). These precursors can be induced to differentiate into p-LCs, p-moDCs (Fig 2, C and D), or macrophages. KLF4 was induced during p-moDC and macrophage but not p-LC differentiation (Fig 2, C). Expectedly, p-moDCs and p-LCs showed inverse expression of CD207, CD209, and CD11b (Fig 2, D). Purified CD34+ cell–derived p-LCs clearly lacked detectable KLF4 (Fig 2, E). Consistently, omission of TGF-β1 from the LC-inducing cytokine mix was accompanied by KLF4 induction (Fig 2, F).
LCs and their precursors lack detectable KLF4, whereas dermal DCs are KLF4+
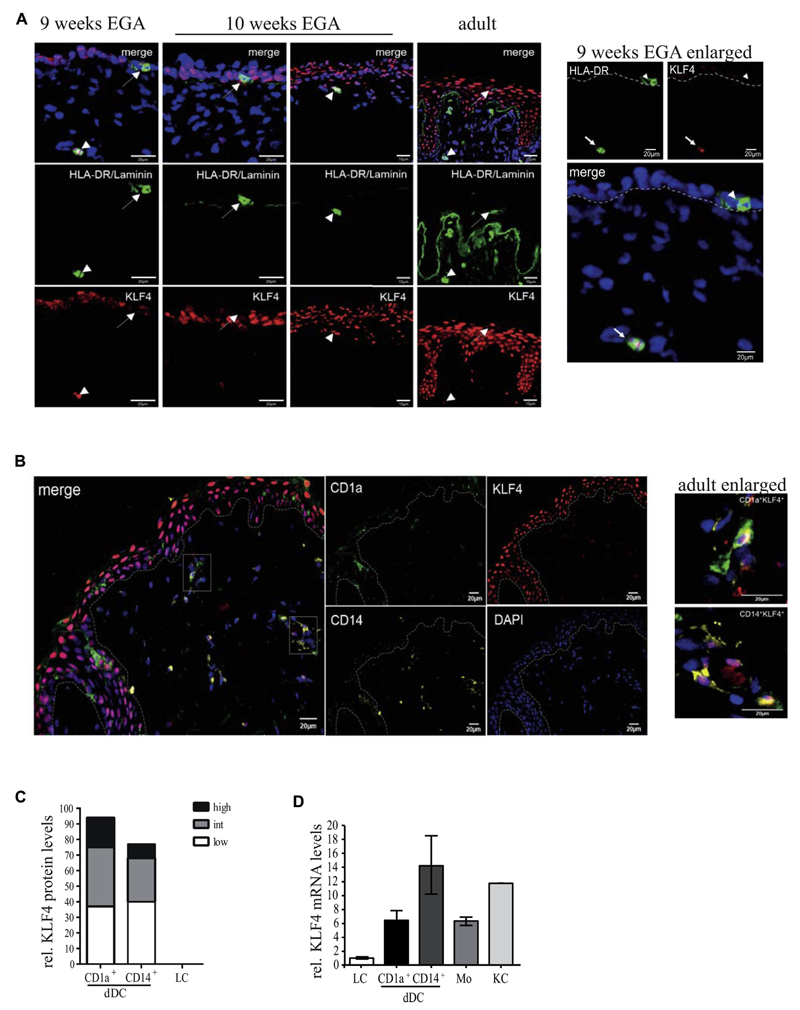
LCs in adult human skin (HLA-DR+CD207+ epidermal cells) lacked detectable KLF4 (Fig 3, A, right panel, arrow), whereas all keratinocyte layers exhibited a nuclear KLF4 expression pattern. Moreover, KLF4 was readily detectable in HLA-DR+ dermal cells (Fig 3, A, right panel, arrowhead). Because LCs develop prenatally from epidermal resident precursors, we also analyzed prenatal human skin. HLA-DR+ cells in embryonic epidermis at 9 weeks of estimated gestational age (EGA)36 lack KLF4 (Fig 3, A, left panel, enlarged right panel), whereas HLA-DR+KLF4+ dermal cells could be identified. KLF4 could be detected at varying levels in most adult CD1a+ and CD14+ dermal DCs, whereas epidermal CD1a+ cells lacked KLF4 (Fig 3, B and C). Grading was performed according to reference levels of KLF4 in keratinocyte layers (outer layers, high; suprabasal, intermediate; and basal, low). Gene profiling of FACS-sorted cell subsets confirmed that both dDC subsets, monocytes, and keratinocytes express KLF4 mRNA at high levels, whereas it is virtually undetectable in LCs (Fig 3, D).
Fig 3. LCs and prenatal LC precursors do not express KLF4.
