Abstract
While next-generation sequencing has accelerated the discovery of human disease genes, progress has been largely limited to the “low hanging fruit” of mutations with obvious exonic coding or canonical splice site impact. In contrast, the lack of high-throughput, unbiased approaches for functional assessment of most noncoding variants has bottlenecked gene discovery. We report the integration of transcriptome sequencing (RNA-seq), which surveys all mRNAs to reveal functional impacts of variants at the transcription level, into the gene discovery framework for a unique human disease, microcephaly-micromelia syndrome (MMS). MMS is an autosomal recessive condition described thus far in only a single First Nations population and causes intrauterine growth restriction, severe microcephaly, craniofacial anomalies, skeletal dysplasia, and neonatal lethality. Linkage analysis of affected families, including a very large pedigree, identified a single locus on Chromosome 21 linked to the disease (LOD > 9). Comprehensive genome sequencing did not reveal any pathogenic coding or canonical splicing mutations within the linkage region but identified several nonconserved noncoding variants. RNA-seq analysis detected aberrant splicing in DONSON due to one of these noncoding variants, showing a causative role for DONSON disruption in MMS. We show that DONSON is expressed in progenitor cells of embryonic human brain and other proliferating tissues, is co-expressed with components of the DNA replication machinery, and that Donson is essential for early embryonic development in mice as well, suggesting an essential conserved role for DONSON in the cell cycle. Our results demonstrate the utility of integrating transcriptomics into the study of human genetic disease when DNA sequencing alone is not sufficient to reveal the underlying pathogenic mutation.
Noncoding mutations, which affect gene expression, regulation, or splicing, are estimated to cause ∼15%–30% of human Mendelian disease (Cartegni et al. 2002; Faustino and Cooper 2003; Lim et al. 2011; Lewandowska 2013). This could, in fact, be an underestimate since most genetic studies focus on coding regions (the exome) and immediately adjacent intronic splice sites whose effects are simpler to predict in silico using the amino acid code and basic splicing consensus sequences (Cartegni et al. 2002; Faustino and Cooper 2003; Pagani and Baralle 2004; Lopez-Bigas et al. 2005; Cooper and Shendure 2011; Lim et al. 2011; Lewandowska 2013). In contrast, noncoding variants are harder to interpret due to the lack of a functional understanding of most noncoding elements in the genome (Pagani and Baralle 2004; MacArthur et al. 2014). Therefore, even though most of the genome and most variants identified by whole-genome sequencing are noncoding, noncoding variants causing Mendelian disease are rarely identified before the affected gene has already been implicated by coding mutations. Although most gene regulation occurs outside of the exome—in noncoding regions such as promoters, enhancers, untranslated regions (UTRs), introns, and intergenic regions—if a genetic disease is caused by a noncoding mutation in a novel gene, it may not be possible to identify it with DNA sequencing alone among the large background of other noncoding variants. Furthermore, mutations in coding regions may also affect gene expression and splicing in addition to the protein sequence (Cartegni et al. 2002; Lopez-Bigas et al. 2005; Lewandowska 2013), effects that cannot be detected by DNA sequencing alone. As a result, even while high-throughput DNA sequencing technologies have led to remarkable progress in identifying mutations causing genetic diseases, important genetic disorders remain unsolved in which this approach fails, presumably because the pathogenic mutation is noncoding or is a coding region mutation affecting transcription.
Transcriptome sequencing (RNA-seq) has been key to revealing the complexity of gene regulation across cell types and states by its ability to profile transcript levels as well as alternative splicing patterns genome-wide (Pan et al. 2008; Wang et al. 2009; de Klerk and ‘t Hoen 2015), processes largely dictated by the noncoding genome. Nevertheless, transcriptome sequencing has not been an integral part of the gene discovery framework of human Mendelian disease studies. This might be because transcriptome sequencing ideally requires RNA from the diseased tissues of affected individuals, which is not always available, and entails the added cost and complexity of RNA sequencing and analysis relative to DNA sequencing alone.
We aimed to test whether RNA-seq could discover, in one experiment, a pathogenic noncoding mutation or transcription-altering exonic mutation causing human disease, without needing to test a large or unfeasible number of possible splicing or gene regulation defects by traditional molecular biology methods. We tested this approach by applying it to microcephaly-micromelia syndrome (MMS), a condition for which we had highly significant statistical linkage to a genetic locus but for which no obvious pathogenic coding or splicing mutation had been found by DNA sequencing.
MMS was first described in 1980 by Ives and Houston (Ives and Houston 1980) in a First Nations population in northern Saskatchewan in Canada. The syndrome's main clinical features are intrauterine growth restriction (IUGR), severe microcephaly, craniofacial dysmorphism, and marked limb malformations. Since the 1950s, an average of two pregnancies or births per year have been diagnosed with MMS in this population and, with the exception of two known children, all were stillborn or died within the first week of life, mostly within 24 h of birth. No cases of MMS have been identified outside of this First Nations population in Saskatchewan. While the clinical phenotype of MMS is distinctive, the constellation of IUGR, microcephaly, and limb anomalies places MMS in the broad category of microcephalic primordial dwarfism (MPD) syndromes, which have been associated with defects in genes involved in genome replication, the DNA damage response, and centrosome function (Klingseisen and Jackson 2011).
In this study, we sought to identify the genetic cause of MMS using a combined RNA-seq plus genome sequencing approach. We took this approach as a proof of principle to gauge the advantages and feasibility of integrating transcriptomics with genomics to reveal pathogenic noncoding mutations in unsolved human Mendelian diseases.
Results
Clinical features of microcephaly-micromelia syndrome
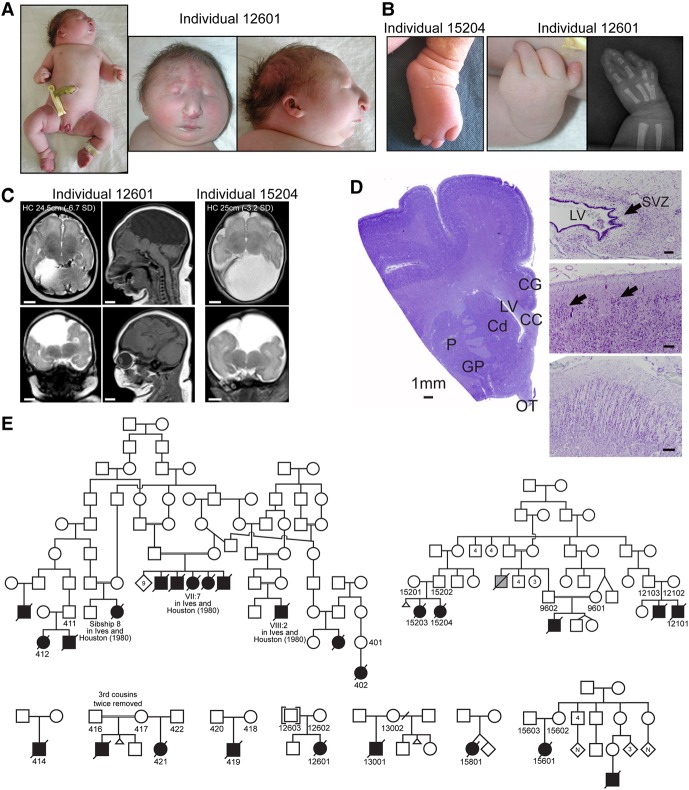
MMS (MIM 251230) is characterized by IUGR, marked microcephaly with distinctive craniofacial features, limb malformations, and nearly uniform perinatal lethality due to respiratory failure. The growth restriction and microcephaly are severe, with term (≥38-wk gestation) birth weights between 0.8–1.6 kg (average 1.2 kg; average z-score −6.5; n = 15), average head circumferences of 24 cm (z-score −7.4; n = 7), and average lengths of 34 cm (z-score −7.4; n = 12). Affected individuals have a characteristic facial appearance with a broad and beaked nose, short palpebral fissures, microstomia, micrognathia, low-set ears, and a short neck (Fig. 1A). Both upper and lower limbs are malformed, with upper limbs more severely affected—forearms are short, with frequent absence or significant underdevelopment of the radius and/or ulna and often humeroradial synostosis (Fig. 1A). Nearly all individuals have bilateral oligodactyly with absent thumbs, and most have absent or poorly developed fifth fingers that sometimes arise from a bifid metacarpal bone (Fig. 1B). Lower limbs are sometimes shortened with an underdeveloped fibula, feet are often clubbed with variable toe syndactyly, the great toes can be short and/or proximally placed, and in some cases, the fourth and fifth metatarsal bones and toes are underdeveloped or absent (Fig. 1B). Additionally, many affected individuals have complete craniosynostosis and other skeletal anomalies such as absence of one or two pairs of ribs.
Figure 1.
