Abstract
Hematopoietic stem cells (HSCs) self-renew in bone marrow niches formed by mesenchymal progenitors and endothelial cells that express the chemokine CXCL12, but whether a separate niche instructs multipotent progenitor (MPP) differentiation remains unclear. Here, we show that MPPs resided in HSC niches where they encountered lineage-instructive differentiation signals. Conditional deletion of the chemokine receptor CXCR4 in MPPs reduced differentiation into common lymphoid progenitors (CLPs), which decreased lymphopoiesis. CXCR4 was required for CLP positioning near Interleukin-7+ (IL-7) cells and for optimal IL-7 receptor signaling. IL-7+ cells expressed CXCL12 and SCF, were mesenchymal progenitors capable of differentiation into osteoblasts and adipocytes, and comprised a minor subset of sinusoidal endothelial cells. Conditional Il7 deletion in mesenchymal progenitors reduced B-lineage committed CLPs, while conditional Cxcl12 or Scf deletion from IL-7+ cells reduced HSC and MPP numbers. Thus, HSC maintenance and multilineage differentiation are distinct cell lineage decisions that are both controlled by HSC niches.
Introduction
In mammals, hematopoietic stem cells (HSCs) are maintained throughout life in specialized niches in bone marrow (BM). The long-term maintenance of HSCs is achieved by a balance between signals promoting quiescence or cell division, and self-renewal or differentiation. These decisions are controlled in part by extracellular signals produced by HSC niche cells. Over the past few years, several studies have characterized BM stromal cells and identified rare mesenchymal stem and progenitor cells (MSPCs), and BM endothelial cells, as cellular components of HSC niches during homeostasis (Morrison and Scadden, 2014). Stromal cells are essential organizers of HSC niches in BM because these cells regulate HSC quiescence and long-term maintenance, at least in part through the production of a potent chemokine, CXCL12, and short-range signals such as membrane-bound stem cell factor (Ding and Morrison, 2013; Ding et al., 2012; Greenbaum et al., 2013; Kunisaki et al., 2013; Mendez-Ferrer et al., 2010; Omatsu et al., 2010). Thus, a model emerged in which CXCL12 attracts HSCs to position near BM stromal cells in order to facilitate their access to critical factors controlling HSC lineage decisions in BM. In favor of such a model, HSCs have been found in proximity to Nestin-expressing MSPCs that express CXCL12 and SCF (Kunisaki et al., 2013; Mendez-Ferrer et al., 2010). Indeed, Nestin+ MSPCs share many morphological and functional similarities with CXCL12-abundant reticular cells (CAR, Sugiyama et al., 2006), including multipotent progenitor differentiation potential and expression of high amounts of SCF (Omatsu et al., 2010), suggesting some overlap exists between these BM stromal cell types.
Upon transplantation, most HSCs home back to the BM where they preferentially localize in vascularized endosteal niches in the calvarium BM (Lo Celso et al., 2009), with downstream multipotent progenitors (MPPs) and differentiated hematopoietic cells residing at undefined sites further away from osteoblasts (Lo Celso et al., 2009). Other studies examining the niches involved in hematopoietic cell differentiation showed that megakaryocyte progenitors reside and differentiate predominantly in vascular niches in the BM parenchyma (Avecilla et al., 2004), whereas lymphoid progenitors may require signals provided by mature osteoblasts and localize to endosteal niches for development (Ding and Morrison, 2013; Terashima et al., 2016; Visnjic et al., 2004; Wu et al., 2008; Zhu et al., 2007). Taken together, these data suggested that separate niches control HSC maintenance and hematopoietic progenitor differentiation.
CXCR4 and its ligand CXCL12 form a chemokine/chemokine receptor pair that controls multiple essential fetal and adult hematopoietic processes. Early studies using mice genetically deficient in CXCR4 or CXCL12 demonstrated a severe reduction in B lymphopoiesis and a mild reduction in myelopoiesis in the fetal liver, and severe impairment in myeloid, lymphoid, and megakaryocyte cell development in fetal BM (Ma et al., 1998; Nagasawa et al., 1996; Zou et al., 1998). Some of these defects were in part explained by defective retention of hematopoietic precursors in BM, and by other findings indicating that CXCR4 is also required for hematopoietic stem cell homing and retention in BM (Ara et al., 2003; Lapidot and Kollet, 2002; Ma et al., 1999; Peled et al., 1999). Furthermore, CXCR4 signaling in HSCs was proposed to be required for HSC quiescence and maintenance through direct regulation of cell cycle gene expression (Nie et al., 2008; Sugiyama et al., 2006; Tzeng et al., 2011).
Amongst the many hematopoietic lineages, B lymphocytes are the most dependent on CXCR4 and CXCL12 (Nie et al., 2008; Sugiyama et al., 2006). This dependence is likely to be at an early hematopoietic stage given the fact that conditional deletion of CXCR4 in proB cells did not impair B cell development in BM (Beck et al., 2014; Nie et al., 2008; Pereira et al., 2009). These findings led us to ask the question of how a single chemoattractant receptor (CXCR4) could control both HSC quiescence and lymphopoiesis. One possibility is that defects in HSC quiescence directly cause hematopoietic differentiation defects regardless of signals provided by BM niches. Alternatively, CXCR4 and CXCL12 guide HSCs and MPPs to BM niches that not only support HSC quiescence but also sustain MPP differentiation.
