Abstract
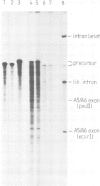
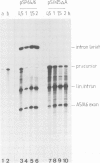
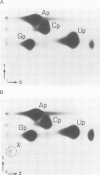
The fifth intron in the gene for cytochrome c oxidase subunit I in yeast mitochondrial DNA is of the group II type and is capable of self-splicing in vitro. The reaction results in lariat formation, concomitant with exon-exon ligation and does not require a guanosine nucleotide for its initiation. It is generally assumed, but not formally proven, that the first step in splicing is a nucleophilic attack of the 2'-hydroxyl of the branchpoint nucleotide (A) on the 5'-exon-intron junction. To investigate the role of intron sequences in recognition of the 5'-splice junction and the ensuing event of cleavage and lariat formation, mutations have been introduced at and around the branchsite. Results obtained show that although branchpoint attack and subsequent lariat formation are strongly preferred events under conditions normally used for self-splicing, addition of a single T residue at intron position 856, a mutation which brings the branchpoint adenosine into a basepair, leads to a conditionally active intron, which at high ionic strength catalyses exon-exon ligation in the absence of lariat formation. Comparable behaviour is also observed with the branchpoint A deletion mutant. The implications of these findings for the mechanism of self-splicing of group II introns are discussed.
Full text
PDF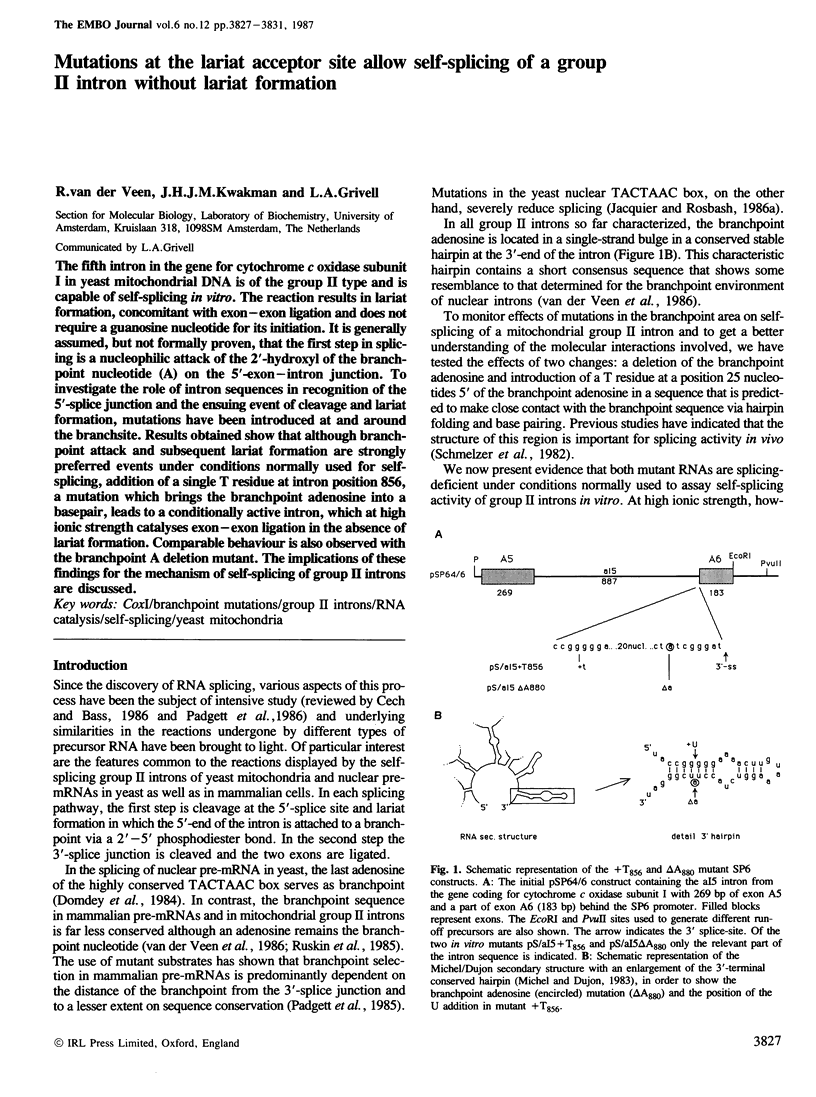



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987 Jul 3;50(1):17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rodriguez J. R., Rosbash M. A quantitative analysis of the effects of 5' junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985 Dec;43(2 Pt 1):423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rosbash M. Efficient trans-splicing of a yeast mitochondrial RNA group II intron implicates a strong 5' exon-intron interaction. Science. 1986 Nov 28;234(4780):1099–1104. doi: 10.1126/science.2430332. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rosbash M. RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5835–5839. doi: 10.1073/pnas.83.16.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Aebi M., Hornig H., Weissmann C., Sharp P. A. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8349–8353. doi: 10.1073/pnas.82.24.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. A self-splicing RNA excises an intron lariat. Cell. 1986 Jan 31;44(2):213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer C., Schmidt C., Schweyen R. J. Identification of splicing signals in introns of yeast mitochondrial split genes: mutational alterations in intron bI1 and secondary structures in related introns. Nucleic Acids Res. 1982 Nov 11;10(21):6797–6808. doi: 10.1093/nar/10.21.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer C., Schweyen R. J. Self-splicing of group II introns in vitro: mapping of the branch point and mutational inhibition of lariat formation. Cell. 1986 Aug 15;46(4):557–565. doi: 10.1016/0092-8674(86)90881-0. [DOI] [PubMed] [Google Scholar]
- Sims P. F., Minter S. J., Stancombe R., Gent M. E., Andrews J., Waring R. B., Towner P., Davies R. W. A modified two primer approach to oligonucleotide-directed in vitro mutagenesis. Biochimie. 1985 Jul-Aug;67(7-8):841–847. doi: 10.1016/s0300-9084(85)80177-2. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Van der Horst G., Osinga K. A., Arnberg A. C. Splicing of large ribosomal precursor RNA and processing of intron RNA in yeast mitochondria. Cell. 1984 Dec;39(3 Pt 2):623–629. doi: 10.1016/0092-8674(84)90469-0. [DOI] [PubMed] [Google Scholar]
- White S. A., Draper D. E. Single base bulges in small RNA hairpins enhance ethidium binding and promote an allosteric transition. Nucleic Acids Res. 1987 May 26;15(10):4049–4064. doi: 10.1093/nar/15.10.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst G., Tabak H. F. Self-splicing of yeast mitochondrial ribosomal and messenger RNA precursors. Cell. 1985 Apr;40(4):759–766. doi: 10.1016/0092-8674(85)90335-6. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Arnberg A. C., Grivell L. A. Self-splicing of a group II intron in yeast mitochondria: dependence on 5' exon sequences. EMBO J. 1987 Apr;6(4):1079–1084. doi: 10.1002/j.1460-2075.1987.tb04861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R., Arnberg A. C., van der Horst G., Bonen L., Tabak H. F., Grivell L. A. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986 Jan 31;44(2):225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]