Microcephaly-micromelia syndrome phenotype and pedigrees. (A) Photographs of an affected individual (12601) illustrating the severe microcephaly, facial dysmorphisms, and limb anomalies characteristic of microcephaly-micromelia syndrome. (B) Photograph of a foot (individual 15204) and photograph and X-ray of a hand (individual 12601) showing both pre-axial (malformed toe and absent thumb) and post-axial (underdeveloped fifth metatarsal bone, and hypoplastic fifth digit arising from bifid fourth metacarpal bone) abnormalities. (C) Brain MRIs of two affected individuals showing the common structural brain abnormalities of MMS: profound microcephaly, simplified gyral pattern, markedly diminished white matter volume and myelination, hypoplastic or absent corpus callosum, aqueductal stenosis, and a small pons. Note the large dorsal interhemispheric cysts in both individuals, which was present in nearly every affected individual examined by MRI or autopsy to date. Cortical thickness is grossly normal and the cerebellar hemispheres are relatively large compared to the rest of the brain. Head circumferences (HC) and z-scores (number of standard deviations [SD] from the mean of newborns of the same gestational age at birth) are shown. MRI sequences were as follows: 12601: axial T2 (top left), mid-sagittal T1 (top right), left-sagittal T1 (bottom right), coronal T2-FIESTA (bottom left); 15204: axial T2 (top), coronal T2-HASTE (bottom). White scale bars = 1 cm. (D) Brain histology of MMS cases. (Left) Low-power Nissl-stained brain section of a child who died at 3 mo of age showing simplified gyral pattern and reduced white matter (CG) cingulate gyrus, (CC) corpus callosum, (LV) lateral ventricle, (Cd) caudate, (P) putamen, (GP) globus pallidus, (OT) optic tract). (Top right) Cresyl violet-stained brain section of a 35-wk-gestation newborn at the angle of the lateral ventricle (LV) showing decreased cells in the subventricular zone (SVZ; arrow). Bar = 100 μm. (Middle right) Hematoxylin- and eosin-stained section of the cerebral cortex of a 41-wk-gestation newborn demonstrating disorganized clusters of neurons (arrows) separated by cell-free zones in superficial layers. Bar = 100 μm. (Bottom right) Cresyl violet-stained section from a full-term newborn cerebral cortex demonstrating the persistence of radial columns of neurons separated by cell-sparse regions. Bar = 500 μm. (E) Pedigrees of the families with MMS profiled in this study. Individual IDs are labeled for individuals whose samples were profiled. The pedigree at the top left can be linked via individuals VII:7 and VIII:2 to the larger pedigree in the original description of the syndrome by Ives and Houston (Ives and Houston 1980). Gray symbol (top right pedigree) represents a child that died in infancy with limb anomalies, but the specific diagnosis of MMS was not confirmed. Deceased status is indicated with crossed-out symbols for affected individuals only and not for unaffected individuals. For simplicity, not all individuals of the pedigrees are illustrated. See Supplemental Data 1 for a list of all case samples in this study.
Brain MRIs of individuals with MMS show several characteristic anomalies, including profound microcephaly with only primary sulci and gyri, diminished white matter, a hypoplastic or absent corpus callosum, aqueductal stenosis, and a large interhemispheric cyst; other gross brain structures are present, and the cerebellum is relatively preserved (Fig. 1C). Histological analysis of the cerebral cortex shows a simplified gyral pattern, reduced white matter, decreased cells in the subventricular zone, and a disorganized distribution of cells with clusters of neurons and vertically oriented columns of neurons separated by cell-sparse zones (Fig. 1D).
The lungs of individuals with MMS are severely hypoplastic with anomalous lobation. As a result, nearly all cases identified to date were either stillborn or died within the first week of life due to respiratory failure; the only known exceptions are two affected children who died at 3 mo and 2 yr of age from respiratory complications. Additionally, cleft palate and cardiac, gastrointestinal, and genitourinary defects are observed in some cases. While IUGR, microcephaly, and dwarfism are also seen in MPD syndromes (Klingseisen and Jackson 2011), the combination of the characteristic craniofacial anomalies, limb malformations, and neonatal lethality is distinct and diagnostic for MMS.
Microcephaly-micromelia syndrome is linked to a locus on Chromosome 21q
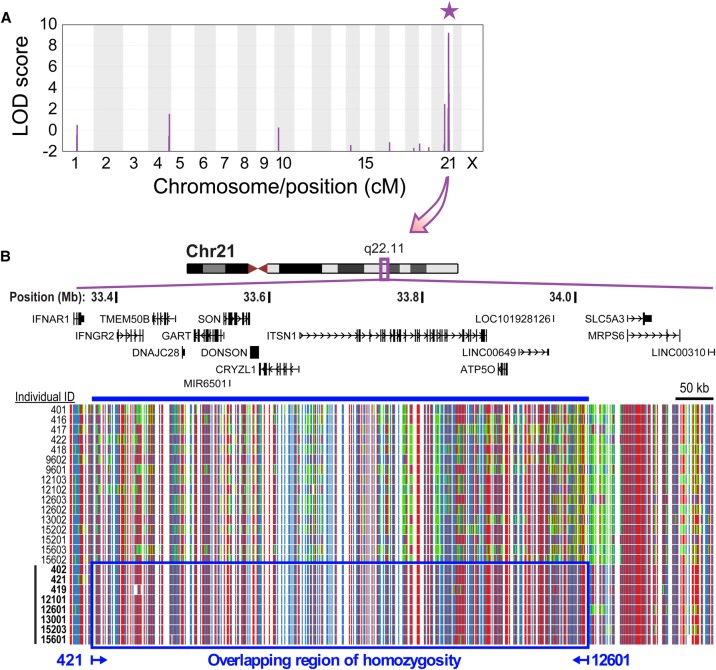
Most individuals with MMS in this study belong to a large and consanguineous pedigree of First Nations origin in Saskatchewan, showing autosomal recessive inheritance (Fig. 1E). Other cases belong to the same population and are known to be descendants of the founders of this pedigree, though their exact relationships are unknown (Fig. 1E; see Supplemental Data 1 for a list of all profiled individuals). High-density genome-wide single nucleotide polymorphism (SNP)-microarray genotyping of seven affected individuals and 15 unaffected parents followed by linkage analysis under a recessive inheritance model identified an 852-kb (1.2 cM) locus on Chromosome 21q22.11 definitively associated with the disease with a maximum combined logarithm of odds (LOD) score of 9.2 (Fig. 2A). Homozygosity and haplotype analysis using SNP-microarray genotypes of affected cases further narrowed this region to a 664-kb minimal overlapping region of homozygosity (ROH) (rs9978569 to rs4443074; Chr 21: 33,364,965–34,029,433; GRCh38/hg38) (Fig. 2B).
Figure 2.
Linkage and homozygosity analysis identifies a locus on Chromosome 21q22.11. (A) Linkage analysis using SNP-microarray genotypes of affected individuals and unaffected parents (see Supplemental Data 1 for list of genotyped individuals) identified a locus associated with the disease at Chromosome 21q22.11 with maximum LOD score of 9.2 (purple star; interval: Chr 21: 33,344,469–34,196,070; GRCh38/hg38). (B) SNP-microarray genotypes in the interval defined by linkage analysis (Fig. 2A). Each line represents an individual (unaffected parents on top and affected individuals labeled in bold on bottom). Each column in the SNP ideogram represents a SNP, with homozygous alleles in red or blue and heterozygous alleles in green. Affected individuals 421 and 12601 define a minimal region of overlapping homozygosity (ROH) at 21q22.11 (blue box and line; Chr 21: 33,364,965–34,029,433; GRCh38/hg38). RefSeq gene annotations are shown above. Low quality SNP calls are omitted. Note: samples 418 and 419 are shown here but these were not used for linkage analysis since they did not pass quality control filters (see Supplemental Methods).
Multimodal genome sequencing of the disease locus fails to identify plausible exonic or canonical splice mutations
We sequenced three affected individuals using three different methods: whole-exome sequencing (WES, individuals 412 and 13001), targeted-capture sequencing of the linkage region (individual 13001), and whole-genome sequencing (WGS, individual 12601). These methods covered 97.0%, 99.8%, and 97.9%, respectively, of the coding exome in the ROH with ≥10× read depth (see Supplemental Fig. 1A for coverage statistics). Targeted-capture and WGS covered 83% and 98% of the entire ROH (i.e., coding exons, introns, UTRs, and intergenic regions), respectively, with ≥10× read depth (Supplemental Fig. 1A). The variants identified by the three methods were highly concordant, though some variants were identified by only one or two of the three methods (Supplemental Fig. 1C,D; Supplemental Data 2). After filtering out common population variants, no rare coding (either synonymous or nonsynonymous) or canonical splice site mutations (i.e., within 2 bp of the intron-exon junction) were identified within the ROH by any of these sequencing approaches (Table 1; Supplemental Fig. 1B). In contrast, 18 rare intronic and 20 rare intergenic noncoding variants were identified in the ROH (Table 1; Supplemental Fig. 1B; Supplemental Data 2). Analysis of highly conserved noncoding elements in the region did not aid in further narrowing the list of noncoding variants.
Table 1.