Here we showed that CXCR4 was intrinsically required in MPPs for differentiation into multiple downstream lineage-restricted hematopoietic precursors, most prominently to the earliest B cell-committed common lymphoid precursor (CLP) that expresses the lymphocyte antigen complex 6 locus D protein (Ly6D, Inlay et al., 2009). We showed that CXCR4 controlled the positioning of Ly6D+ CLPs in the vicinity of IL-7+ cells in BM, and that defective positioning near IL-7+ cells resulted in reduced IL-7R signaling. We further show that IL-7 is primarily made by a subset of CAR cells, which also contained multipotent mesenchymal differentiation capacity. CXCL12 or SCF deletion from IL-7+ cells significantly reduced the number of HSCs and MPPs in BM. Our studies demonstrate that positional cues play critical roles in enabling hematopoietic precursors to access short-range lineage-instructive cytokines. Furthermore, our studies favor a model in which HSC quiescence and multilineage differentiation are locally controlled by HSC niches in the BM.
Results
CXCR4 controls CLP development in BM
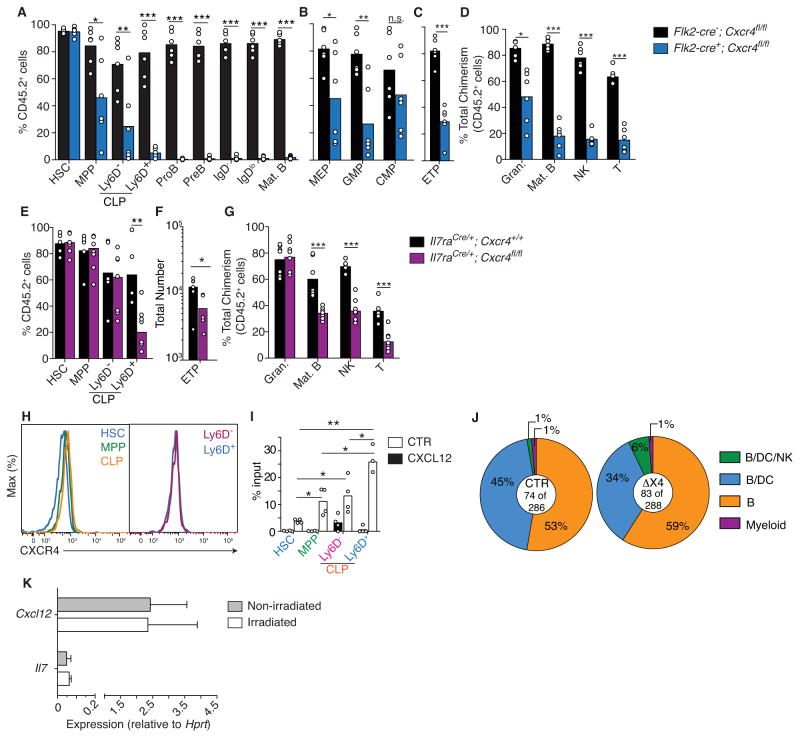
In order to dissociate the role played by CXCR4 in HSC quiescence from its role in MPP differentiation, we conditionally deleted Cxcr4 exclusively in hematopoietic MPPs using a Flk2-cre transgene (Boyer et al., 2011). To further restrict multilineage differentiation from adult BM niches we reconstituted lethally irradiated, congenically marked, wild-type (WT) mice (CD45.1+ C57BL/6) with a mixture of 90% BM cells from Flk2-cre+; Cxcr4fl/fl or Flk2-cre−;Cxcr4fl/fl mice (both CD45.2+) and 10% BM cells from WT mice (CD45.1+ C57BL/6), and analyzed hematopoietic cell lineages in BM, blood, spleen, lymph nodes, peritoneal cavity, Peyer’s patch, liver and lung, 6 to 20 weeks after reconstitution (gating strategy for hematopoietic cells analyzes is depicted in Fig. S1.) The frequency of hematopoietic cell subsets that differentiated from Flk2-cre−; Cxcr4fl/fl (WT, CD45.2+) or from competitor precursors (WT, CD45.1+) was similar to engrafted cells (Fig. 1A). In contrast, CXCR4-deficient MPPs were reduced by 2-fold, multipotent CLPs were reduced by 3-fold, B-lineage restricted Ly6D+ CLPs (Inlay et al., 2009) were reduced by more than 8-fold, and developing B cell subsets were nearly absent from BM (Fig. 1A and Fig. S2A and B). Using this approach CXCR4 deletion was detectable at the MPP and CLP stages, and was evident at the preB cell stage (Fig. S2H). CXCR4-deficient megakaryocyte and erythroid progenitors (MEPs), and granulocyte and monocyte progenitors (GMPs) were also significantly reduced by 2-fold in BM (Fig. 1B and Fig. S2C). The 3-fold reduction in multipotent CLPs led to a similar reduction in early thymic precursors (ETPs; Fig. 1C and Fig. S2D). We also detected reduced numbers of lymphoid precursors and lymphocyte subsets in BM of non-irradiated Flk2-cre+; Cxcr4fl/fl when compared to Flk2-cre−; Cxcr4fl/fl adult mice, albeit the fold change was less prominent than that seen in mixed BM chimeras (Fig. S2I and J). Although hematopoietic progenitors, including CLP and early B cell subsets, were mobilized to the spleen (Fig. S2K) as expected (Nie et al., 2008), it nevertheless resulted in a profound reduction in total T, B, and NK cell production (Fig. 1D and Fig. S2E). Because CXCR4 was required for MPP differentiation into CLP subsets (Fig. 1A), the profound defect in lymphoid lineage production could have been caused by defective CLP differentiation. To address this possibility we conditionally deleted Cxcr4 from CLPs using Il7raCre/+ mice (Fig. S2L and M) and performed similar BM mixed chimeras. Although multipotent (Ly6D−) CLPs were normally represented in BM (Fig. 1E and S2F), ETPs were significantly reduced (Fig. 1F) possibly due to reduced CLP entry into the thymus, and B-lineage committed Ly6D+ CLPs were significantly reduced by 3-fold in BM (Fig. 1E and S2F). These defects also resulted in a significant 2–3 fold reduction in total T, B and Natural Killer (NK) cells produced (Fig. 1G and S2G), despite the mobilization of lymphoid precursors into the spleen (Fig. S2M). These data showed that CXCR4 was essential at the MPP and CLP stages for their retention in BM, and for the generation of a full lymphocyte compartment. These data also revealed a requirement for CXCR4 in MPPs for their efficient differentiation into other hematopoietic cell lineage-restricted progenitors (e.g. MEPs and GMPs).
