Abstract
Mitochondrially encoded large and small ribosomal RNAs (mtlrRNA and mtsrRNA) are transported out of mitochondria to polar granules, the distinctive organelles of germ plasm in Drosophila. Reduction of the extramitochondrial mtlrRNA amount leads to the failure of embryos to form the germ-line progenitors, or pole cells, suggesting that mtlrRNA, along with mtsrRNA, functions on the polar granules to specify the germ line. In this study, we provide several lines of evidence showing that there are mitochondria-type ribosomes on the polar granules during a short period before pole cell formation. Our ultrastructural analysis reveals that these ribosomes include both mitochondrial rRNAs and at least two mitochondrial ribosomal proteins (S12 and L7/L12). Furthermore, these ribosomes are integrated into well developed polysomes on the surface of polar granules. We propose that translation dependent on mitochondria-type ribosomes is an important mechanism underlying germ-line formation.
How the germ line segregates from the soma is a century-old issue in cell and developmental biology. In many animal species, the factor required for germ-line establishment has been postulated to be localized in a histologically distinct region in egg cytoplasm, or germ plasm (1, 2). Ultrastructural studies have shown that the germ plasm is basically composed of germinal granules and mitochondria (1, 2). The germinal granules are electron-dense structures, which act as a repository for the factors required for germ-line formation. Although the primary roles of the mitochondria are oxidative phosphorylation and biosynthesis of many metabolites, it has now become evident that they are also involved in germ-line formation.
In Drosophila, formation of the germ-line progenitors, or pole cells, requires the function of mitochondrial ribosomal RNA in germ plasm (3, 4). We have previously reported that mitochondrial large rRNA (mtlrRNA) and small rRNA (mtsrRNA) are both transported from mitochondria to the germinal granules, referred to as polar granules (5–7). This transportation occurs during early embryogenesis, when mitochondria are tightly associated with polar granules in germ plasm (6, 7), and it depends on the function of the maternally acting gene, tudor, which is known to be required for pole cell formation (8, 9). Mitochondrial rRNAs remain on the polar granules until pole cell formation and are no longer discernible on the granules within pole cells. Reduction of the extra-mitochondrial mtlrRNA amount results in the failure to form pole cells (4), and injection of mtlrRNA is able to induce pole cells in embryos whose ability to form these cells has been abolished by UV irradiation (3). These observations clearly show that the extra-mitochondrial mtlrRNA on polar granules has an essential role in pole cell formation, presumably cooperating with mtsrRNA.
Because both mtlrRNA and mtsrRNA are major components of ribosomes within mitochondria, we speculated that these rRNAs function to form ribosomes on the polar granules. Here, we report that mtlrRNA and mtsrRNA are both localized in the polysomes formed on the surface of polar granules during a short period (embryonic stage 2) before pole cell formation. Furthermore, mitochondrial ribosomal proteins (S12 and L7/L12) are enriched in the polysomes on the polar granules as well as in mitochondria. In the polysomes around polar granules are smaller ribosomes, of which size is almost identical to that of mitochondrial ribosomes but is smaller than that of cytosolic ones. We conclude that mitochondrial rRNAs form a mitochondrial type of ribosomes on polar granules, cooperating with mitochondrial ribosomal proteins.
Materials and Methods
Tagging of Ribosomal Protein.