A, LCs and prenatal HLA-DR+ LCs are indicated by arrows, and dermal HLA-DR+ cells are indicated by arrowheads (n = 4 per developmental stage). B, Staining of dermal CD14+ or CD1a+ DCs of adult human skin. C, Grading of KLF4 expression of dDCs (n = 6). D, KLF4 mRNA levels of LCs, monocytes (Mo), CD1a+ dDCs, or CD14+ dDCs and keratinocytes (KC) isolated from healthy adult human skin (n = 3).
Notch signaling is activated in LCs
Notch activation in response to Delta-1 represses KLF4, as well as the monocyte lineage markers CD14 and CD11b, in blood monocytes. Moreover, Delta-1 is required for TGF-β1–induced moLC differentiation (Fig 2, A). CD1a+ epidermal LCs stain strongly positive for an antibody specific for active intracellular NOTCH1 (aN1)37–39; conversely, they lack KLF4 (Fig 4, A, upper panel). Consistently, CD207+ cells generated from monocytes or CD34+ cells are aN1+ but KLF4− (Fig 4, A, lower panel). Inversely, moDCs lack aN1 expression but are strongly KLF4+. Positive aN1 reactivity of LCs was confirmed by using a chromogen-based immunohistochemistry method (see Fig E1 in this article’s Online Repository at www.jacionline.org).
Fig 4. KLF4 is repressed by Notch signaling.
A, Sections of healthy human skin (top) and cytospun in vitro–generated p-LCs, moLCs, and moDCs (bottom). B, TGF-β1–dependent upregulation/downregulation of Notch target genes after 6 and 24 hours (n = 3). C, NOTCH1 and JAG2 surface expression of CD34+ progenitors and p-LC generation cultures (n = 4). D, FACS analysis of intracellular active NOTCH1 (aN1) of day 5 moLCs and moDCs and day 7 p-LCs (n = 4).
The above-described microarray screen (Fig 1, B) revealed induction of Notch signaling pathway members by TGF-β1 within 6 hours. The Notch signature gene HES1 was induced along with NOTCH1 receptor and JAG2 ligand, as well as components of the γ-secretase complex (PSEN1 and PSEN2; Fig 4, B, and see Table E6 in this article’s Online Repository at www.jacionline.org). Consistently, NOTCH1 was induced in response to TGF-β1 signaling in hematopoietic progenitors within 24 to 72 hours (Fig 4, C, upper panel). Moreover, a portion of day 5 p-LC precursors expressed JAG2, which was further upregulated in day 7 generated CD1a+CD207+ p-LCs (Fig 4, C, lower panel). Notch signaling occurs through a cell contact–dependent mechanism.40 Consistently, p-LC generation cultures exhibited typical cell clustering involving E-cadherin adhesion41,42 and aN1 positivity (Fig 4, D). These characteristics were not observed in GM-CSF/IL-4–dependent p-moDC differentiation cultures, irrespective of the presence or absence of exogenous TGF-β1.
Ectopic KLF4 represses LC differentiation from monocyte/LC precursors
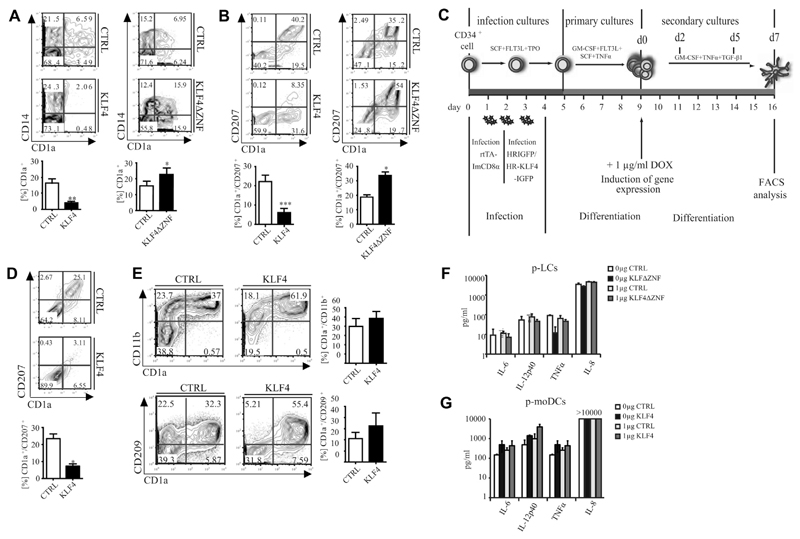
CD34+ progenitors were transduced with KLF4–internal ribosomal entry site (IRES)–green fluorescent protein (GFP) or empty control vector and subsequently induced to differentiate for 5 days into monocyte/LC precursors (schematically shown in Fig 2, C).22 Ectopic KLF4 reduced percentages of CD1a+ cells in favor of CD14+CD1a− cells (Fig 5, A), whereas a KLF4 mutant (KLF4ΔZNF-IRES-GFP), lacking DNA-binding activity but still retaining the transactivation/transrepression domain,35,43 showed the opposite effect (Fig 5, A, right panel). Consistently, KLF4 overexpression strongly inhibited whereas KLF4ΔZNF promoted p-LC generation from monocyte/LC precursors (Fig 5, B).