Figure 1. Intrinsic expression of CXCR4 in MPPs and CLPs is critical for lymphoid cell development.
A–D, Frequency of CD45.2+ hematopoietic cell subsets in BM (A and B), thymus (C), and in all lymphoid organs (BM, blood, spleen, thymus, lymph nodes, Peyer’s patch, peritoneal cavity, lung and liver, D) of mice reconstituted with 90% CD45.2+ Flk2-cre−;Cxcr4fl/fl or Flk2-cre+;Cxcr4fl/fl cells mixed with 10% CD45.1+ WT cells (data pooled from three independent experiments). E, Frequency of CD45.2+ HSCs, MPPs and CLP subsets in BM of mice reconstituted with 90% CD45.2+ Il7raCre/+;Cxcr4fl/fl or Il7raCre/+;Cxcr4+/+ cells and 10% WT CD45.1+ cells. F, ETP number in Il7raCre/+;Cxcr4fl/fl and control mice. G, Frequency of CD45.2+ hematopoietic cell subsets isolated from all lymphoid organs of mice reconstituted with 90% CD45.2+ Il7raCre/+; Cxcr4fl/fl or Il7raCre/+; Cxcr4+/+ cells and 10% WT CD45.1+ cells. Data are representative of 3 independent experiments with 3–6 mice in each group and each experiment. Bars indicate the average; circles depict individual mice. H, CXCR4 surface expression in HSCs, MPPs, total CLPs, and in Ly6D− and Ly6D+ CLPs. I, In vitro cell chemotaxis through 5-μm transwells: nil, CXCL12 (300 ng/mL). Bars indicate mean, circles depict individual experiments. J, In vitro single cell CLP differentiation on OP-9 stromal cells for 12 days in the presence of IL-7 (50ng/mL). K, Il7 and Cxcl12 mRNA expression in non-irradiated and irradiated OP-9 stromal cells. Expression is relative to Hprt. Data in panels H–K are representative of at least 3 independent experiments. * P<0.05, **P<0.01, ***P<0.001 (unpaired, two-tailed Student’s t-test). See also Figures S1 and S2.
CXCR4 expression and function is increased at the CLP stage when compared to earlier HSC and MPP stages (Fig. 1H and I). CXCR4 signaling has been suggested to regulate cell cycle entry in HSCs presumably independently of its role as a chemoattractant receptor (Nie et al., 2008; Sugiyama et al., 2006). Thus, we asked whether CXCR4 signaling was also intrinsically required for CLP proliferation and differentiation into B-lineage cells. To test this possibility we measured the efficiency of CXCR4 sufficient and deficient single cell-sorted CLPs to differentiate into B and NK cells in vitro. We found that CXCR4-sufficient and -deficient CLPs differentiated similarly into B and NK cells when cultured on OP-9 stromal cell layers with exogenous IL-7 (Fig. 1J). As OP-9 cells express CXCL12 (Fig. 1K), the ability of CXCR4-sufficient and -deficient CLPs to differentiate equally into B and NK cells suggests that CXCR4 signaling is not required for lymphoid cell development from CLPs per se.
CXCR4 guides hematopoietic precursors towards IL-7+ cells in BM
Next, we asked how CXCR4 controls lymphoid lineage differentiation. The developmental block at the MPP to CLP transition, and particularly to the Ly6D+ CLP stage, was reminiscent of the phenotype of Il7 deficient mice (Carvalho et al., 2001; Dias et al., 2005; Tsapogas et al., 2011). This similarity led us to hypothesize that CXCR4 was required at the CLP stage for optimal IL-7-mediated IL-7R signaling. To test this hypothesis we analyzed the distribution of Ly6D+ CLPs relative to IL-7-expressing cells in BM. Even though IL-7 has been detected by immunofluorescence microscopy in situ (Tokoyoda et al., 2004), its visualization is difficult and may be restricted to cells that express IL-7 at the highest levels. Thus, we visualized Ly6D+ CLPs in femur BM of Il7-ECFP (enhanced cyan fluorescent protein) transgenic mice (Mazzucchelli et al., 2009), and of Il7GFP/+ knock-in mice (Miller et al., 2013). In both Il7 reporter mice, IL-7+ cells could only be visualized after signal amplification with anti-GFP antibodies (Fig. S3A and B), which strongly suggests that Il7 was expressed at very low amounts in vivo. B-lineage committed CLPs were identified in situ as being negative for hematopoietic cell lineage markers (see Supplementary Methods for details), IL-7Rα+, and Ly6D+ (Fig. 2A), a cell gate that was ~93% enriched in Ly6D+ CLPs (Fig. S3C and D). Similar CLP frequencies were detected by immunofluorescence microscopy and by flow cytometry (Fig. S3E). We found that more than 90% of Ly6D+ CLPs were positioned within 15 μm from IL-7+ cells, whereas most IgD+ B lymphocytes were localized at greater distances from IL-7+ cells (Fig. 2B and C). Interestingly, Ly6D+ CLP proximity to IL-7+ cells was dependent on intrinsic CXCR4 expression (Fig. 2D and E) and, thus, their positioning was sensitive to a 3-day treatment with the CXCR4 antagonist AMD3100 (Fig. 2F and G). Defective CLP positioning near IL-7+ stromal niches resulted in a significant reduction in IL-7Rα signaling, as measured by STAT5α phosphorylation (Fig. 2H and I). Combined, these data suggested that CXCR4 controls lymphopoiesis by promoting CLP positioning near IL-7+ cells in BM, which enabled IL-7Rα signaling and STAT5α phosphorylation.