We added the sequence encoding enhanced green fluorescent protein (EGFP) to the C terminus of S12 and L7/L12 as follows. S12 and L7/L12 cDNAs were amplified from a cDNA library and an EST clone (GM03767), respectively, by PCR by using pairs of primers 5′-GGGGGTACCCTCCGAGAGCATGAATTTTCTG-3′ and 5′-TGGCTCTTCTTGACGACGTGC-3′ for S12 and 5′-GGGGGTACCTACCAATAAGATGCACATCACAC-3′ (L7/L12 F) and 5′-TCGATCTCGATGATGGCGCC-3′ for L7/L12. After blunting and KpnI digestion, these cDNAs were cloned between SmaI and KpnI sites in pBluescript, and the resulting DNAs were digested again with KpnI and BamHI. A BamHI–NotI fragment encoding EGFP was obtained from pEGFP-N1 (CLONTECH). The KpnI–BamHI fragment encoding S12 or L7/L12 and the BamHI–NotI fragment were cloned together between KpnI and NotI sites in the upstream activation sequence (UASp) vector (10) to generate the UASp-S12-EGFP or UASp-L7/L12-EGFP. To add the hemagglutinin (HA) tag to the C terminus of L7/L12, a cDNA was amplified by PCR using the L7/L12 F primer and 5′-CTAGGCGGCGGCGTAATCGGGCACATCGTAGGGGTACTCGATCTCGATGATGGCGC-3′ primer. This results in the addition of DNA encoding YPYDVPDYAAA amino acids (underlined) in frame to the DNA encoding L7/L12 protein. After blunting and KpnI digestion, these cDNAs were cloned between SmaI and KpnI sites in pBluescript, and the resulting DNAs were digested again with KpnI and BamHI. The KpnI–BamHI fragment encoding L7/L12 was cloned between KpnI and BamHI sites in UASp vector to generate the UASp-L7/L12-HA.
Transformation.
The UASp constructs were transformed into yw flies (11) and homozygous lines were established. Distribution of the tagged proteins was examined in embryos and oocytes from the females carrying the UASp construct and the nanos-Gal4:VP16 transgene, which can drive the expression of the genes inserted downstream of the UASp in the female germ line throughout oogenesis (10, 12). Control embryos were obtained from the females carrying only UASp construct.
Antibody Against S12.
A cDNA fragment encoding the full length of S12 protein was amplified by PCR from a cDNA library using 5′-GGGAATTCATGAATTTTCTGCGGCAATC C-3′ and 5′-GGGAATTCCTATTGGCTCTTCTTGACGAC-3′ as a pair of primers and was subcloned into pMALc2 (New England Biolabs) and pProEX HTa (GIBCO/BRL). Recombinant MBP-S12 protein was purified on Amylose resin (New England Biolabs) and was used as an antigen. Polyclonal rabbit antiserum was generated by Takara Shuzo (Kyoto, Japan). Immune serum was affinity purified using His-tagged S12 protein, which was purified on Ni-NTA agarose (Qiagen). For immunohistochemical detection in embryos, affinity-purified anti-S12 antibody was diluted 1:300, and a biotinylated goat anti-rabbit IgG (1:400 dilution, Vector Laboratories) was used as a second antibody. The signal was enhanced by using the Vectastain Elite ABC-AP kit (Vector Laboratories) and was then detected with 5-bromo-4-chloroindolyl phosphate/nitroblue tetrazolium substrates (Boehringer Mannheim).
Immunohistochemistry at the Light Microscopic Level.
Immunohistochemical detection of the EGFP- and HA-tagged proteins in embryos was carried out according to Kobayashi et al. (13). A rabbit polyclonal anti-GFP antibody (CLONTECH) was diluted 1:300, and an Alexa 568-conjugated goat anti-rabbit IgG (1:1000 dilution, Molecular Probes) was used as a second antibody. A rat monoclonal anti-HA antibody (3F10) (Boehringer Mannheim) was diluted 1:300, and a biotinylated rabbit anti-Rat IgG (1:400 dilution, Vector Laboratories) was used as a second antibody. The signal was enhanced by using the Vectastain Elite ABC kit (Vector Laboratories) and was detected by using HistoMark Orange (Kirkegaard & Perry Laboratories).
Electron Microscopy.
Dechorionated wild-type (Oregon R) embryos were fixed and embedded according to Amikura et al. (6). The stages of the embryos were referred to according to Campos-Ortega and Hartenstein (14), determined by the observation of the nuclei of embryos in thick sections stained by toluidine blue. Ultrathin sections were stained by uranyl acetate and lead citrate and observed under an electron microscope (JEM-2000EXII, JEOL).
In Situ Hybridization at the Electron Microscopic Level.
We performed in situ hybridization at electron microscopic level according to Kobayashi et al. (13).
Immunohistochemistry at the Electron Microscopic Level.