Fig 5. Ectopic KLF4 interferes with LC differentiation.
A, FACS of GFP+ day 5 progenitors (n = 6). *P < .05 and **P < .01, t test. B, Day 5 progenitors subcultured with TGF-β1 (n = 6). *P < .05 and ***P < .001, t test. C, Schematic representation of experimental set-up for KLF4 induction. D, CD1a/CD207 expression of day 7 GFP+ cells cultured according to Fig 5, C (n = 3). *P < .05, t test. E, FACS of day 5 progenitors subcultured under p-moDC conditions (n = 6). F and G, Cytokine secretion of day 7 GFP+CD1a+CD207+ p-LCs (Fig 5, F) or GFP+CD1a+CD11b+ p-moDCs (Fig 5, G; ± 1 μg of Pam3CSK4; n = 3).
We next generated day 5 monocyte/LC precursors (according to Fig 2, C, FACS plot day 0) and analyzed consequences of KLF4 overexpression on p-LC differentiation by using an inducible retroviral system (Fig 5, C).26 Expanded progenitor cells were transduced with 2 retroviral vectors (inducible “tet-on” system26), followed by the generation of DC precursors in primary cultures. Thereafter, cells were subcultured under secondary LC generation conditions in the presence of doxycycline to induce KLF4-IRES-GFP or empty control–GFP (Fig 5, C). FACS analysis of GFP+ cells was then performed at day 7 in LC generation cultures. The induction of KLF4 at the stage of DC precursors was sufficient to inhibit p-LC generation (Fig 5, D). Conversely, ectopic KLF4 expression in CD34+ cells did not inhibit but rather promoted generation of p-moDCs (CD207−CD209+CD11b+CD1a+ cells; Fig 5, E, and see Fig E2, A, in this article’s Online Repository at www.jacionline.org).
Apart from phenotypic characteristics, LCs and moDCs also differ in functional properties. p-LCs expressed substantially lower levels of proinflammatory cytokines than p-moDCs (Fig 5, F and G), confirming previous observations.14,44 KLF4-transduced p-moDCs showed slightly higher mean production levels of proinflammatory cytokines relative to controls (Fig 5, F). However, these data did not reach statistical significance in paired t test statistics. Consistent with the observed repression of KLF4 during LC differentiation, KLF4ΔZNF transduced p-LCs equaled control transduced cells in low levels of cytokine production.
RUNX3 and KLF4 are inversely regulated during LC and moDC differentiation
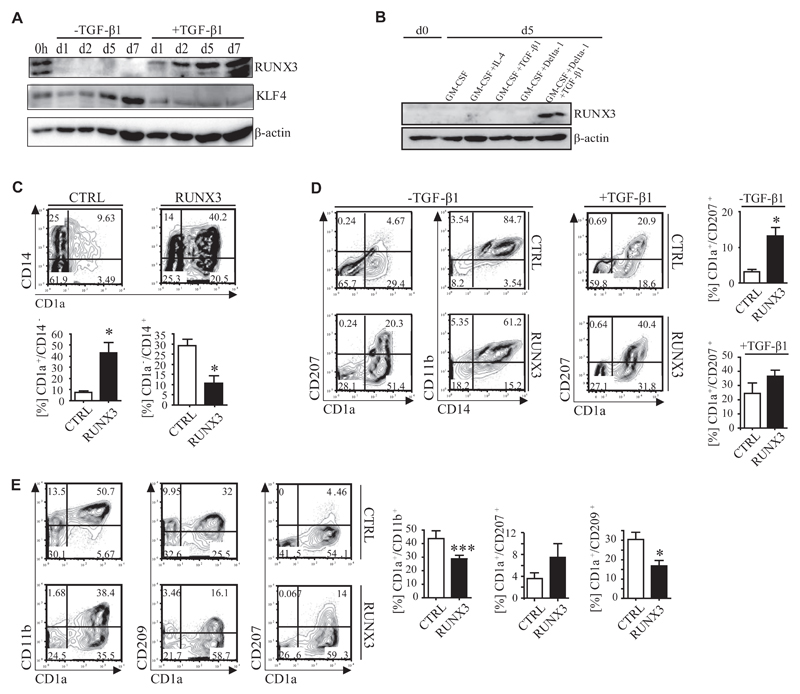
RUNX3–/– mice were shown to lack LCs.18 RUNX3 mRNA is strongly induced concomitant with LC differentiation (Fig 1, C). RUNX3 protein was induced during p-LC (+TGF-β1) but remained low/undetectable under monocyte (−TGF-β1) differentiation conditions, whereas KLF4 showed an inverse expression pattern (Fig 6, A). Furthermore, RUNX3 was expressed by moLCs but by neither moDCs nor macrophages generated from peripheral blood monocytes (Fig 6, B, and see also Fig 2, A).