Variants identified in the MMS region of homozygosity by genomic DNA sequencing
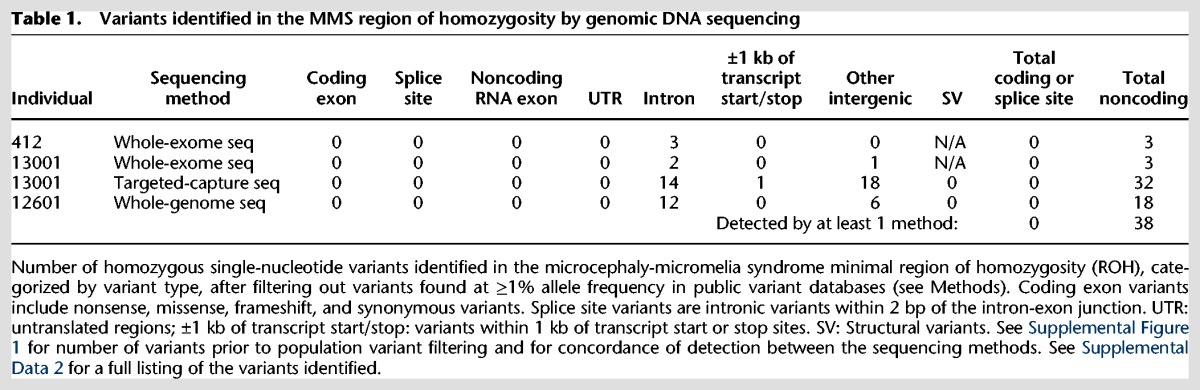
Furthermore, copy number variant (CNV) and structural variant analyses were performed using the targeted-capture and WGS data. CNV analysis was also performed by a targeted array comparative genomic hybridization experiment. None of these methods identified a CNV or structural variant in the ROH (Table 1; Supplemental Fig. 1B).
RNA-seq identifies a splicing defect associated with an intronic variant in DONSON
Since comprehensive genome-sequencing did not reveal a coding or canonical splicing mutation in the MMS ROH, we hypothesized that one of the 38 noncoding variants detected in the ROH may be the cause of the disease and that transcriptome profiling of affected samples could reveal which of these noncoding mutations causes MMS via cis effects on gene expression or splicing. We therefore performed RNA-seq of wild-type control (n = 13), heterozygous parent (n = 6), and homozygous MMS case samples (n = 11) derived from a variety of cell lines and tissues.
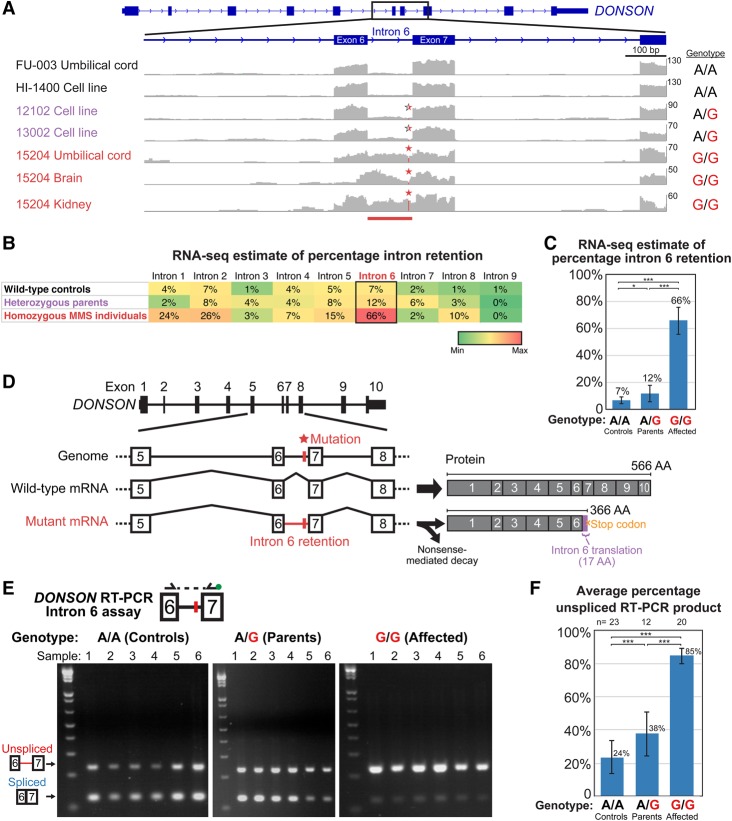
RNA-seq analyses of gene expression and splicing in the 30 samples from affected and unaffected individuals identified only one abnormality within the ROH in MMS samples: significantly increased retention of intron 6 of DONSON in MMS samples relative to heterozygous parent and wild-type control samples (Fig. 3A). Importantly, intron 6 of DONSON contained one of the 38 noncoding variants identified previously by genome sequencing, an A to G transition (in the transcript sense strand) located 9 bp upstream of the intron 6–exon 7 junction (DONSON [NM_017613.3]:c.1047-9A>G; Chr 21[NC_000021.8]:g.33582064T>C [GRCh38;hg38]) (Supplemental Data 2). The only two other variants in DONSON were located distantly in other introns, 1.2 kb (intron 5) and 2.4 kb (intron 4) away (Supplemental Data 2). Furthermore, MMS samples showed no evidence of any novel DONSON isoforms, such as might arise from activation of cryptic 5′ splice donor or 3′ splice acceptor sites within intron 6. Retention of intron 6 creates a premature stop codon after 52 bp of the 109-bp intron, which is predicted to lead to nonsense-mediated decay (NMD) of the mutant transcript (Wong et al. 2016) or to a truncated protein (366 amino acids versus wild-type 566 amino acids) that ends with 17 aberrant amino acids due to translation of the first half of intron 6 (Fig. 3D). Overall, DONSON expression levels in RNA-seq showed a trend toward lower expression in heterozygous parents and homozygous MMS cases, but these differences were not statistically significant (controls: 5.2 ± 2.8 fragments per kilobase per million mapped reads [FPKM mean ± std dev], parents: 3.9 ± 2.4 FPKM, MMS cases: 3.4 ± 1.7 FPKM; P-value >0.05 for all comparisons). However, a more sensitive TaqMan qPCR assay confirmed significantly decreased DONSON transcript levels in the presence of the MMS variant (56% of wild-type levels in heterozygous cell lines; 95% confidence interval: 44%–72%) (Supplemental Fig. 2C), consistent with NMD of the mutant transcript. Altogether, these results suggest that MMS is caused by the c.1047-9A>G noncoding variant in DONSON via an intron retention mechanism.
Figure 3.
RNA-seq identifies an intron-retention splicing defect associated with an intronic variant in DONSON. (A) RNA-seq read coverage for representative samples shows aberrant retention of intron 6 of DONSON (red bar) in affected individuals associated with the c.1047-9A>G noncoding mutation (Chr 21: g.33582064:T>C; red asterisk). The interval shown is Chr 21: 33,580,994–33,583,594 (GRCh38/hg38), illustrated in the reverse strand direction. The genotype of each sample is shown on the right. Homozygous MMS, heterozygous parent, and wild-type control sample names are colored red, purple, and black, respectively. Read coverage graph Y-axes are scaled (numbers on right side of Y-axis) to show the maximum coverage of each sample in the interval. (B) RNA-seq quantification of intron retention for each intron of DONSON, based upon pooling all the RNA-seq samples of each genotype (see Supplemental Methods for details). Cells are shaded green to red according to the percentile between the minimum and maximum values in the table. The aberrant retention of intron 6 is highlighted. The table also shows the mild increase in intron 6 retention in heterozygous parents relative to wild-type controls and the baseline low-level retention of intron 6 in controls relative to other introns. MMS individuals also showed a trend of increased retention of other introns upstream of intron 6, suggesting that impaired splicing of intron 6 might affect splicing of other introns; however, the mechanism by which this would occur is unclear. (C) RNA-seq quantification of intron 6 retention calculated as in Figure 3B. Error bars are 95% confidence intervals (see Supplemental Methods). All group comparisons were significant: controls versus parents: P = 0.03; controls versus affected: P < 10−15; parents versus affected: P < 10−15 (Fisher's exact test with Holm multiple comparisons adjustment). (D) Schematic of the intron retention splicing defect caused by the c.1047-9A>G (Chr 21: 33582064 T>C) mutation in intron 6 in microcephaly-micromelia syndrome. Retention of intron 6 would lead to either nonsense-mediated decay of the transcript due to the stop codon within intron 6 or to a truncated protein. On the right are the predicted wild-type and truncated mutant proteins and their amino acid (AA) lengths. Translation of the first part of the aberrantly retained intron 6 creates 17 amino acids followed by a premature stop codon. (E) RT-PCR spanning from exon 6 to exon 7 of DONSON (top schematic) in various tissues confirms increased retention of intron 6 in MMS samples, which are homozygous for the Chr 21: 33582064 T>C mutation, compared to heterozygous parents and wild-type controls (unspliced transcript with intron 6: 230 bp; spliced transcript: 121 bp). Shown here are six representative samples for each genotype. (Note that the variant is A>G in the DONSON transcript strand and T>C in the genomic plus strand). See Supplemental Figure 2A for RT-PCR gel images of all assayed samples. The exon 7 PCR primer contains a FAM fluorescent label (green circle) for quantification of PCR products (Fig. 3E). Wild-type (T/T) samples: 1- FU-009 umbilical cord; 2- FU-006 umbilical cord; 3- FU-004 umbilical cord; 4- fetal liver; 5- fetal brain; 6- cerebellum. Heterozygous (T/C) parent samples: 1- 15603 cell line; 2- 15602 cell line; 3- 15202 cell line; 4- 15201 cell line; 5- 15202 blood sample a; 6- 15202 blood sample b. Homozygous (C/C) MMS samples: 1- 15204 brain (RNAlater); 2- 15204 brain (fresh-frozen sample a); 3- 15204 brain (fresh-frozen sample b); 4- 15204 heart (RNAlater); 5- 15204 heart (fresh-frozen); 6- 15204 kidney (RNAlater). (F) Quantification of the RT-PCR intron 6 retention assay products. RT-PCR was performed with the PCR primer for exon 7 containing a FAM-fluorescent label allowing quantification of the RT-PCR products with a capillary electrophoresis DNA analyzer (see Methods). Percentage unspliced RT-PCR product was calculated as [Area of unspliced band]/[Area of unspliced band + Area of spliced band], and averaged across all samples of each genotype (number of samples in each group is shown on top). Groups were significantly different from each other (Controls versus Parents, P = 0.005; Controls vs. Affected, P < 10−22; Parents vs. Affected, P < 10−7; two-tailed unpaired t-test). Importantly, note that this measurement can be used to evaluate relative splicing differences between genotypes but is not an absolute measurement of splicing, since the PCR amplification efficiencies of the unspliced and spliced products differ. See Supplemental Figure 2A for percentage unspliced RT-PCR product of all assayed samples and Supplemental Figure 2B for percentage unspliced RT-PCR product summarized by tissue type for brain, umbilical cord, and cell lines/blood leukocytes.