Immunohistochemical detection at electron microscopic level was performed as described (13). As the primary antibodies, we used a rabbit polyclonal anti-GFP antibody (1:30 dilution) for S12-EGFP and L7/L12-EGFP, a rat monoclonal anti-HA antibody (1:20 dilution) for L7/L12-HA, and the rabbit polyclonal anti-S12 antibody (1:150 dilution) for S12. Gold-conjugated antibodies (15 nm, 1:50 dilution; British BioCel, Cardiff, U.K.) were used as secondary antibodies.
Freeze Substitution.
The samples for electron microscopy were made by a freeze substitution method. Dechorionated embryos were dipped in n-octane for 2 min. Then, the embryos were dipped in 15% butanediol in PBT (phosphate-buffered saline/0.1% Tween 20) for 10 min at 4°C for cryoprotection (15). The embryos collected on a copper grid were rapidly frozen in liquid propane at −190°C. The embryos were stored in liquid nitrogen until use. After freeze substitution (16), the embryos were processed for electron microscopy according to Amikura et al. (6). Diameters of ribosomes in areas within 200 nm from polar granules, within mitochondria, in the anterior pole region and on rough endoplasmic reticulum were measured in electron micrographs at the magnification of ×288,000. The diameter of each ribosome was calculated by averaging the size of the longer and shorter axes.
Results and Discussion
Because both mtlrRNA and mtsrRNA are major components of ribosomes within mitochondria, we speculated that these rRNAs function to form ribosomes on the polar granules. This idea is compatible with an early model that mRNAs encoding proteins for pole cell formation are stored in polar granules and are translated on the polysomes developed on the surface of these granules (17–19). At stage 1 of embryogenesis, immediately after oviposition, polysomes are rarely detectable on the polar granules, but their number gradually increases as the embryos proceed to stage 2 (Fig. 1A) (17, 18). After pole cell formation at stage 3, well developed polysomes are no longer discernible (17, 18). Here, we present several lines of evidence showing that polar-granule polysomes are composed of mitochondrial rRNAs and mitochondrial ribosomal proteins.
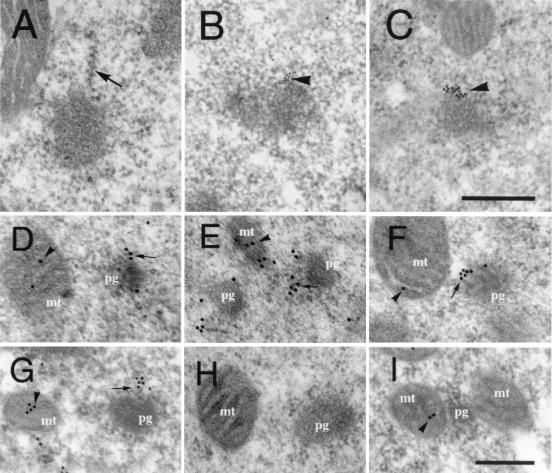
Figure 1.
Presence of mitochondrial rRNAs and ribosomal proteins in the polar granule polysomes at stage 2. (A) An electron micrograph showing well developed polysomes (arrow) on the surface of polar granules. (B and C) Electron micrographs of sections hybridized with probes for mtlrRNA (B) and mtsrRNA (C). Signals were arranged linearly from the surface of polar granules (arrowheads). (Scale bar in C = 0.25 μm.) (D–G) Distribution of S12-EGFP (D), L7/L12-EGFP (E), L7/L12-HA (F), and endogenous S12 (G). Signals were detected at the periphery of polar granules (arrows), as well as within mitochondria (arrowheads). (I) Distribution of endogenous S12 in polar plasm of early stage-1 embryo. S12 was detected within mitochondria (arrow) but not on polar granules. This distribution pattern was essentially identical to the patterns of the tagged ribosomal proteins (data not shown). Without induction of the tagged proteins, we could not detect signals for L7/L12-EGFP (H) and S12 EGFP and L7/L12-HA (data not shown). mt, mitochondria; pg, polar granules. (Scale bar in I = 0.2 μm.)