Fig 6. RUNX3 and KLF4 are inversely regulated during LC and moDC differentiation.
A and B, KLF4 and RUNX3 protein expression of day 5 p-LC precursors (Fig 6, A) and CD14+ monocytes (Fig 6, B). C, FACS of GFP+ day 5 precursors (n = 5). *P < .05, t test. D and E, FACS of GFP+ day 5 precursors subcultured under p-LC–promoting (+/-TGF-β1; Fig 6, D) or p-moDC–promoting (Fig 6, E; GM-CSF plus IL-4) conditions at day 7 (n = 6). *P < .05, t test; ***P < .001.
Ectopic RUNX3 promotes LC but inhibits moDC differentiation
Ectopic expression of RUNX3 in CD34+ cells promoted the generation of day 5 p-LC precursors (CD1a+ cells) at the expense of monocyte precursors (CD14+CD1a− cells; Fig 6, C). Accordingly, RUNX3 promoted the generation of CD1a+CD207+ p-LCs in secondary cultures in the absence or presence of TGF-β1 (Fig 6, D). Furthermore, RUNX3 reduced percentages of CD11b+CD209+CD1a+ p-moDCs generated in the presence of GM-CSF plus IL-4 in favor of CD11b−CD209−CD1a+CD207+ p-LCs (Fig 6, E, and see Fig E2, B).
Inhibition of TGF-β1–mediated RUNX3 induction by ectopic KLF4 is counteracted by RUNX3 overexpression
CD34+ cells were transduced with KLF4-IRES-GFP or empty control vector and induced to differentiate into p-LCs or monocytes (schematically shown in Fig 2, C). TGF-β1–mediated RUNX3 induction was substantially impaired in KLF4-transduced cells (Fig 7, A and B). In comparison, other TGF-β1–induced genes from the screen, such as TIEG1 and SMAD7, were not repressed by KLF4 (Fig 7, C). Consistently, KLF4 binds to the RUNX3 promoter in KLF4+ p-moDCs (Fig 7, D), and ectopic RUNX3 overcame the inhibitory effect of KLF4 partially on the generation of CD1a+CD14− and total CD1a+ cells (Fig 7, E). It could re-establish the generation of CD1a+CD207+ p-LCs from KLF4-transduced progenitor cells (Fig 7, F).
Fig 7. RUNX3 restores LC differentiation from KLF4-transduced progenitors.
A-C, RUNX3 protein (Fig 7, A) and RUNX3, TIEG, and SMAD7 mRNA (Fig 7, B and C) expression of GFP+ day 5 precursors cultivated with or without TGF-β1. D, Semiquantitative PCR analysis of KLF4 chromatin immunoprecipitation from CD1a+ moDCs (n = 3). E and F, FACS analysis of cotransduced GFP+NGFR+-gated day 5 precursors (Fig 7, E) and p-LCs (Fig 7, F; + TGF-β1; n = 6). *P < .05, **P < .01, and ***P < .001, t test.
Discussion
Despite DCs having been recognized as involved in inflammatory skin diseases, the mechanisms of human DC differentiation in steady state versus inflammation remained poorly defined. Here we demonstrated that Notch signaling–dependent repression of KLF4 is critical for LC commitment of monocytes. Loss of KLF4 enables LC commitment at least partially through derepression of RUNX3. Moreover, we identified KLF4 as a key switch factor regulating differentiation of monocytes into LCs versus moDCs/dDCs. Our study highlights the role of signals within the epidermal/epithelial microenvironment for instructing LC commitment of monocytes (see graphic abstract).