Figure 2. CXCR4 controls CLP positioning near IL-7+ cells in BM.
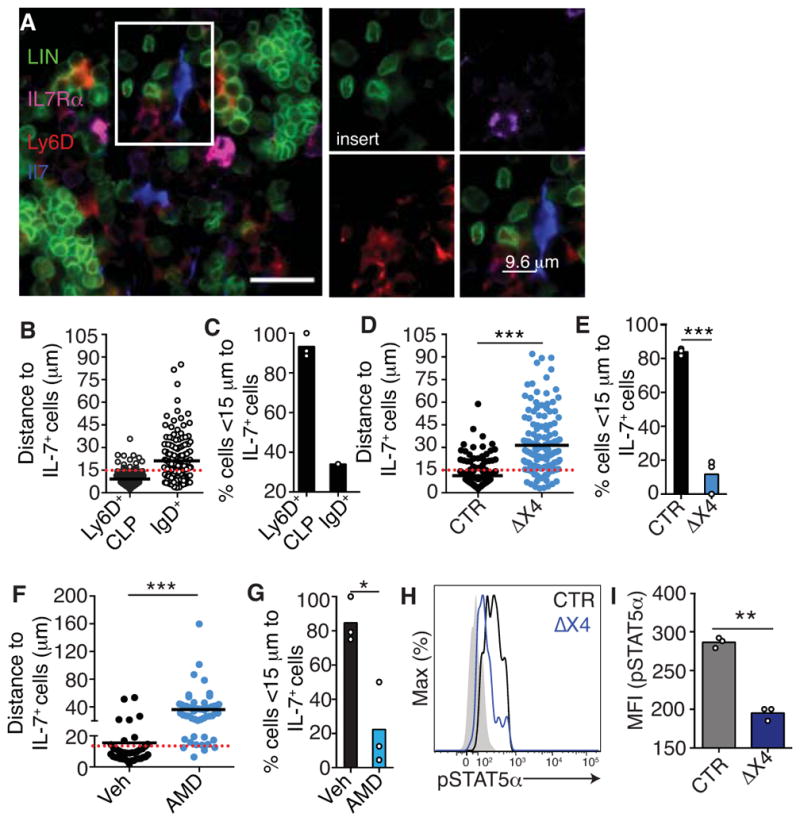
A, 7μm-thick section of Il7-ECFP transgenic femur. Lin., IL-7Rα, Ly6D and Il7. Scale bar, 20 μm. B, Ly6D+ CLP (n=166), IgD+ (n=164), and IL-7+ cell distribution in BM. C, Ly6D+ CLP and IgD+ cell frequency near IL-7+ cells (<15 μm). D and E, Distribution of Flk2-cre+;Cxcr4fl/fl (ΔX4) or Flk2-cre−;Cxcr4fl/fl (CTR) Ly6D+ CLPs, and IL-7+ cells (D); cell frequency near IL-7+ cells (<15 μm) (E). F and G, Distribution of Ly6D+ CLPs and IL-7+ cells after AMD3100 (AMD) or vehicle (Veh) treatment for 3 days (F). Cell frequency near IL-7+ cells (<15 μm) (G). Data in panels A–G are representative of 2–3 independent experiments. H and I, pSTAT5a in Il7raCre/+;Cxcr4fl/+ (CTR) and Il7raCre/+;Cxcr4fl/fl (ΔX4), Ly6D+ CLPs. Shaded histogram shows pSTAT5α in Ly6D+ CLPs isolated from Il7−/− mice. (H). pSTAT5α geometric mean intensity (I). Data in panels H and I are representative of two independent experiments with at least 3 mice in each group per experiment. Bars indicate mean; circles depict individual mice. * P<0.05, **P<0.01, ***P<0.001 (unpaired, two-tailed Student’s t-test). See also Figure S3.