Genotyping of the intron 6 noncoding variant across all available samples from the extended MMS pedigrees confirmed it was homozygous in all affected individuals and heterozygous in all parents (Supplemental Data 1). The variant was absent from the NHLBI Exome Sequencing Project (6503 individuals), the 1000 Genomes Project, a set of 69 Complete Genomics control genomes, and 1464 unrelated exomes previously sequenced by our laboratory. The variant was found in the heterozygous state in one of 736 additional neurologically normal control samples that we genotyped and in six of 121,390 chromosomes in the Exome Aggregation Consoritum (ExAC) database, all in the heterozygous state in European individuals (Supplemental Data 2). The locus of this intronic variant is captured well by exome sequencing due to its relative proximity to an intron-exon junction (e.g., the locus was called in 121,390 of 121,412 chromosomes in ExAC), so its low frequency in control exomes was not due to inefficient capture. The extremely low allele frequency of the variant across these studies (5 × 10−5), along with the unique hypomorphic nature of the allele and the essential requirement for DONSON in body development (see below analyses), is consistent with the extreme rarity of the microcephaly-micromelia syndrome.
In order to independently confirm and quantify the splice defect identified by RNA-seq, we designed a reverse transcriptase-polymerase chain reaction (RT-PCR) assay for intron 6 retention using primers flanking intron 6, from exon 6 to exon 7. The assay confirmed the intron 6 retention defect in all MMS samples and showed a mild but detectable increase in intron retention in heterozygous parents relative to control samples (Fig. 3E; Supplemental Fig. 2A). Interestingly, the assay also showed that splicing of intron 6 is not perfectly efficient in control samples, with low and variable levels of intron 6 retention in normal tissues, though significantly less than in the MMS samples (Fig. 3E; Supplemental Fig. 2A). The RT-PCR assay employed a fluorescent primer (Fig. 3E schematic), allowing quantification of the two RT-PCR products (spliced product and intron 6 retention product). The intron 6 retention amplicon was, on average, 85 ± 5% (standard deviation) of the total RT-PCR product in homozygous MMS samples, 38 ± 13% in heterozygous parent samples, and 24 ± 10% in wild-type control samples (Fig. 3F). All differences in intron 6 splicing between the groups of samples were statistically significant (controls vs. parents, P = 0.005; controls vs. affected, P < 10−20; parents vs. affected, P < 10−5; two-tailed unpaired t-test with Holm multiple comparisons adjustment) (Fig. 3F). These RT-PCR results confirm a significant intron 6 retention splice defect of DONSON in MMS samples, a small but detectable increase in intron 6 retention in heterozygous parents, and that intron 6 splicing is not perfectly efficient in normal tissues.
The above RT-PCR assay allows robust comparisons of the relative efficiency of intron 6 splicing between samples; however, because the RT-PCR amplification efficiency of the spliced and unspliced products may differ due to their different sizes, the assay might not be a fully accurate measure of absolute splicing efficiency. We therefore also estimated intron 6 retention using the RNA-seq data by counting the number of intron–exon junction spanning reads (reflecting intron retention) versus exon 6–exon 7 splice reads (reflecting intron splicing). In MMS samples, 66% of these reads at the intron 6 locus were intron–exon junction spanning reads that arose from DONSON transcripts retaining intron 6, versus 12% in heterozygous parents and 7% in controls (Fig. 3A–C). The differences between each category of samples were statistically significant (controls versus parents: P = 0.03; controls versus affected: P < 10−15; parents versus affected: P < 10−15; Fisher's exact test with Holm multiple comparisons adjustment). Furthermore, in heterozygous parent samples, more intron 6-retaining transcripts were derived from the mutant allele than the wild-type allele: 69% of RNA-seq reads at the intron 6 mutation locus (pooled from all heterozygous samples) contained the mutation (P = 0.02; binomial test versus the expected 50% if there were no association between the intron retention and the mutant allele). The above RNA-seq results again confirm that (1) individuals with MMS have a hypomorphic DONSON allele in which most transcripts retain intron 6, while a small fraction of transcripts are correctly spliced, (2) there is a small increase in intron 6 retention in heterozygous parents relative to controls, (3) the intron 6 variant is associated in cis with intron 6 retention, and (4) there is baseline low-level retention of intron 6 in wild-type tissues.
Notably, plotting of intron retention estimated by RNA-seq across all introns of DONSON and all sample types showed not only the specific intron 6 retention defect, but also that in wild-type control tissues, intron 6 is one of the gene's least efficiently spliced introns (Fig. 3B). Therefore, the low level of intron 6 retention seen in control samples by RNA-seq and RT-PCR is not due to overall splicing inefficiency in the assayed samples or global capture of unspliced transcripts during RNA-seq sample preparation, but rather a specific feature of intron 6. The baseline decreased efficiency of intron 6 splicing compared to other introns might explain the intron's susceptibility to the MMS c.1047-9A>G intronic mutation.
DONSON is essential for early embryonic development and is associated with components of the DNA replication machinery
In order to determine how the MMS phenotype relates to DONSON function during development, we assessed DONSON expression in human and mouse embryos by in situ hybridization. In both human and mouse, we found that DONSON is expressed in multiple organs of the developing embryo, including the brain, heart, lungs, gastrointestinal tract, limbs, and kidneys (Supplemental Figs. 3A–C), which grossly corresponds to the organs affected in MMS.
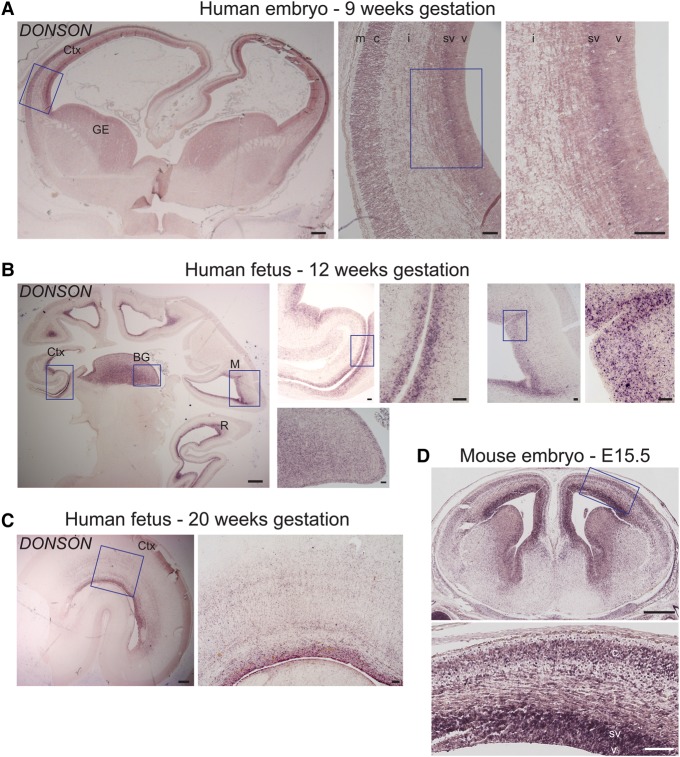
Because MMS has a severe microcephaly phenotype, we assessed DONSON expression in greater detail in human and mouse embryonic and fetal brains. These in situ hybridization studies found that DONSON is highly enriched in the ventricular and subventricular zones of the neocortex, which contain proliferating progenitor cells, as well as in the cortical plate containing newborn neurons, and the ganglionic eminences (Fig. 4A–D). This pattern of expression is consistent with the observed simplified gyral pattern, reduced white matter, and disorganized cortical columns seen in histology of MMS brains (Fig. 1D). We also investigated DONSON expression in a prior transcriptomic study of laser-microdissected regions of the developing human brain (Miller et al. 2014) and found a similar pattern of expression: in 15- and 16-wk-post-conception (wpc) human fetal brains, DONSON is highly expressed in the ventricular and subventricular zones, respectively; it is highly expressed in the ganglionic eminence of 15-wpc brain and subplate of 16-wpc brain, and in 21-wpc neocortex, DONSON expression is highest in the cortical plate (Supplemental Fig. 4A). Loss of DONSON function in early neocortical progenitor cells is consistent with the profound microcephaly and brain malformations of MMS. Furthermore, assays of DONSON expression across a broad range of prenatal and adult time points as part of the Human Brain Transcriptome project (Kang et al. 2011) show that DONSON expression is higher in prenatal brain relative to adult brain across all profiled brain regions (Supplemental Fig. 4B), supporting an important role for DONSON specifically during brain development.