To address the question whether mtlrRNA and mtsrRNA are both components of the polar-granule polysomes, we examined the distribution of these rRNAs in polar plasm of embryos in stage 2 at the ultrastructural level by in situ hybridization. At stage 2, signals of mtlrRNA and mtsrRNA were arranged linearly on the polar-granule polysomes (Fig. 1 B and C); 86% of mtlrRNA and 81% of mtsrRNA signals on polar granules showed this type of distribution (total number of mtlrRNA and mtsrRNA signals counted in a 40-μm2 area were 350 and 313, respectively). In contrast, at stage 1, almost all signals of both rRNAs did not show the linear arrangements from the surface of the granules but localized at the boundaries between the granules and mitochondria (6, 7). These ultrastructural data show that mtlrRNA and mtsrRNA are both components of the polar-granule polysomes at stage 2.
As the next step, we examined the distribution of S12 and L7/L12, which are components of small and large subunits of mitochondrial ribosomes, respectively (20, 21). These ribosomal proteins were tagged by inserting sequences encoding an EGFP and an immunologically detectable HA tag into the 5′ end of their ORFs. The fusion genes were transformed into Drosophila and were expressed by using the Gal4/UASp system during oogenesis. The tagged ribosomal proteins were deposited into oocytes and then into embryos (data not shown).
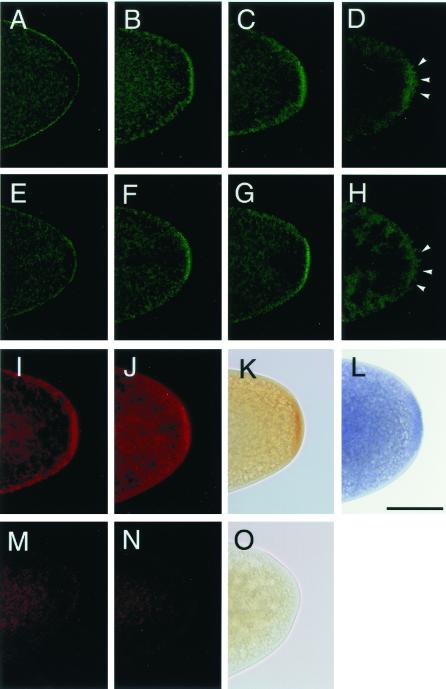
Under light microscopy, the distribution of the EGFP-tagged S12 protein (S12-EGFP) in the oocytes and embryos was detected by EGFP fluorescence. In mature oocytes, S12-EGFP was distributed evenly all over the oocytes in a punctate pattern (Fig. 2A). This distribution pattern represents S12-EGFP within mitochondria (see Fig. 1D). Once these oocytes were fertilized and oviposited, S12-EGFP began to be concentrated in polar plasm (Fig. 2B). Posterior enrichment reached a maximum at stage 2 (Fig. 2 C and I) and became undetectable after pole cell formation (Fig. 2D). We also examined the distribution of endogenous S12 in oocytes and embryos by using an antibody against S12. The distribution of endogenous S12 protein was identical to that of S12-EGFP (Fig. 2L), indicating that addition of the EGFP tag did not interfere with the distribution of mitochondrial ribosomal proteins. We further examined the distribution of EGFP- and HA-tagged L7/L12 protein (L7/L12-EGFP and L7/L12-HA) in oocytes and embryos by using EGFP fluorescence and antibodies against GFP and HA. The distribution pattern of the tagged L7/L12 protein was indistinguishable from those of S12-EGFP and endogenous S12 (compare Fig. 2 E–H, J, and K with Fig. 2 A–D, I, and L). Control experiments show that signals were never observed without induction of S12-EGFP, L7/L12-EGFP, and L7/L12-HA by Gal4 (Fig. 2 M–O).
Figure 2.