Osteoblast- and hematopoietic-derived IL-7 is not required for B-lymphopoiesis
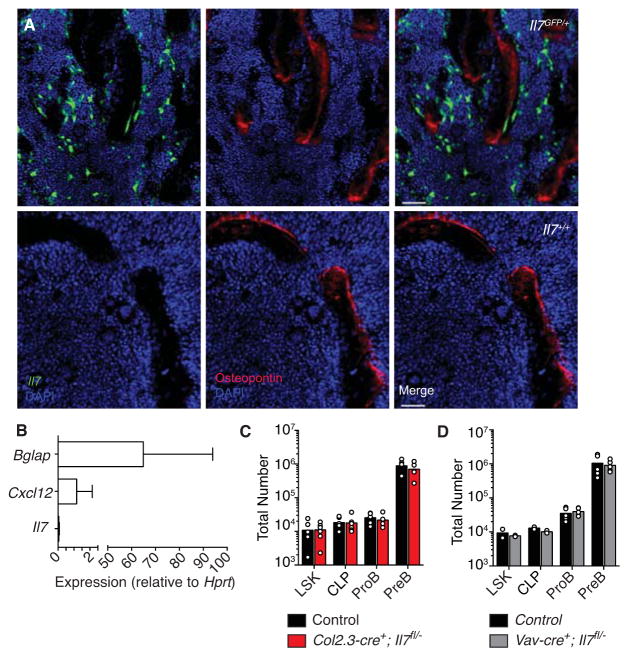
Previous studies suggested that osteoblasts form a specialized BM niche supporting B-lineage cell development (Ding and Morrison, 2013; Visnjic et al., 2004; Wu et al., 2008; Zhu et al., 2007). Osteoblasts differentiated in vitro can express IL-7, and CXCL12 has also been detected in osteoblasts in vivo, albeit at much lower amounts than in CXCL12-abundant cells (Ding and Morrison, 2013; Sugiyama et al., 2006). Furthermore, a significant percentage of IL-7Rα+ lineage-negative cells, of which ~ 8% are CLPs (Fig. S4A), was suggested to be positioned near the endosteum (Ding and Morrison, 2013). Thus, we asked whether Ly6D+ CLPs were preferentially distributed near the endosteum. To address this question, we quantified Ly6D+ CLP distance to the nearest bone surface and found that more than 80% of these cells were positioned > 30 μm away from the endosteum (Fig. S4B and C). We then asked if Il7 was expressed in osteoblasts in vivo and in vitro. To address this question, we stained 25 μm-thick femur whole mount sections from Il7GFP/+ or from control mice with anti-osteopontin or anti-osteocalcin antibodies but could not detect Il7-driven GFP expression in osteoblasts in situ (Fig. 3A, and Fig. S4D and E, respectively). Occasionally, we could find low Il7-driven GFP signal adjacent to osteopontin or osteocalcin-positive cells, but upon careful 3-dimensional analysis we failed to detect signal co-localization (Fig. S4D). In order to analyze Il7 expression in osteoblasts in a quantitative manner we measured Il7 mRNA expression in cultured osteoblasts from adult femur BM precursors, and from neonatal calvaria BM precursors, by quantitative PCR. We found that Il7 expression is essentially undetectable in these cells, whereas osteocalcin (Bglap, a gene highly expressed in pre-osteoblasts and mature osteoblasts) was readily detected, and Cxcl12 expression was also detected (Fig. 3B, and Fig. S4F), as expected (Ding and Morrison, 2013; Sugiyama et al., 2006). To convincingly rule out the possibility that an undetectable amount of Il7 expressed by osteoblasts contributed to B lymphopoiesis we conditionally deleted Il7 in osteoblasts using the Col1a1 2.3Kb promoter driven-cre recombinase approach (Col2.3-cre) (Liu et al., 2004). We found no evidence for a significant contribution of osteoblast-expressed IL-7 to B lymphopoiesis (Fig. 3C and Fig. S4G). Furthermore, we could not detect Il7 expression in hematopoietic cell subsets (not shown), nor did Il7 deletion from hematopoietic cells using the Vav-cre transgene resulted in measurable defects in B cell production (Fig. 3D). In summary, these data demonstrate that osteoblasts are not a physiologically relevant source of IL-7 for lymphopoiesis.
Figure 3. Osteoblast-derived IL-7 is not required for B-lymphopoiesis.
A, Distribution of IL-7+ and osteopontin+ cells in BM. B, Bglap (osteocalcin), Cxcl12, and Il7 expression in osteoblasts by qPCR. Gene expression is relative to Hprt. Data in A and B are representative of 3–5 independent experiments. C, Total number of hematopoietic cell subsets in BM of Il7fl/−; Col2.3-cre+ mice and control littermates. D, Total number of hematopoietic cell subsets in BM of Il7fl/−; Vav-cre+ mice and control littermates. Bars indicate average, circles depict individual mice. Data panels C and D are representative of at least 3 independent experiments with 4–6 mice per group. See also Figure S4.
IL-7+ cells are a subset of BM CAR cells and are critical for B-lymphopoiesis
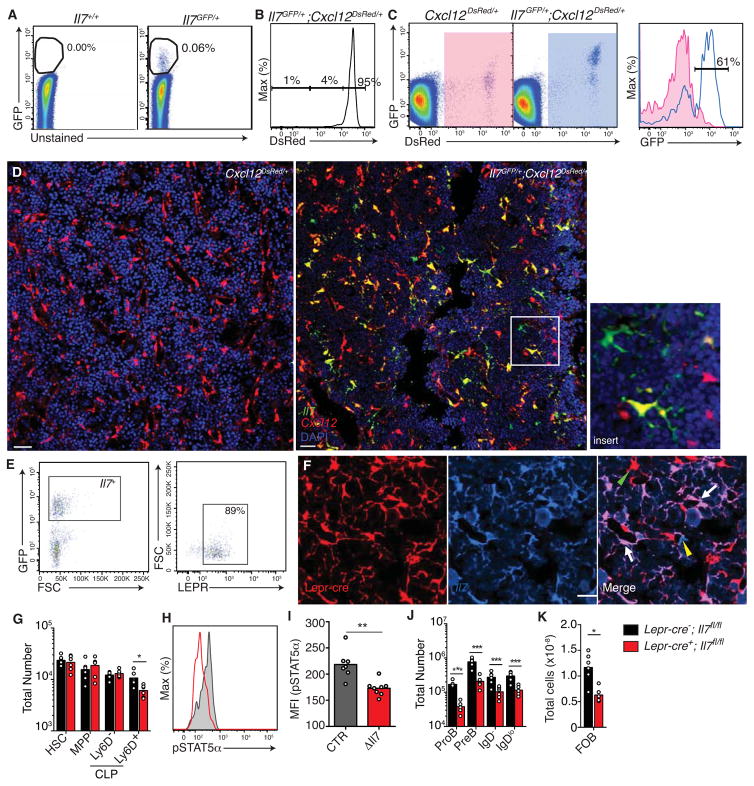
The fact that CXCR4 was critical for CLP positioning near or in contact with IL-7-producing reticular cells suggested that IL-7+ cells express significant amounts of CXCL12. However, distinct stromal cell subsets were suggested to produce either CXCL12 or IL-7, but not both (Tokoyoda et al., 2004). To gain insight into the BM niches that support lymphoid lineage differentiation, we analyzed the cellular composition and distribution of IL-7+ cells in BM of Il7GFP/+; Cxcl12DsRed/+ double reporter mice. We carefully extracted and digested BM cells with collagenase (see Experimental Procedures for details) and analyzed cells by flow cytometry. We found that 0.05±0.01% of total BM cells expressed Il7 (Fig. 4A). Of these cells, 90.3±6.0% expressed Cxcl12 at very high levels (Fig. 4B), and were large reticular cells distributed in BM parenchyma and perivascular areas (Fig. 4D). A few IL-7+ cells did not express Cxcl12 (2.8±3.2%, Fig. 4B and D), and these cells were also large reticular cells distributed in parenchyma and perivascular areas (not shown). Although >97% of IL-7+ cells expressed Cxcl12 (at intermediate and high amounts), only 61.5±3.9% of the CXCL12+ cells were marked by Il7-driven GFP signal (Fig. 4C).