Figure 4.
DONSON expression in human and mouse brain development. (A) DONSON expression by in situ hybridization in a coronal section of a 9-wk-gestation human fetal brain. Expression is prominent in the neocortex subventricular zone, which contains progenitor cells, and in the cortical plate, where newly born neurons reside. Expression is also seen in the ganglionic eminences, which give rise to the basal ganglia and interneurons that migrate into the neocortex. Scale bar for main image (left) is 500 µm; scale bars for other images are 100 µm. Hybridization was performed with antisense probe 1 (see Supplemental Methods). (Ctx) Neocortex, (GE) ganglionic eminence, (v) ventricular zone, (sv) subventricular zone, (i) intermediate zone, (c) cortical plate, (m) marginal zone. (B) DONSON expression by in situ hybridization in a sagittal section of a 12-wk-gestation human fetal brain. Expression is prominent in the basal ganglia (BG) and the ventricular and subventricular zones and cortical plate of the neocortex (Ctx), mesencephalon (M, midbrain), and rhombencephalon (R, hindbrain). Scale bar for main image (left) is 1000 µm; scale bars for other images are 100 µm. Hybridization was performed with antisense probe 1. (C) DONSON expression by in situ hybridization in a coronal section of a hemisphere of a 20-wk-gestation human fetal brain. Expression is evident in the ventricular and subventricular zones, intermediate zone, and cortical plate of the neocortex (Ctx). Scale bar for main image (left) is 1000 µm; scale bar for other image is 100 µm. Hybridization was performed with antisense probe 2. (D) DONSON expression by in situ hybridization in a coronal section of an E15.5 mouse brain. Expression is evident in the ventricular (v) and subventricular zones (sv), intermediate zone (i), and cortical plate (c) of the neocortex. Scale bar for top and bottom images are 500 and 100 µm, respectively. Hybridization was performed with human antisense probe 3. For all above tissue sections, negligible signal was observed with sense sequence probes in adjacent sections (Supplemental Fig. 3D), confirming specificity of the antisense probe staining.
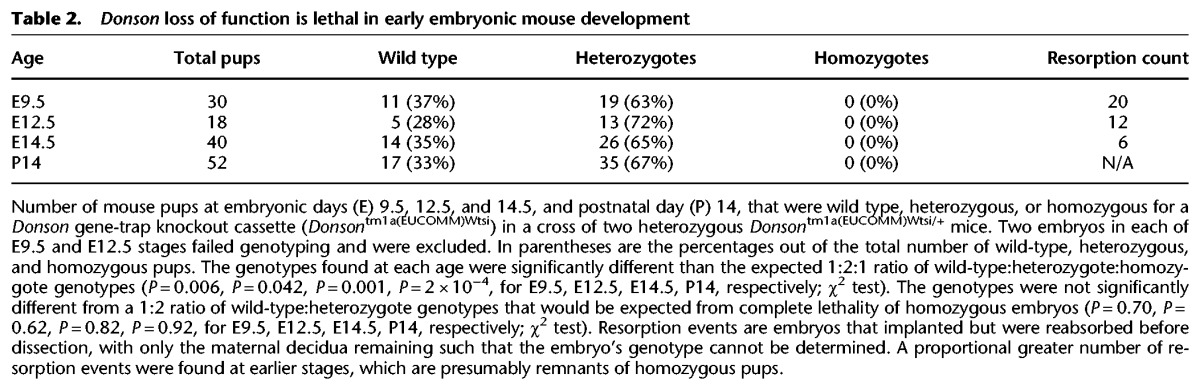
To confirm the essential role of DONSON in prenatal development, we also analyzed the phenotype of Donson knockout mice, which were created as part of the International Knockout Mouse Consortium. Matings between mice heterozygous for a Donson loss-of-function allele did not yield any homozygous pups, and genotyping of E9.5, E12.5, and E14.5 embryos did not detect any homozygous embryos (Table 2), indicating that complete loss of DONSON function is lethal during early murine embryonic development. Heterozygous knockout mice were phenotyped on >150 anatomic and laboratory measures (including brain weight) via the standardized International Mouse Phenotyping Consortium protocol (Supplemental Methods; White et al. 2013), with no major discernible abnormalities detected, though it is possible that subtle brain phenotypes could be revealed by additional detailed neuroanatomic studies. The essential role of DONSON in organismal development is also supported by phylogenetic analyses—DONSON is conserved across all multicellular eukaryotes, with orthologs found in mammals, birds, reptiles, insects, roundworms, plants, and fungi (Supplemental Fig. 5B; Bandura et al. 2005).
Table 2.
Donson loss of function is lethal in early embryonic mouse development
We utilized public proteomic and gene expression databases to identify complexes and proteins with which DONSON may function in the cell cycle. The Drosophila Protein Interaction Map project, which used tagged proteins to profile the Drosophila proteome, found that Humpty Dumpty, DONSON's ortholog in fly, is the highest-ranked interactor of Asf1 (Guruharsha et al. 2011; Chatr-Aryamontri et al. 2015). Asf1 is a histone chaperone that coordinates nucleosome assembly on newly replicated DNA with chromatin unwinding by the MCM2-7 prereplicative and replication helicase complexes. This points to a role for DONSON in the genome replication machinery.
Next, we mined the COXPRESdb database (Okamura et al. 2015), which collates thousands of microarray and RNA-seq samples from various species to create robust lists of co-expressed genes, and found further independent validation of an association between DONSON and components of the DNA replication and replication fork machinery. In Drosophila, more than half of the top 20 genes most highly co-expressed with humpty dumpty have known roles in the DNA replication complexes, including (1) DNA polymerase subunits involved in initiation of replication: DNApol-alpha73 [POLA2] and DNApol-alpha60 [PRIM2] (human homologs in brackets), (2) DNA polymerase processivity factors: RfC38 [RFC3], CG8142 [RFC4], and CG11788 [DSCC1] that help load the PCNA sliding clamp, which mediates polymerase processivity, onto primed DNA (Bermudez et al. 2003; Bowman et al. 2004), (3) DNA ligase 1 (DNA-ligI [LIG1]), which joins Okazaki fragments during DNA replication and is also associated with PCNA, (4) components of the DNA replication pre-initiation and replication fork complexes: CG3430 [MCMBP], Mcm3 [MCM3], Cdc45 [CDC45], (5) Orc2 [ORC2], a component of the origin recognition complex, and (6) regulators of the cell cycle: Lethal-(2)-denticleless [DTL]—which codes for a ubiquitin ligase targeting key cell cycle regulators and associated with PCNA and the DNA replication licensing factor CDT1 (Higa et al. 2006), and Cortex—a member of the Cdc20/fizzy family of cell cycle regulators (Nadeau et al. 2016).
In the human COXPRESdb data, the top genes most highly co-expressed with DONSON were again strikingly enriched for genes coding for components of the DNA replication machinery, including (1) the ORC1 origin recognition complex subunit 1, (2) CDC6, which together with CDT1 loads the MCM2-7 replication helicase onto replication origins, (3) GINS1, a component of the active CMG (CDC45/MCM2-7/GINS) replication helicase, (4) MCM4 and MCM6, components of the MCM2-7 helicase, (5) POLE2, a subunit of DNA polymerase-epsilon, (6) MCM10, an essential component of the replication fork that mediates the association of MCM2-7 and DNA polymerase-alpha with replication origins (Homesley et al. 2000; Ricke and Bielinsky 2004), (7) CHAF1B, a subunit of the chromatin assembly factor that loads histones onto newly replicated DNA (Volk and Crispino 2015), and (8) cell cycle regulators: CDK1, DTL, cyclin E2, and cyclin A2 (a regulator of origin of replication firing).