KLF4 is a monocyte lineage identity factor known to promote monocyte commitment of myeloid progenitors.31 As such, KLF4 induces a set of monocyte lineage-associated molecules. Among these, CD11b is well known as a critical marker for classifying DC subsets. CD11b is strongly expressed by in vitro–generated moDCs, and high levels of CD11b are a phenotypic hallmark of IDECs, the latter representing putative monocyte-derived dendritic cells in atopic dermatitis/eczema lesions.45 Additionally, CD11b is expressed by the 2 major subsets of dDCs (CD1a+ and CD14+). We detected KLF4 in dDCs and in ex vivo–generated moDCs, which were previously shown to be phenotypically similar to IDECs.45 The previous demonstration that ectopic wild-type KLF4 induces CD11b in human myeloid progenitor cells31 is therefore supportive to the here observed positive correlation between KLF4 and CD11b among skin DC subsets. Consistently, stage-dependent loss of LC but not moDC differentiation potential during successive steps of monocyte/macrophage differentiation marked by CD11b was previously noted in cultures of CD34+ precursors.46
Our observation that KLF4 promotes CD11b+ moDC/dDC differentiation is reminiscent of previously observed functions of RelB.25 RelB promotes CD11b+ moDC/dDC differentiation from human CD34+ progenitors through inducing the generation of monocytic CD11b+ intermediates. Interestingly, CD11b+ classical DCs dependent on KLF4 or RelB have also been described in the murine system.47,48
Here we describe epithelial Notch activation–dependent KLF4 repression, which might allow LC differentiation from monocyte-committed cells under inflammatory conditions. Human CD14+ dDCs express KLF4; these cells can reconstitute LCs in vivo.49 Similarly, murine Gr1hi monocytes are dependent on KLF432 and can reconstitute LCs in vivo.9 We also detected KLF4 in fetal dermal HLA-DR+ cells, opening the possibility that such cells might give rise to LCs in prenatal epidermis. Reminiscent of similar observations in endothelial cells,50 here we demonstrated that TGF-β1 induces Notch signaling pathway members in monocytopoietic cells undergoing LC commitment.
Interestingly, human CD1c+ blood DCs differentiate into LCs without exogenous Notch ligand (ie, GM-CSF plus either TGF-β1 or BMP7).51 Whether Notch signaling is activated during CD1c+ blood DC–derived LC differentiation remains to be analyzed.
In support of our findings, cleaved NOTCH1 was previously shown to bind to a specific site within the KLF4 promoter,52 and antagonism between KLF4 and Notch signaling has been documented in nonhematopoietic cells.52 Moreover, it was previously described that KLF4 can inhibit the negative effect of TGF-β1 on proinflammatory cytokine expression.53 Because it has been demonstrated that LCs can cause induction of regulatory T cells,54,55 lack of KLF4 by LCs might indicate their involvement in tolerogenic functions.
Our study confirms previous observations claiming that the Notch ligand Delta-1 facilitates DC differentiation from monocytes by inhibiting default macrophage differentiation.56 Delta-1 alone repressed KLF4 in monocytes along with the induction of certain DC characteristics, such as CD1a expression. A portion of cells generated under these conditions still expressed low CD14 levels. It is interesting to speculate that these cells represent intermediate stages of LC differentiation from monocytes. Downregulation of KLF4 through Delta-1 in these cells might allow TGF-β1–induced terminal differentiation of LCs within the epidermis. KLF4 downregulation is not generally required for skin DC development because, unlike LCs, dDCs and moDCs express KLF4.
RUNX3-deficient mice lack LCs, and similarly, LCs are missing in mice in which RUNX3 has been conditionally deleted in CD11c+ cells.20 Here presented data support the concept that RUNX3 is a critical positive regulator of LC differentiation. In addition to RUNX3, LC development in vivo requires ID2.57 Unlike observed for RUNX3, ectopic ID2 did not promote LC differentiation.19 However, ID2 mRNA levels are suppressed by KLF4 overexpression in LCs (see Fig E2, C). Our screen identified additional factors induced during LC differentiation, which might also be involved in a transcription factor network regulating LC differentiation (eg, VDR,16 as well as DEC1, DEC2, BLIMP-1, and PPARγ).
Supplementary Material
Key messages.
KLF4 positively regulates differentiation of CD11b+ inflammatory DCs.
KLF4 represses RUNX3 and thereby inhibits tolerogenic LC commitment.
Notch signaling inhibits KLF4 during monocyte to LC lineage conversion.
Acknowledgments
We thank G. Zlabinger and M. Merio for cytokine measurements, M. Jaritz for heat map illustration with Spotfire, W. Ellmeier and J. Stöckl for critically reading the manuscript, and T. Bauer for designing the graphic abstract.
Supported by research grants of the Austrian Science Fund (FWF; P19474-B13 to A.E.-B., P22441-B13 and P23228-B19 to D.M., and P22058, P19245, P25720, and SFB-2304 to H.S.) and the PhD programs W1212 “Inflammation and Immunity,” W1248-B13 “Molecular, Cellular and Clinical Allergology,” and W1241 “Molecular Fundamentals of Inflammation.”