Distribution of mitochondrial ribosomal proteins in oocytes and early embryos. (A–H) Developmental changes in the distribution of S12-EGFP (A–D) and L7/L12-EGFP (E–H) detected by EGFP fluorescence. (A and E) Mature oocytes, (B and F) stage-1 embryos, (C and G) stage-2 embryos, and (D and H) stage-3 embryos. (I–L) Immunohistochemical detection of the Gal4-induced S12-EGFP (I) and L7/L12-EGFP (J) by an anti-GFP antibody and L7/L12-HA by an anti-HA antibody (K). Without induction by Gal4, no staining for S12-EGFP (M), L7/L12-EGFP (N), or L7/L12-HA (O) was observed. (L) A wild-type embryo stained with anti-S12 antibody. (I–O) Stage-2 embryos. The stages of the embryos were determined by the number and the location of nuclei stained by propidium iodide (22) referred to by Campos-Ortega and Hartenstein (13). (Scale bar = 50 μm.)
The above observations show that at least two ribosomal proteins, S12 and L7/L12, are enriched in polar plasm during a short period from stage 1 to 2. Given that mitochondrial rRNAs show an essentially identical distribution pattern to these mitochondrial ribosomal proteins under light microscopy, we speculated that these proteins are also components of the polysomes on the surface of polar granules. To address this issue, we examined the distribution of the tagged ribosomal proteins in polar plasm of embryos in stage 2 at an ultrastructural level. Fig. 1 (D–F) clearly demonstrates that signals for proteins S12-EGFP, L7/L12-EGFP, and L7/L12-HA were at the periphery of polar granules, as well as within mitochondria. On the granules, signals were arranged linearly from the surface of the granules to cytosol (Fig. 1 D–F), as was the case for mitochondrial rRNAs (Fig. 1 B and C). Endogenous S12 protein detected by the anti-S12 antibody shows the identical distribution pattern to these tagged ribosomal proteins (Fig. 1G). When we examined the distribution of S12, S12-EGFP, L7/L12-EGFP, and L7/L12-HA, 57–77% of total signals in a 40-μm2 area were arranged linearly from polar granules (39–184 signals were counted). Without inducing these tagged proteins, signals on polar granules and within mitochondria were not detected (Fig. 1H). Taken together, our ultrastructural observations reveal that mitochondrial ribosomal protein, S12 and L7/L12, like mitochondrial rRNAs, are components of the polysomes on the surface of polar granules.
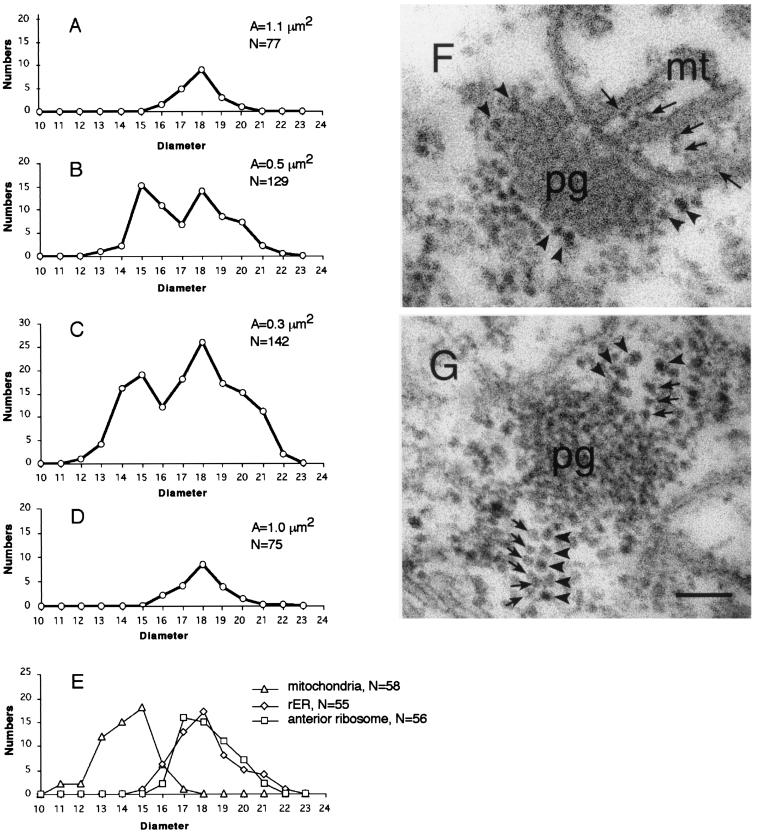
The above observations led to the idea that mitochondrial ribosomal RNAs and proteins form mitochondria-type ribosomes on polar granules. To test this idea, we measured diameters of ribosomes integrated into the polar-granule polysomes in stage-2 embryos because mitochondrial ribosomes are smaller than cytosolic ribosomes (23). Fig. 3 (C and G) demonstrates that the polar-granule polysomes contained two types of ribosomes; one was ca. 15 nm and the other was ca. 18 nm in diameter. The larger size corresponds to that of ribosomes on rough endoplasmic reticulum and of polysomes in the anterior pole region of embryos (Fig. 3E). In contrast, the smaller diameter was identical to that of ribosomes within mitochondria (Fig. 3 E and F). These ultrastructural data demonstrate that the mitochondria-type ribosomes are integrated into polar-granule polysomes in stage-2 embryos.