A similarly remarkable enrichment of replication complex genes was seen in mouse and rat COXPRESdb data, including Orc6, Cdt1, Cdc6, Mcm2, Mcm3, Mcm4, Pcna, Dscc1, Fen1 (the endonuclease processing Okazaki fragments), and cyclin E1—all of which were within at least one of the two species’ top 20 genes most highly co-expressed with Donson.
Finally, to confirm an effect of DONSON loss on cell cycle progression, we assayed the expression of a panel of cell cycle genes (i.e., cyclins, cyclin-dependent kinases, checkpoint regulators) by qPCR after siRNA knockdown of DONSON in HeLa cells. After DONSON knockdown, the most significantly up-regulated gene in the panel was CDKN1A (p21), and the most significantly down-regulated genes were cyclin D2 and cyclin E2 (Supplemental Fig. 4C). Since p21 inhibits, and cyclins D2 and E2 mediate, the G1/S phase transition (Vermeulen et al. 2003), these results are consistent with arrest prior to or slowed progression through S phase seen with DONSON loss in fly and HeLa cells in prior studies (Bandura et al. 2005; Fuchs et al. 2010). It also suggests that p21 up-regulation mediates the cell cycle arrest triggered by DONSON loss. Further evidence that DONSON loss impairs cell proliferation is that, despite multiple attempts, we have been unable to culture cell lines from affected MMS patients—the cells die after their derivation and cannot be expanded in vitro, while multiple cell lines from their unaffected parents grow normally.
Altogether, the above data strongly suggest that DONSON is essential for genome replication, and therefore cell proliferation and early embryonic development, via a regulative or integral function in the prereplication and/or replication fork complexes, which notably, are disrupted in other MPD syndromes.
Discussion
Here, we present a noncoding mutation in DONSON as the cause of microcephaly-micromelia syndrome and link DONSON to the key complexes mediating genome replication that are disrupted in other microcephalic primordial dwarfism syndromes. Identification of this unusual hypomorphic mutation—in an essential gene whose complete loss of function we predict would otherwise be embryonic-lethal—was facilitated by a genomic plus transcriptomic integrated approach that serves as a model for discovering causes of other Mendelian diseases for which genome sequencing alone has been unsuccessful.
A transcriptomic approach facilitates the discovery of variants causing Mendelian disease
Our study illustrates how transcriptome sequencing (RNA-seq) can provide in vivo functional evidence for rapid identification of pathogenic variants, in particular those that alter gene expression or splicing. While our study identified an unexpected noncoding mutation in a novel disease gene, several prior studies have also shown the potential of RNA-seq for discovering splicing perturbations in known human disease genes (Chandrasekharappa et al. 2013), splicing defects caused by variants in canonical (i.e., predictable) splice sites of genes not previously associated with disease (Wang et al. 2013), fusion transcripts in cancer (Pierron et al. 2012; Morin et al. 2013), and RNA-seq confirmation of known transcription-altering mutations from genetic screens in model organisms (Miller et al. 2013). These studies also identified decreased transcript levels due to nonsense-mediated decay and other unexpected splice defects such as missense and synonymous exonic mutations causing exon skipping (Chandrasekharappa et al. 2013; Miller et al. 2013). We have also previously used RNA-seq to confirm a splicing defect that we had first identified by traditional molecular biology methods in the ZNF335 gene causing microcephaly (Yang et al. 2012). RNA-seq showed that the pathogenic missense mutation in the last base of one of the gene's exons caused retention not just of the immediately following intron, but, unexpectedly, also the preceding intron. These examples emphasize the complex and unpredictable nature of splicing mutations and the utility of RNA-seq as a high-throughput adjunct assay to DNA sequencing in assessing the transcriptional impact of both coding and noncoding variants.
RNA-seq has several additional important advantages. First, RNA-seq might allow direct identification of the pathogenic mutation and transcriptional defect without prior variant filtering or linkage analysis. While in our study of DONSON, the retained intron 6 is short and cannot provide such genome-wide power of detection, in some cases, RNA-seq of a single proband could be sufficient for identifying the disease-causing variant. For example, in our earlier study of ZNF335, the retained long intron ranked as the fourth most significant differentially transcribed intron out of >350,000 RefSeq-annotated introns, and the other top entries were false-positives due to unannotated exons (Yang et al. 2012). Situations that could feasibly provide such power for genome-wide detection would be the retention of long introns and aberrant splice donor or acceptor sites, which provide greater signal to background ratios than other defects such as exon-skipping that are more common normal alternative splice events.
Second, RNA-seq can reveal allelic association of variants to abnormal transcripts, providing additional functional evidence of causality. In heterozygous samples, biased expression of an aberrant transcript from the variant allele, as seen with the DONSON mutation, indicates that the variant causes the abnormal transcript in cis. Third, RNA-seq provides a functional read-out of the entire in vivo context of variants, which is not fully recapitulated in minigene and other reporter assays.
On the other hand, RNA-seq is limited by its dependence on whether the affected gene is normally expressed in the available tissues at sufficient levels to allow detection of abnormal transcripts. The feasibility of a transcriptomic approach therefore depends on the studied disease and the accessibility of affected tissues. One possible approach when affected tissues are not available may be to create induced pluripotent stem cells from patient samples and to differentiate them into the cell types affected by the disease, thereby allowing RNA-seq analysis of the disease-relevant mRNA species.
Intron retention as a mechanism of genetic disease
While progress has been made in understanding the mechanisms of RNA splicing and how splicing patterns relate to DNA sequence (Barash et al. 2010; Xiong et al. 2015), much of the genomic code mediating splicing remains unknown (Lewandowska 2013; Guigo and Valcarcel 2015; Lee and Rio 2015). Intron retention in particular has been the least studied alternative splicing pattern. Recent work, however, has shown that intron retention might be a conserved and more common mode of transcript regulation than previously appreciated (Galante et al. 2004; Braunschweig et al. 2014; Boutz et al. 2015; Mudvari et al. 2015; Wong et al. 2016), with examples found in hematopoiesis (Wong et al. 2013; Edwards et al. 2016), T-cell activation (Ni et al. 2016), and neuronal development and activity (Bell et al. 2010; Yang et al. 2012; Yap et al. 2012). Inappropriate intron retention has also been identified in diverse genetic diseases, including cancer (Seifert et al. 2006; Tanackovic et al. 2011; Dvinge and Bradley 2015; Jung et al. 2015; Ortega-Recalde et al. 2015; Kallabi et al. 2016). Although there are tools that can predict the effects of sequence variants on alternative exon use or alternative 5′ donor or 3′ acceptor splice sites (Jian et al. 2014), to our knowledge, there are no computational tools to predict intron retention from sequence data. The increasingly appreciated role of intron retention in normal physiology and disease therefore supports the need for empirical methods such as transcriptomics to address this gap.
Intron retention may regulate normal gene expression or cause disease not only by introducing additional protein sequence, but also by NMD or by preventing export of the transcript from the nucleus if the retained intron introduces a premature stop codon (Ge and Porse 2014; Wong et al. 2016). The trend toward lower DONSON levels in MMS samples suggests that the intron retention leads to at least some NMD. Interestingly, in normal tissues, intron 6 of DONSON is both less efficiently spliced out compared to other introns and is variably spliced across tissues. This suggests that perhaps, more generally, inefficiently spliced introns might be more vulnerable to disruption by genetic mutation.
A nonconserved noncoding variant causing disease
Evolutionary conservation is often used to predict the likelihood of variant pathogenicity. However, while DONSON is conserved across all multicellular eukaryotes (Supplemental Fig. 5A,B), the wild-type allele at the location of the MMS intronic mutation is not well conserved (Supplemental Fig. 5A). This illustrates that conservation analyses cannot be exclusively relied upon to predict the clinical significance of noncoding variants.
DONSON is associated with genome replication complexes disrupted in other microcephalic primordial dwarfisms
Clinically, MMS is a microcephalic primordial dwarfism, a class of heterogeneous disorders characterized by prenatal and postnatal growth restriction along with microcephaly (Klingseisen and Jackson 2011; Khetarpal et al. 2016). MPD syndromes are classified into four types based on their clinical features and affected cellular pathways, all of which are involved in some aspect of cell cycle progression (Klingseisen and Jackson 2011; Fenwick et al. 2016; Khetarpal et al. 2016). Seckel syndrome is caused by mutations in DNA damage response signaling and centriole biogenesis factors (ATR, ATRIP, CEP152, CENPJ). Microcephalic osteodysplastic primordial dwarfism (MOPD) type I/III is caused by mutation of RNU4ATAC, a component of the minor spliceosome mediating U12-intron splicing in many genes including some involved in DNA replication. MOPD type II is caused by mutations in PCNT encoding a key centrosomal protein. Meier-Gorlin syndrome (MGS) is caused by mutations in components of the prereplication and pre-initiation DNA replication complexes (ORC1, ORC4, ORC6, CDT1, CDC6, GMNN, CDC45), which license replication origins and mediate loading and functioning of the replication fork helicase (Bicknell et al. 2011; Burrage et al. 2015; Fenwick et al. 2016). Although MMS is distinct from these four classic types of MPD syndromes, in this study we have presented genetic, transcriptomic, and proteomic evidence linking MMS and DONSON to other MPD syndromes, in particular, MGS.