Abbreviation used
- aN1
Active intracellular NOTCH1
- DC
Dendritic cell
- FITC
Fluorescein isothiocyanate
- FLT3L
FMS-like tyrosine kinase 3 ligand
- GFP
Green fluorescent protein
- IDEC
Inflammatory dendritic epidermal cell
- IRES
Internal ribosomal entry site
- KLF4
Kruppel-like factor 4
- LC
Langerhans cell
- moDC
Monocyte-derived dendritic cell
- moLC
Peripheral blood CD14+ monocyte-derived Langerhans cell
- p-LC
CD34+ progenitor cell–derived Langerhans cell
- p-moDC
CD34+ progenitor cell–derived monocyte-derived dendritic cell
- PPAR
Peroxisome proliferator-activated receptor
- RUNX3
Runt-related transcription factor 3
- SCF
Stem cell factor
Footnotes
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
References
- 1.Kaplan DH. In vivo function of Langerhans cells and dermal DC. Trends Immunol. 2010;31:446–51. doi: 10.1016/j.it.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merad M, Ginhoux F, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol. 2008;8:935–47. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- 3.Eisenwort G, Jurkin J, Yasmin N, Bauer T, Gesslbauer B, Strobl H. Identification of TROP2 (TACSTD2), an EpCAM-like molecule, as a specific marker for TGF-β1-dependent human epidermal Langerhans cells. J Invest Dermatol. 2011;131:2049–57. doi: 10.1038/jid.2011.164. [DOI] [PubMed] [Google Scholar]
- 4.Gaiser MR, Lämmermann T, Feng X, Igyarto BZ, Kaplan DH, Tessarollo L, et al. Cancer-associated epithelial cell adhesion molecule (EpCAM; CD326) enables epidermal Langerhans cell motility and migration in vivo. Proc Natl Acad Sci U S A. 2012;109:E889–97. doi: 10.1073/pnas.1117674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmerli SC, Hauser C. Langerhans cells and lymph node dendritic cells express the tight junction component claudin-1. J Invest Dermatol. 2007;127:2381–90. doi: 10.1038/sj.jid.5700882. [DOI] [PubMed] [Google Scholar]
- 6.Seneschal J, Clark RA, Gehad A, Baecher-Allan CM, Kupper TS. Human epidermal Langerhans cells maintain immune homeostasis in skin by activating skin resident regulatory T cells. Immunity. 2012;36:873–84. doi: 10.1016/j.immuni.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E, et al. Human dermal CD14+ cells are a transient population of monocyte-derived macrophages. Immunity. 2014;41:465–77. doi: 10.1016/j.immuni.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strobl H, Bello-Fernandez C, Riedl E, Pickl WF, Majdic O, Lyman SD, et al. flt3 ligand in cooperation with transforming growth factor-beta1 potentiates in vitro development of Langerhans-type dendritic cells and allows single-cell dendritic cell cluster formation under serum-free conditions. Blood. 1997;90:1425–34. [PubMed] [Google Scholar]
- 9.Ginhoux F, Tacke F, Angeli V, Bogunovic M, Loubeau M, Dai X-M, et al. Langerhans cells arise from monocytes in vivo. Nat Immunol. 2006;7:265–73. doi: 10.1038/ni1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seré K, Baek JH, Ober-Blöbaum J, Müller-Newen G, Tacke F, Yokota Y, et al. Two distinct types of Langerhans cells populate the skin during steady state and inflammation. Immunity. 2012;37:905–16. doi: 10.1016/j.immuni.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 11.Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. Transforming growth factor beta1, in the presence of granulocyte/macrophage colony-stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J Exp Med. 1998;187:961–6. doi: 10.1084/jem.187.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoshino N, Katayama N, Shibasaki T, Ohishi K, Nishioka J, Masuya M, et al. A novel role for Notch ligand Delta-1 as a regulator of human Langerhans cell development from blood monocytes. J Leukoc Biol. 2005;78:921–9. doi: 10.1189/jlb.1204746. [DOI] [PubMed] [Google Scholar]
- 13.Pickl WF, Majdic O, Kohl P, Stöckl J, Riedl E, Scheinecker C, et al. Molecular and functional characteristics of dendritic cells generated from highly purified CD14+ peripheral blood monocytes. J Immunol. 1996;157:3850–9. [PubMed] [Google Scholar]
- 14.Ueno H, Klechevsky E, Morita R, Aspord C, Cao T, Matsui T, et al. Dendritic cell subsets in health and disease. Immunol Rev. 2007;219:118–42. doi: 10.1111/j.1600-065X.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- 15.Klechevsky E, Liu M, Morita R, Thompson-snipes L, Palucka AK, Banchereau J, et al. Understanding human myeloid dendritic cell subsets for the rational design of novel vaccines. Hum Immunol. 2009;70:281–8. doi: 10.1016/j.humimm.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Göbel F, Taschner S, Jurkin J, Konradi S, Vaculik C, Richter S, et al. Reciprocal role of GATA-1 and vitamin D receptor in human myeloid dendritic cell differentiation. Blood. 2009;114:3813–21. doi: 10.1182/blood-2009-03-210484. [DOI] [PubMed] [Google Scholar]