Figure 3.
Two types of ribosomes in the polar-granule polysomes. (A–D) The average number of ribosomes in a unit area (0.3 μm2) is plotted against its diameter. Diameters of the ribosomes around polar granules were measured in stage-14 oocytes (A), stage-1 embryos (B), stage-2 embryos (C), and in pole cells of stage-4 embryos (D). In A–D, total area examined (A) and total number of the ribosomes counted (N) are shown. (E) The number of ribosomes within mitochondria, in the anterior region and on the surface of rough endoplasmic reticulum, are plotted against their diameters. (F and G) Electron micrographs of sections through polar granules. (F) In stage-1 embryos, polar granules are intimately associated with mitochondria, and no ribosomes were found at the boundary between these organelles. Arrows point to ribosomes within a mitochondrion. (G) In stage-2 embryos, the smaller ribosomes (arrows) are integrated into the polar granule-polysomes. Arrowheads (F and G) point to the larger ribosomes. pg, polar granules; mt, mitochondria. (Scale bar = 0.1 μm.)
The smaller ribosomes were undetectable around polar granules in polar plasm of mature oocytes (Fig. 3A). After fertilization, they became detectable on the granules at stage 1 (Fig. 3B). As the embryos proceeded to stage 2, the smaller ribosomes were frequently observed in the polar-granule polysomes (Fig. 3C). Once pole cell formation was completed, the smaller ribosomes were no longer discernible on the granules in pole cells (Fig. 3D). These observations are consistent with our ultrastructural data that mitochondrial ribosomal RNAs and proteins are present on polar granules during stages 1 and 2, but not before and after these stages (Fig. 1 B–G) (6, 7).
Our results help to clarify the role of mitochondrial rRNAs in pole cell formation. Several lines of evidence from our analyses definitively support our conclusion that mitochondrial rRNAs form mitochondria-type ribosomes on polar granules during a short period before pole cell formation. First, mtlrRNA and mtsrRNA are localized on the polar-granule polysomes at stage 2. Second, mitochondrial ribosomal proteins, at least S12 and L7/L12, show the identical distribution pattern to those of mitochondrial rRNAs. Third, on polar granules there are small ribosomes, whose size is identical to that of ribosomes within mitochondria. Finally, the small ribosomes are seen on polar granules only when the mitochondrial ribosomal RNAs and proteins are present on the granules. This report demonstrates that mitochondria-type ribosomes are present outside mitochondria in eukaryotic cells.
We have previously reported that the transport of mitochondrial rRNAs from mitochondria to polar granules occurs in early stage-1 embryos (5–7). At this stage, both rRNAs are localized at the boundaries between mitochondria and polar granules (5–7, 9). In contrast, the localization, if any, of mitochondrial ribosomal protein S12 and L7/L12 at the boundaries between these organelles is yet to be observed (Fig. 1I). Furthermore, small ribosomes, or mitochondria-type ribosomes, are never seen at the boundaries (Fig. 3F). These observations lead to the idea that mitochondrial rRNAs and proteins are transported to the polar granules using two distinct mechanisms and are later assembled as ribosomes on the surface of the granules. Thus, we propose that polar granules are the site for assembly of mitochondria-type ribosomes.