Multiple independent lines of evidence strongly suggest an essential role for DONSON in the same DNA prereplication and replication fork complexes affected in MGS. First, expression of both human DONSON and its fly ortholog Humpty Dumpty (Hd) peaks in late G1 and S-phase when origins of replication complexes assemble and genome replication takes place (Whitfield et al. 2002; Bandura 2005; Bandura et al. 2005; Fuchs et al. 2010). Second, endogenous Hd localizes to the nucleus in foci that overlap, though not exclusively, with origins of replication (Bandura et al. 2005). Third, genetic loss of Hd in fly and siRNA knockdown of human DONSON leads to impaired genome replication, arrest prior to or slowed progression through S-phase of the cell cycle, and impaired cell proliferation (Bandura 2005; Bandura et al. 2005; Fuchs et al. 2010). Hd and DONSON depletion also lead to an increase in histone variants marking DNA double-strand breaks, which could be a consequence of stalled replication forks (Bandura et al. 2005; Fuchs et al. 2010). Notably, Hd-null fly mutants, which survive until the metamorphosis stage due to maternally supplied transcripts, have small brains and absent imaginal discs (Bandura et al. 2005), the structures that give rise to the legs, wings, antennae, and other external structures—a phenotype reminiscent of MMS despite the vast phylogenetic distance between fly and human. Fourth, we identified numerous components of the prereplication and replication fork complexes that are highly co-expressed with DONSON in multiple species spanning from fly to human. Remarkably, our co-expression analysis identified five of the seven known MGS genes (all except ORC2 and GMNN), and GMNN (geminin), which was not identified in the co-expression analysis, is a genetic enhancer of the humpty dumpty phenotype in fly double mutants (Bandura 2005). Fifth, at the protein level Hd interacts with Asf1, a histone chaperone associated with the MCM2-7 replication helicase.
Mechanistically, it is not unexpected that impaired genome replication due to the reduction of DONSON leads to impaired cellular proliferation and decreased overall growth, especially of organs such as the brain whose development depends on a high number of progenitor cell mitoses. DONSON is essential for normal embryonic and fetal development and its complete absence is lethal, so the survivability of MMS is presumably due to the hypomorphic nature of its allele. Between two-thirds and ∼90% of DONSON transcripts are not properly spliced in MMS, suggesting that additional MPD cases, particularly MGS-like syndromes, might be caused by other DONSON mutations that are less severe.
Indeed, during review of this manuscript, Reynolds et al. (2017) published a series of cases of recessive, hypomorphic mutations in DONSON causing primary microcephaly, microcephaly with short stature, and MPD. Most of the phenotypes they report are nonlethal and milder than MMS, implying that the mutations had milder effects on DONSON function than the MMS mutation. However, they also reported one family (P21) from Saudi Arabia that resembles the phenotype of MMS in the First Nations population and which, remarkably, contains the same homozygous mutation, suggesting that a range of hypomorphic DONSON mutations cause a wide spectrum of MPD and microcephalic syndromes. Reynolds et al. (2017) further present extensive cell biological evidence for an essential role for DONSON in the replication fork mediating genome replication. Altogether, the unique MMS cases we have studied and the above related syndromes define DONSON as a novel human disease gene and will help inform future investigations of its essential function in cellular replication and organismal development.
Methods
Human subjects and samples
All human studies were reviewed and approved by the Research Ethics Committee of the University of Saskatchewan and the Committee on Clinical Investigation of Boston Children's Hospital. The study was also supported throughout the project by the Northern Medical Services of the University of Saskatchewan and leaders of the local First Nations community. See Supplemental Methods for further details. All samples in the study are listed in Supplemental Data 1.
Linkage analysis
Multipoint parametric linkage analysis was performed using Illumina Omni 2.5 SNP array data analyzed with Plink (Purcell et al. 2007) and Merlin (Abecasis et al. 2002) under a recessive model. See Supplemental Methods for full details.
Genome sequencing
Targeted-capture sequencing was performed using a Roche NimbleGen custom 385K capture array. Paired-end sequencing libraries were generated after capture and sequenced on an Illumina sequencer. Whole-exome sequencing libraries were prepared using the Sure-Select Human All Exon v2 kit (Agilent) and sequenced on an Illumina sequencer. Whole-genome sequencing was performed by Complete Genomics. Targeted-capture and whole-exome data were analyzed using GATK (Van der Auwera et al. 2013). Whole-genome sequencing data were analyzed using Complete Genomics software. Variants were annotated with ANNOVAR (Wang et al. 2010) and filtered if their allele frequency was ≥1% in any of the following public variant databases: ExAC (Lek et al. 2016), 1000 Genomes Project (The 1000 Genomes Project Consortium 2015), NHLBI Exome Sequencing Project (Tennessen et al. 2012), and Complete Genomics 69 control genomes (Drmanac et al. 2010). See Supplemental Methods for further details.
RNA sequencing
RNA-sequencing libraries were prepared with the Illumina TruSeq Stranded mRNA kit and sequenced on HiSeq 2000 Illumina sequencers. Reads were aligned to the human reference genome (GRCh38/hg38) with HISAT2 (Kim et al. 2015) using standard settings. See Supplemental Methods for full details.
RT-PCR validation and relative splicing quantification
Residual DNA was eliminated from RNA samples using TURBO DNA-free (Ambion), and cDNA was synthesized using oligo-dT primers. RT-PCR was performed with primers in exon 6 and exon 7, flanking intron 6. The exon 7 primer was labeled with FAM to allow RT-PCR product quantification and calculation of percentage unspliced product on a 3730 DNA Analyzer capillary electrophoresis instrument (Applied Biosystems). See Supplemental Methods for full details.
In situ hybridization in human and mouse embryos
Human embryo and fetus sections were obtained by the Joint MRC/Wellcome Trust (Grant #099175/Z/12/Z) Human Developmental Biology Resource (www.hdbr.org). In situ hybridization probes were generated by PCR-cloning DONSON genomic sequence into plasmids for in vitro transcription with digoxigenin-UTP. Hybridized probes were visualized using anti-digoxigenin alkaline phosphatase-conjugated antibody and NBT/BCIP (Roche). Specificity of the in situ hybridization was confirmed with sense probes. See Supplemental Methods for full method details.
Knockout mice
Mice heterozygous for a Donson knockout allele (Donsontm1a(EUCOMM)Wtsi) were produced as part of the European Conditional Mouse Mutagenesis Program (EUCOMM) and the International Knockout Mouse Consortium and phenotyped at the Wellcome Trust Sanger Institute. See Supplemental Methods for further details.
siRNA knockdown and qPCR
cDNA of HeLa cells transfected with DONSON or GFP siRNA was assayed using a qPCR panel of 91 cell cycle regulation genes (Roche) on a LightCycler 480 instrument (Roche). cDNA of MMS patient samples and controls was assayed using 18S rRNA and DONSON TaqMan qPCR assays (Thermo Fisher). See Supplemental Methods for full method details.
Data access
DNA and RNA sequencing data from this study (all samples except HI-1400 and HI-2185) have been submitted to the NCBI Database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap) under accession number phs000492.v2.p1. Data for Autism Genetic Resource Exchange (AGRE) cell lines (HI-1400 and HI-2185) have been submitted to the AGRE data repository (https://research.agre.org). Sanger sequencing from this study has been submitted to the NCBI Trace Archive (https://trace.ncbi.nlm.nih.gov/Traces/trace.cgi) under TI numbers 2344113440–2344113468.
Supplementary Material
Acknowledgments
We thank the families who participated in this research and the leaders of the First Nations communities of Northern Saskatchewan for their support. This work was also supported by the genetic counselors at the Royal University Hospital in Saskatoon, Canada, including Tara Scriver, Lacey Benoit, Candice Jackel-Cram, Katherine Osczevski, and Pamela Blumenschein. We gratefully acknowledge the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families for the provision of cell lines. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). G.D.E. was supported by National Institutes of Health (NIH) MSTP grant T32GM007753 and the Louis Lange III Scholarship in Translational Research. D.R.C. was supported by The Eunice Kennedy Shriver National Institute of Child Health and Human Development Women's Reproductive Health Research Career Development grant K12HD01255, the Eleanor and Miles Shore Scholars in Medicine award, and the William Randolph Hearst perinatal research award in neurodevelopmental disorders. J.S. was supported by the Victoria and Stuart Quan Fellowship and a grant from NIDCD (R03 DC013866). C.A.W. was supported by the Manton Center for Orphan Disease Research and a grant from the National Institute of Neurological Disorders and Stroke (R01 NS035129). C.A.W. is a Distinguished Investigator of the Paul G. Allen Family Foundation and an Investigator of the Howard Hughes Medical Institute.