- 17.Yasmin N, Konradi S, Eisenwort G, Schichl YM, Seyerl M, Bauer T, et al. β-Catenin promotes the differentiation of epidermal Langerhans dendritic cells. J Invest Dermatol. 2013;133:1250–9. doi: 10.1038/jid.2012.481. [DOI] [PubMed] [Google Scholar]
- 18.Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, Goldenberg D, et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–79. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinz LX, Platzer B, Reisner PM, Jörgl A, Taschner S, Göbel F, et al. Differential involvement of PU.1 and Id2 downstream of TGF-β 1 during Langerhans-cell commitment. Blood. 2006;107:1445–53. doi: 10.1182/blood-2005-04-1721. [DOI] [PubMed] [Google Scholar]
- 20.Chopin M, Seillet C, Chevrier S, Wu L, Wang H, Morse HC, 3rd, et al. Langerhans cells are generated by two distinct PU.1-dependent transcriptional networks. J Exp Med. 2013;210:2967–80. doi: 10.1084/jem.20130930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratzinger G, Baggers J, de Cos M, Yuan J, Dao T, Reagan JL, et al. Mature human Langerhans cells derived from CD34+ hematopoietic progenitors stimulate greater cytolytic T lymphocyte activity in the absence of bioactive IL-12p70, by either single peptide presentation or cross-priming, than do dermal-interstitial or monocyte-derived dendritic cells. J Immunol. 2004;173:2780–91. doi: 10.4049/jimmunol.173.4.2780. [DOI] [PubMed] [Google Scholar]
- 22.Caux C, Massacrier C, Vanbervliet B, Dubois B, Durand I, Cella M, et al. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to granulocyte-macrophage colony-stimulating factor plus tumor necrosis factor alpha: II. Functional analysis. Blood. 1997;90:1458–70. [PubMed] [Google Scholar]
- 23.Gatti E, Velleca M, Biedermann BC, Ma W, Unternaehrer J, Ebersold MW, et al. Large-scale culture and selective maturation of human Langerhans cells from granulocyte colony-stimulating factor-mobilized CD34+ progenitors. J Immunol. 2000;164:3600–7. doi: 10.4049/jimmunol.164.7.3600. [DOI] [PubMed] [Google Scholar]
- 24.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;113:4313–8. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 25.Platzer B, Jörgl A, Taschner S, Höcher B, Strobl H. RelB regulates human dendritic cell subset development by promoting monocyte intermediates. Blood. 2004;104:3655–63. doi: 10.1182/blood-2004-02-0412. [DOI] [PubMed] [Google Scholar]
- 26.Jörgl A, Platzer B, Taschner S, Heinz LX, Hocher B, Reisner PM, et al. Human Langerhans-cell activation triggered in vitro by conditionally expressed MKK6 is counterregulated by the downstream effector RelB. Blood. 2007;109:185–93. doi: 10.1182/blood-2006-05-022954. [DOI] [PubMed] [Google Scholar]
- 27.Lansdorp PM, Sutherland HJ, Eaves CJ. Selective expression of CD45 isoforms on functional subpopulations of CD34 + hemopoietic cells from human bone marrow. Cytometry. 1990;172:363–6. doi: 10.1084/jem.172.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canque B, Camus S, Dalloul A, Kahn E, Yagello M, Dezutter-Dambuyant C, et al. Characterization of dendritic cell differentiation pathways from cord blood CD34(+)CD7(+)CD45RA(+) hematopoietic progenitor cells. Blood. 2000;96:3748–56. [PubMed] [Google Scholar]
- 29.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci U S A. 2002;99:11872–7. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haddad R, Guardiola P, Izac B, Thibault C, Radich J, Delezoide A, et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood. 2004;104:3918–26. doi: 10.1182/blood-2004-05-1845. [DOI] [PubMed] [Google Scholar]
- 31.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–48. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, et al. Kruppel-like factor 4 is essential for in ammatory monocyte differentiation in vivo. J Immunol. 2008;180:5645–52. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Spittau B, Krieglstein K. Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 2012;347:65–72. doi: 10.1007/s00441-011-1186-6. [DOI] [PubMed] [Google Scholar]
- 35.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, SenBanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–58. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 36.Schuster C, Vaculik C, Fiala C, Meindl S, Brandt O, Imhof M, et al. HLA-DR1+ leukocytes acquire CD1 antigens in embryonic and fetal human skin and contain functional antigen-presenting cells. J Exp Med. 2009;206:169–81. doi: 10.1084/jem.20081747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin S, Mutvei AP, Chivukula IV, Andersson ER, Ramsköld D, Sandberg R, et al. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKα/IKKβ. Oncogene. 2013;32:4892–902. doi: 10.1038/onc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirata N, Yamada S, Shoda T, Kurihara M, Sekino Y, Kanda Y. Sphingosine-1-phosphate promotes expansion of cancer stem cells via S1PR3 by a ligand-independent Notch activation. Nat Commun. 2014;5:4806. doi: 10.1038/ncomms5806. [DOI] [PubMed] [Google Scholar]