The question of how the mitochondrial ribosomal proteins, S12 and L7/L12, are transported to polar granules is yet to be addressed. These proteins are encoded by the nuclear genome, and their mRNAs are translated in the cytosol (20, 21). The ribosomal proteins contain sequences at their N-terminal regions that target these proteins to mitochondria, which are cleaved during import (21). We show that S12, S12-EGFP, L7/12-EGFP, and L7/L12-HA are all incorporated within mitochondria (Fig. 1 D–G). It is likely that the ribosomal proteins within mitochondria are transported to polar granules. Alternatively, it is also possible that some of the proteins produced in the cytosol are directly transferred to the granules. Experiments to test whether the ribosomal proteins on the granules retain the targeting sequences at their N-terminal region are required to address this issue.
We found that mitochondria-type ribosomes were arranged linearly on the polar-granule polysomes (Fig. 3G). Considering that polysomes are sites of active translation (24) and mtlrRNA is essential to form pole cells (4), we propose that the mitochondria-type ribosomes on polar granules are specifically required to produce proteins for pole cell formation, although more rigorous biochemical evidences are required. The number of ribosomes that appear to be aligned on single polysomes varies from 2 to 8. They are less than the predicted number (≈20) of total ribosomes on an mRNA (18), suggesting that the mitochondria-type ribosomes are restricted to only a part of single polysomes. Unfortunately, we could not directly test this idea because we could not observe single strands of mRNA under the conditions we used and because the large size of polysomes usually caused them to extend out of the plane of ultrathin sections. To know the role of mitochondria-type ribosomes in the polar-granule polysomes, we need further biochemical investigations to test whether the factors for translational initiation and polypeptide elongation within mitochondria also participate in this translation system and whether the typical mitochondrial genetic code is used for the polypeptide elongation by these ribosomes.
In Xenopus, mtlrRNA and mtsrRNA are also present on the distinctive organelles of germ plasm, called germinal granules, which are very similar to polar granules of Drosophila (25, 26). We have in fact observed small ribosomes on the surface of germinal granules as well (26). These ribosomes are only seen during a short period from 4-cell to blastula stages, when the mitochondrial rRNAs are present on the granules (25, 26). In addition, mitochondrial rRNAs are present outside mitochondria in a discrete region of egg cytoplasm in ascidian, sea urchin, and planarian embryos (27–29). Although the function of the rRNAs in these animals remains elusive, we propose that mitochondria-type ribosomes have a widespread role to produce proteins required for germ-line development. In this context, it would be of great interest to identify the mRNA(s) that is translated by these mitochondria-type ribosomes.
Acknowledgments
We thank Dr. Paul Lasko for critical reading of the manuscript and Dr. Masukichi Okada for valuable comments. This work was supported in part by a grant-in-aid from the Ministry of Education, Science, Sports, and Culture (Japan), by Tsukuba Advanced Research Alliance Project, by a research project for a future program from the Japan Society for the Promotion of Science, and by Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology Corporation. M.K. was a Research Fellow of the Japan Society for the Promotion of Science.