Figure 4. Most IL-7+ cells are a subset of CXCL12-abundant reticular cells.
A, Frequency of IL-7+ cells in BM. B, Cxcl12 expression in IL-7+ CD45− cells. C, Cxcl12 and Il7 expression in CD45− cells (left and middle). Il7 expression in Cxcl12+ cells (right). D, 25μm-thick section of Il7GFP/+;Cxcl12DsRed/+ mouse femur stained to detect IL-7+ cells. CXCL12+ cells were directly visualized. Scale bar is 50 μm. Insert, example of an IL-7+ CXCL12− cell, and of an IL-7+ CXCL12+ cell. E Left, gated IL-7+ cells; right, LEPR expression in gated IL-7+ cells. Data in panels A–D is representative of more than 5 mice analyzed. F, Lineage mapping of Il7 expressing cells using Lepr-cre transgenic mice. 8 μm-thick femur section of Lepr-cre; Rosa26tdtomato/+; Il7GFP/+ mice. Scale bar is 50 μm. Arrow and arrowhead indicate Lepr-cre-derived IL-7+ cells and Lepr-cre-derived IL-7− cells respectively. Yellow arrow indicates IL-7+ Lepr-cre− cells. Data are representative of 2 independent experiments. G, HSC, MPP, and CLP number in BM of Lepr-cre+; Il7fl/fl and Lepr-cre−; Il7fl/fl mice. H, histogram of pSTAT5α in Ly6D+ CLPs from Lepr-cre+; Il7fl/fl and Lepr-cre−; Il7fl/fl mice. I, pSTAT5α geometric mean intensity in Ly6D+ CLPs in Lepr-cre−; Il7fl/fl mice and in Lepr-cre+; Il7fl/fl (ΔIl7) mice. J and K, Developing B cell numbers in BM (J); splenic B cells (K) in Lepr-cre+; Il7fl/fl and Lepr-cre−; Il7fl/fl mice. In panels G, I, J and K, bars indicate mean, circles depict individual mice. Data are representative of 2 independent experiments with 4–6 mice per group. * P<0.05, **P<0.01, ***P<0.001 (unpaired, two-tailed Student’s t-test). See also Figure S5.
CAR cells share functional similarities with a subset of mesenchymal progenitor cells that expresses Leptin receptor (Omatsu et al., 2010; Zhou et al., 2014). The majority of IL-7+ cells (88.3±8.6%) expressed LEPR, (Fig 4E); Lepr-cre driven tandem dimer Tomato expression marked IL-7+ BM cells (Fig. 4F), and Lepr-cre driven Zoanthus sp Green fluorescent protein (ZsGreen) expression marked 97.6% of CAR cells ((ZsGreen expression driven by the Rosa26 promoter repressed by a loxp-flanked transcriptional repressor; Fig. S5A and B). Next, we asked if Il7 expressed by LEPR+ cells was required for B-lymphopoiesis. We found that conditional deletion of Il7 from LEPR+ cells resulted in a significant reduction in Ly6D+ CLPs in BM (Fig. 4G), and reduced phosphorylated (p)STAT5α in these cells (Fig. 4H and I). Consequently, the number of developing B cell subsets was dramatically reduced (Fig. 4J), which led to peripheral B lymphopenia (Fig. 4K, and Fig. S5C). The number of monocytes and granulocytes were unchanged (Fig. S6C). In the thymus, even though early thymic precursors (ETPs) were significantly reduced (Fig. S5D), it did not impact downstream thymocyte populations, nor peripheral T cell numbers (Fig. S5E–G). Finally, deletion of Il7 from mesenchymal progenitor cells using Prx1-cre (PRX1 is a transcription factor expressed in the limb bud mesoderm) (Logan et al., 2002)) also led to a significant reduction in developing B cell subsets in BM (Fig. S5H and I).