Author contributions: Identification and recruitment of research subjects, and collection and analysis of clinical information, was performed by D.R.C., J.S., J.N.P., M.J.M., B.S., C.A.R., R.C., D.D., R.F., S.W., A.J.B., J.I., E.G.L., and P.B. Linkage analysis was performed by J.S., D.R.C., and R.S.H. D.R.C. and T.W.Y. performed the genome sequencing and identified the disease variant. G.D.E. performed the RNA experiments and identified the splicing defect. D.G., D.G.D., and C.S. performed human and mouse embryo in situ hybridization. A.G. and M.E.C. analyzed the knockout mice, which were created by the European Conditional Mouse Mutagenesis Program as part of the International Knockout Mouse Consortium. U.T. and S.W. performed the siRNA experiments. G.D.E., D.R.C., J.S., J.N.P., and C.A.W. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.219899.116.
Freely available online through the Genome Research Open Access option.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. 2002. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30: 97–101. [DOI] [PubMed] [Google Scholar]
- Bandura JL. 2005. “Humpty dumpty defines a new gene family required for S phase.” PhD thesis, University of Pennsylvania. [Google Scholar]
- Bandura JL, Beall EL, Bell M, Silver HR, Botchan MR, Calvi BR. 2005. humpty dumpty is required for developmental DNA amplification and cell proliferation in Drosophila. Curr Biol 15: 755–759. [DOI] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. 2010. Deciphering the splicing code. Nature 465: 53–59. [DOI] [PubMed] [Google Scholar]
- Bell TJ, Miyashiro KY, Sul JY, Buckley PT, Lee MT, McCullough R, Jochems J, Kim J, Cantor CR, Parsons TD, et al. 2010. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc Natl Acad Sci 107: 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J. 2003. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci 100: 10237–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PA, et al. 2011. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 43: 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Bhutkar A, Sharp PA. 2015. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 29: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD, O'Donnell M, Kuriyan J. 2004. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429: 724–730. [DOI] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 24: 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrage LC, Charng WL, Eldomery MK, Willer JR, Davis EE, Lugtenberg D, Zhu W, Leduc MS, Akdemir ZC, Azamian M, et al. 2015. De novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin syndrome. Am J Hum Genet 97: 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Chandrasekharappa SC, Lach FP, Kimble DC, Kamat A, Teer JK, Donovan FX, Flynn E, Sen SK, Thongthip S, Sanborn E, et al. 2013. Massively parallel sequencing, aCGH, and RNA-Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood 121: e138–e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O'Donnell L, et al. 2015. The BioGRID interaction database: 2015 update. Nucleic Acids Res 43: D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Shendure J. 2011. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet 12: 628–640. [DOI] [PubMed] [Google Scholar]
- de Klerk E, ‘t Hoen PA. 2015. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet 31: 128–139. [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81. [DOI] [PubMed] [Google Scholar]
- Dvinge H, Bradley RK. 2015. Widespread intron retention diversifies most cancer transcriptomes. Genome Med 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CR, Ritchie W, Wong JJ, Schmitz U, Middleton R, An X, Mohandas N, Rasko JE, Blobel GA. 2016. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood 10.1182/blood-2016-01-692764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. 2003. Pre-mRNA splicing and human disease. Genes Dev 17: 419–437. [DOI] [PubMed] [Google Scholar]
- Fenwick AL, Kliszczak M, Cooper F, Murray J, Sanchez-Pulido L, Twigg SR, Goriely A, McGowan SJ, Miller KA, Taylor IB, et al. 2016. Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier-Gorlin syndrome and craniosynostosis. Am J Hum Genet 99: 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs F, Pau G, Kranz D, Sklyar O, Budjan C, Steinbrink S, Horn T, Pedal A, Huber W, Boutros M. 2010. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol Syst Biol 6: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. 2004. Detection and evaluation of intron retention events in the human transcriptome. RNA 10: 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Porse BT. 2014. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays 36: 236–243. [DOI] [PubMed] [Google Scholar]
- Guigo R, Valcarcel J. 2015. RNA. Prescribing splicing. Science 347: 124–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. 2006. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5: 1675–1680. [DOI] [PubMed] [Google Scholar]
- Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK. 2000. Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev 14: 913–926. [PMC free article] [PubMed] [Google Scholar]
- Ives EJ, Houston CS. 1980. Autosomal recessive microcephaly and micromelia in Cree Indians. Am J Med Genet 7: 351–360. [DOI] [PubMed] [Google Scholar]
- Jian X, Boerwinkle E, Liu X. 2014. In silico tools for splicing defect prediction: a survey from the viewpoint of end users. Genet Med 16: 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. 2015. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet 47: 1242–1248. [DOI] [PubMed] [Google Scholar]
- Kallabi F, Ben Rhouma B, Baklouti S, Ghorbel R, Felhi R, Keskes L, Kamoun H. 2016. Splicing defects in the AAAS gene leading to both exon skipping and partial intron retention in a Tunisian patient with Allgrove syndrome. Horm Res Paediatr 86: 90–93. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature 478: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetarpal P, Das S, Panigrahi I, Munshi A. 2016. Primordial dwarfism: overview of clinical and genetic aspects. Mol Genet Genomics 291: 1–15. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingseisen A, Jackson AP. 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 25: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska MA. 2013. The missing puzzle piece: splicing mutations. Int J Clin Exp Pathol 6: 2675–2682. [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. 2011. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci 108: 11093–11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. 2005. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett 579: 1900–1903. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, et al. 2014. Guidelines for investigating causality of sequence variants in human disease. Nature 508: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AC, Obholzer ND, Shah AN, Megason SG, Moens CB. 2013. RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res 23: 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. 2014. Transcriptional landscape of the prenatal human brain. Nature 508: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff R, Scott DW, Ding J, Roth A, Chiu R, et al. 2013. Mutational and structural analysis of diffuse large B-cell lymphoma using whole genome sequencing. Blood 122: 1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudvari P, Movassagh M, Kowsari K, Seyfi A, Kokkinaki M, Edwards NJ, Golestaneh N, Horvath A. 2015. SNPlice: variants that modulate Intron retention from RNA-sequencing data. Bioinformatics 31: 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Pardo-Diaz C, Whibley A, Supple MA, Saenko SV, Wallbank RW, Wu GC, Maroja L, Ferguson L, Hanly JJ, et al. 2016. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534: 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Yang W, Han M, Zhang Y, Shen T, Nie H, Zhou Z, Dai Y, Yang Y, Liu P, et al. 2016. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res 10.1093/nar/gkw591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Aoki Y, Obayashi T, Tadaka S, Ito S, Narise T, Kinoshita K. 2015. COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res 43: D82–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Recalde O, Moreno MB, Vergara JI, Fonseca DJ, Rojas RF, Mosquera H, Medina CL, Restrepo CM, Laissue P. 2015. A novel TGM1 mutation, leading to multiple splicing rearrangements, is associated with autosomal recessive congenital ichthyosis. Clin Exp Dermatol 40: 757–760. [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE. 2004. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet 5: 389–396. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. 2012. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet 44: 461–466. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JJ, Bicknell LS, Carroll P, Higgs MR, Shaheen R, Murray JE, Papadopoulos DK, Leitch A, Murina O, Tarnauskaite Z, et al. 2017. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat Genet 49: 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK. 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol Cell 16: 173–185. [DOI] [PubMed] [Google Scholar]
- Seifert W, Holder-Espinasse M, Spranger S, Hoeltzenbein M, Rossier E, Dollfus H, Lacombe D, Verloes A, Chrzanowska KH, Maegawa GH, et al. 2006. Mutational spectrum of COH1 and clinical heterogeneity in Cohen syndrome. J Med Genet 43: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Ayuso C, Harper S, Berson EL, Rivolta C. 2011. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am J Hum Genet 88: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11.10.1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen K, Van Bockstaele DR, Berneman ZN. 2003. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif 36: 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk A, Crispino JD. 2015. The role of the chromatin assembly complex (CAF-1) and its p60 subunit (CHAF1b) in homeostasis and disease. Biochim Biophys Acta 1849: 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Kim C, Bradfield J, Guo Y, Toskala E, Otieno FG, Hou C, Thomas K, Cardinale C, Lyon GJ, et al. 2013. Whole-genome DNA/RNA sequencing identifies truncating mutations in RBCK1 in a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Med 5: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13: 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595. [DOI] [PubMed] [Google Scholar]
- Wong JJ, Au AY, Ritchie W, Rasko JE. 2016. Intron retention in mRNA: no longer nonsense: known and putative roles of intron retention in normal and disease biology. Bioessays 38: 41–49. [DOI] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, et al. 2015. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 347: 1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Baltus AE, Mathew RS, Murphy EA, Evrony GD, Gonzalez DM, Wang EP, Marshall-Walker CA, Barry BJ, Murn J, et al. 2012. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell 151: 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. 2012. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 26: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.