Abbreviations
- mtlrRNA
mitochondrial large rRNA
- mtsrRNA
mitochondrial small rRNA
- EGFP
enhanced green fluorescent protein
- UASp
upstream activation sequence
- HA
hemagglutinin
References
- 1.Beam H W, Kessel R G. Int Rev Cytol. 1974;39:413–479. doi: 10.1016/s0074-7696(08)60944-4. [DOI] [PubMed] [Google Scholar]
- 2.Eddy E M. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi S, Okada M. Development (Cambridge, UK) 1989;107:733–774. doi: 10.1242/dev.107.4.733. [DOI] [PubMed] [Google Scholar]
- 4.Iida T, Kobayashi T. Proc Natl Acad Sci USA. 1998;95:11274–11278. doi: 10.1073/pnas.95.19.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi S, Amikura R, Okada M. Science. 1993;260:1521–1524. doi: 10.1126/science.7684857. [DOI] [PubMed] [Google Scholar]
- 6.Amikura R, Kobayashi S, Saito H, Okada M. Dev Growth Differ. 1996;38:489–498. doi: 10.1046/j.1440-169X.1996.t01-4-00005.x. [DOI] [PubMed] [Google Scholar]
- 7.Kashikawa M, Amikura R, Nakamura A, Kobayashi S. Dev Growth Differ. 1999;41:495–502. doi: 10.1046/j.1440-169x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 8.Boswell R E, Mahowald A P. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 9.Amikura, R., Hanyu, K., Kashikawa, M. & Kobayashi, S. (2001) Mech. Dev., in press. [DOI] [PubMed]
- 10.Rørth P. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 11.Spradling A C. In: Drosophila. Roberts D B, editor. Oxford: IRL; 1986. pp. 175–197. [Google Scholar]
- 12.Van Doren M, Williamson A L, Lehmann R. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi S, Amikura R, Nakamura A, Lasko P F. In: Advances in Molecular Biology: A Comparative Methods Approach to the Study of Oocytes and Embryos. Richer J D, editor. New York: Oxford Univ. Press; 1999. pp. 426–445. [Google Scholar]
- 14.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Heidelberg: Springer; 1985. pp. 9–84. [Google Scholar]
- 15.Campbell S S, Crawford B J, Reimer C L. J Microsc (Oxford) 1991;64:197–215. [Google Scholar]
- 16.Newman G R, Habot J A. Resin Microscopy and On-Section Immunocytochemistry. Heidelberg: Springer; 1993. p. 53. [Google Scholar]
- 17.Mahowald A P. J Exp Zool. 1968;167:232–267. doi: 10.1002/jez.1401670211. [DOI] [PubMed] [Google Scholar]
- 18.Mahowald A P. J Exp Zool. 1971;176:345–352. doi: 10.1002/jez.1401760309. [DOI] [PubMed] [Google Scholar]
- 19.Mahowald A P. Science. 1992;255:1216–1217. doi: 10.1126/science.1372132. [DOI] [PubMed] [Google Scholar]
- 20.Royden C S, Pirrotta V, Yan L Y. Cell. 1987;51:165–173. doi: 10.1016/0092-8674(87)90144-9. [DOI] [PubMed] [Google Scholar]
- 21.Marty L, Fort P. J Biol Chem. 1996;271:11468–11476. doi: 10.1074/jbc.271.19.11468. [DOI] [PubMed] [Google Scholar]
- 22.Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Nat Cell Biol. 1999;7:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 23.Tzagoloff A. In: Mitochondria. Siekevitz P, editor. New York: Plenum; 1982. p. 238. [Google Scholar]
- 24.Davidson E H. Gene Activity in Early Development. New York: Academic; 1986. pp. 46–120. [Google Scholar]
- 25.Kobayashi S, Amikura R, Mukai M. Curr Biol. 1998;8:1117–1120. doi: 10.1016/s0960-9822(98)70466-x. [DOI] [PubMed] [Google Scholar]
- 26.Kashikawa M, Amikura R, Kobayashi S. Mech Dev. 2001;101:71–77. doi: 10.1016/s0925-4773(00)00553-0. [DOI] [PubMed] [Google Scholar]
- 27.Oka T, Amikura R, Kobayashi S, Yamamoto H, Nishida H. Dev Growth Differ. 1998;41:1–8. doi: 10.1046/j.1440-169x.1999.00409.x. [DOI] [PubMed] [Google Scholar]
- 28.Ogawa M, Amikura R, Akasaka K, Kinoshita T, Kobayashi S, Shimada H. Zool Sci. 1999;16:445–451. [Google Scholar]
- 29.Sato K, Sugita T, Kobayashi K, Fujita K, Fujii T, Matsumoto Y, Mikami T, Nishizuka N, Nishizuka S, Shojima K, et al. Dev Growth Differ. 2001;43:107–114. doi: 10.1046/j.1440-169x.2001.00558.x. [DOI] [PubMed] [Google Scholar]