- 39.Hutter C, Kauer M, Simonitsch-Klupp I, Jug G, Schwentner R, Leitner J, et al. Notch is active in Langerhans cell histiocytosis and confers pathognomonic features on dendritic cells. Blood. 2012;120:5199–208. doi: 10.1182/blood-2012-02-410241. [DOI] [PubMed] [Google Scholar]
- 40.Bray S. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 41.Riedl E, Stöckl J, Majdic O, Scheinecker C, Rappersberger K, Knapp W, et al. Functional involvement of E-cadherin in TGF-beta 1-induced cell cluster formation of in vitro developing human Langerhans-type dendritic cells. J Immunol. 2000;165:1381–6. doi: 10.4049/jimmunol.165.3.1381. [DOI] [PubMed] [Google Scholar]
- 42.Riedl E, Stöckl J, Majdic O, Scheinecker C, Knapp W, Strobl H. Ligation of E-cadherin on in vitro-generated immature Langerhans-type dendritic cells inhibits their maturation. Blood. 2000;96:4276–84. [PubMed] [Google Scholar]
- 43.Yusuf I, Kharas MG, Chen J, Peralta RQ, Maruniak A, Sareen P, et al. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–81. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 44.Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, Thompson-Snipes L, et al. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 2008;29:497–510. doi: 10.1016/j.immuni.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wollenberg A, Mommaas M, Oppel T, Schottdorf EM, Gunther S, Moderer M. Expression and function of the mannose receptor CD206 on epidermal dendritic cells in inflammatory skin diseases. J Invest Dermatol. 2002;118:327–34. doi: 10.1046/j.0022-202x.2001.01665.x. [DOI] [PubMed] [Google Scholar]
- 46.Jaksits S, Kriehuber E, Charbonnier AS, Rappersberger K, Stingl G, Maurer D. CD34+ cell-derived CD14+ precursor cells develop into Langerhans cells in a TGF-beta 1-dependent manner. J Immunol. 1999;163:4869–77. [PubMed] [Google Scholar]
- 47.Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40:642–56. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–28. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larregina AT, Morelli AE, Spencer LA, Logar AJ, Watkins SC, Thomson AW, et al. Dermal-resident CD14+ cells differentiate into Langerhans cells. Nat Immunol. 2001;2:1151–8. doi: 10.1038/ni731. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y, Xue S, Zhou J, Voorhees JJ, Fisher GJ. Notch and TGF-β pathways cooperatively regulate receptor protein tyrosine phosphatase-κ (PTPRK) gene expression in human primary keratinocytes. Mol Biol Cell. 2015;26:1199–206. doi: 10.1091/mbc.E14-12-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milne P, Bigley V, Gunawan M, Haniffa M, Collin M. CD1c+ blood dendritic cells have Langerhans cell potential. Blood. 2015;125:470–3. doi: 10.1182/blood-2014-08-593582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng H, Pritchard DM, Yang X, Bennett E, Liu G, Liu C, et al. KLF4 gene expression is inhibited by the notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2009;296:G490–8. doi: 10.1152/ajpgi.90393.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu D, Wan Y. Regulation of Kruppel-like factor 4 by the anaphase promoting complex pathway is involved in TGF-beta signaling. J Biol Chem. 2011;286:6890–901. doi: 10.1074/jbc.M110.179952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senechal B, Elain G, Jeziorski E, Grondin V, De Serre NPM, Jaubert F, et al. Expansion of regulatory T cells in patients with Langerhans cell histiocytosis. PLoS Med. 2007;4:1374–84. doi: 10.1371/journal.pmed.0040253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stary G, Klein I, Bauer W, Koszik F, Reininger B, Kohlhofer S, et al. Glucocorticosteroids modify Langerhans cells to produce TGF-β and expand regulatory T cells. J Immunol. 2011;186:103–12. doi: 10.4049/jimmunol.1002485. [DOI] [PubMed] [Google Scholar]
- 56.Ohishi K, Varnum-Finney B, Serda RE, Anasetti C, Bernstein ID. The Notch ligand, Delta-1, inhibits the differentiation of monocytes into macrophages but permits their differentiation into dendritic cells. Blood. 2001;98:1402–7. doi: 10.1182/blood.v98.5.1402. [DOI] [PubMed] [Google Scholar]
- 57.Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, Kuhl C, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–6. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.