Endothelial cell-derived IL-7 contributes to B-lymphopoiesis
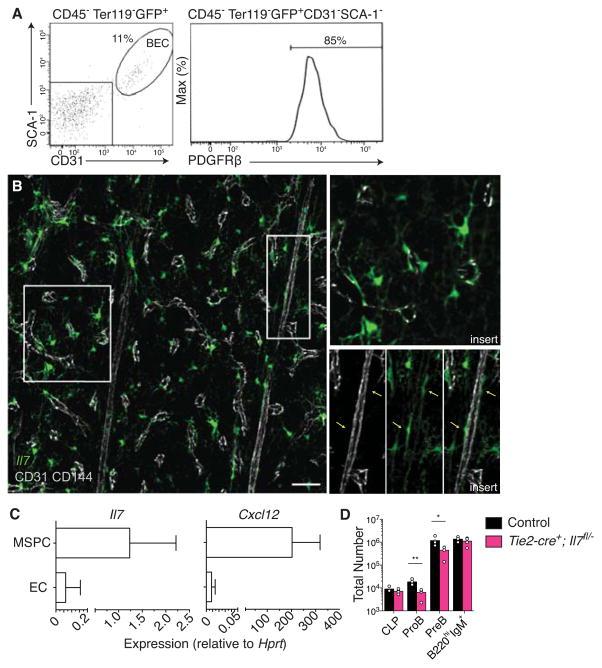
A previous study indicated that some IL-7+ cells in BM express endothelial cell markers (Hara et al., 2012). Likewise, we found that a small fraction (20.6±7.2%) of IL-7+ cells expressed CD31 (PECAM-1) and Stem Cells Aantigen-1 (SCA-1, Fig. 5A). To rule out the possibility of an in vitro staining artifact, we analyzed IL-7+ cells in relation to BM endothelial cells by confocal microscopy. We found that in some BM sinusoidal endothelial cells, vascular endothelial growth factor-3 (VEGFR3), CD31 and CD144 co-localize with GFP expression driven by Il7 (Fig. 5B and S6A). Occasionally, we found extremely large and elongated IL-7+ cells adjacent to arterioles, which resembled a population of Nestin-GFP bright cells, Nestinperi, (Kunisaki et al., 2013), that also expressed the pericyte marker neural-glial antigen-2 (NG2, Fig. 5B and Fig. S6B and C). We then sorted CD144+ CD31+ CD45− Ter119− BM endothelial cells, and LEPR+ CD144− CD31− CD45− Ter119− MSPCs, and compared Il7 and Cxcl12 expression by qPCR. We found that even though Il7 (and Cxcl12) was mostly expressed by MSPCs, Il7 expression could also be readily detected in BM endothelial cells (Fig. 5C). To analyze the contribution of endothelial cell-derived IL-7 to B cell development, we crossed Il7−/−; Tie2-cre male mice with Il7fl/fl females, and analyzed CLPs and B cell development in the BM. Il7 deletion from endothelial cells using a Tie2-cre transgene resulted in a small, selective, but significant reduction in proB and preB cell numbers in BM (Fig. 5D), suggesting that proB and preB cell proliferation was sensitive to endothelial sources of IL-7. In control experiments using Tie2-cre male mice crossed with Rosa26tdTomato/+ female mice (tdTomato expression driven by the Rosa26 promoter repressed by a loxp-flanked transcriptional repressor), we also noted that Tie2-cre activity was significantly higher in BM endothelial cells isolated from female progeny as compared to male progeny (Fig. S6D). In summary, a small fraction of sinusoidal endothelial cells provide a minor source of IL-7 for B lymphocyte development.
Figure 5. Endothelial cell-derived IL-7 contributes to B cell development.
A, Flow cytometric analysis of SCA-1 and CD31 expression in IL-7+ BM cells. Blood endothelial cells (BECs). B, Il7 colocalization with CD31 and VE-Cadherin. Scale bar is 50 μm. Data are representative of more than 5 mice analyzed. C, Il7 and Cxcl12 expression in FACS sorted CD45−Ter119−CD144−CD31−LEPR+ MSPCs and endothelial cells (ECs, CD45−Ter119−CD144+CD31+LEPR−). Gene expression is relative to Hprt. Bars indicate average ± SD of 3-independent cell sorting experiments. D, Total number of hematopoietic cell subsets in BM of Il7fl/−; Tie2-cre+ mice and control littermates. Bars indicate average, circles depict individual mice. Data are representative of 2 independent experiments with 4–6 mice per group. * P<0.05, (unpaired, two-tailed Student’s t-test). See also Figure S6.
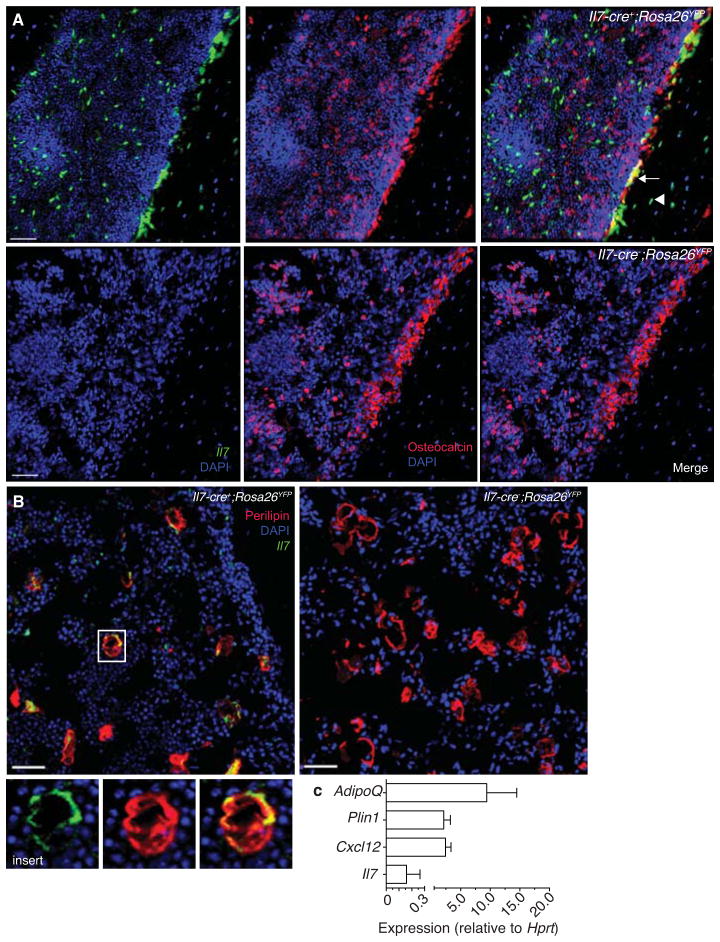
IL-7+ cells are mesenchymal progenitor cells
CAR cells include mesenchymal progenitor cells that can differentiate into multiple cell lineages including osteoblasts, osteocytes, chondrocytes and adipocytes, and possibly stromal cells (Omatsu et al., 2010). As most IL-7+ cells form a subset of CAR cells it raised the possibility that IL-7+ cells also overlap with mesenchymal progenitor cells. Alternatively, IL-7+ CAR cells could represent a differentiated mesenchymal subset that no longer retains multipotent differentiation potential. To distinguish between these possibilities, we traced cell lineages that differentiated from IL-7+ cells by crossing Il7-cre transgenic mice (Repass et al., 2009) with Rosa26YFP mice (YFP expression driven by the Rosa26 promoter repressed by a loxp-flanked transcriptional repressor), and examined the cellular composition and distribution of YFP+ cells in BM. We found that a large fraction of bone-lining osteocalcin+ osteoblasts were YFP+, and a few bone-embedded osteocytes were also YFP+ (Fig. 6A). To determine if adipocytes could differentiate from IL-7+ cells, we induced adipogenesis by sub-lethal irradiation (Bryon et al., 1979) and examined femurs and tibias of Il7-cre+; Rosa26YFP mice for YFP expression in perilipin+ adipocytes. We found that irradiation-induced adipogenesis occurred partly from IL-7+ cells (Fig. 6B), while adipocytes expressed marginal amounts of Il7 and high amounts of adipocyte-specific adiponectin and perilipin (Fig. 6C). Combined, these data show that IL-7+ stromal cells can differentiate into adipocytes and osteoblasts that express low and undetectable amounts of Il7 mRNA, respectively. In summary, IL-7+ cells contain mesenchymal progenitor activity with multilineage differentiation capacity.
Figure 6. IL-7+ BM cells are mesenchymal progenitor cells.
A, 25μm-thick sections of Il7-cre; Rosa26YFP/+ or control mouse femurs stained to detect osteocalcin+ cells and YFP+ cells. Arrow indicates osteoblasts derived from IL-7+ cells, arrowhead indicates osteocyte derived from IL-7+ cell. Scale bar is 50 μm. B, 25μm-thick sections of irradiated Il7-cre; Rosa26YFP/+ or control mouse femurs stained to detect perilipin+ cells and YFP+ cells. Scale bar is 50 μm. Insert shows Il7-cre-derived adipocyte. C, Adipoq, Plin1, Cxcl12, and Il7 mRNA expression in adipocytes. mRNA expression is relative to Hprt. Data in all panels are representative of two independent experiments.
IL-7+ cells are a component of HSC niches and regulate HSC and MPP numbers in BM
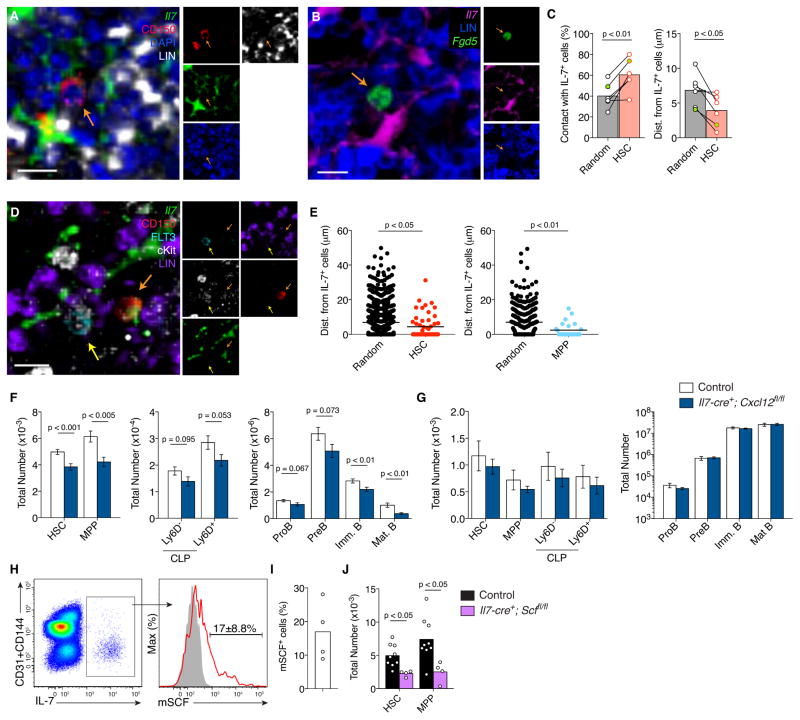
The fact that IL-7+ cells share many functional and phenotypic similarities with HSC niche cells led us to examine the distribution of HSCs and MPPs relative to IL-7+ cells in femur whole-mounts by confocal microscopy. Even though IL-7+ cells comprise < 0.1% of total BM cells, these cells are large, reticular, and form a vast cellular network that interacts with many cells (Fig. 4D and Fig. S3A), thus raising the concern that HSC distribution relative to IL-7+ cells might be random. To address this concern, we measured the distribution of randomly placed cells relative to IL-7+ cells. In tissues collected from 5 mice, we detected approximately 60% of HSCs identified as Lin− CD41− CD48− CD150+, or as Lin− CD41− CD48− Fgd5-ZsGreen+ (Gazit et al., 2014), in direct contact with IL-7+ cells (Fig. 7A – C, and Fig. S7A and B), and the average distance between HSCs and the nearest IL-7+ cell was 3.9 μm (Fig. 7C). We found that the HSC distribution was significantly different from random cell positioning in BM. The distribution of MPPs (identified as Lin− CD41− CD48− cKIT+ CD150− FLT3+) relative to IL-7+ cells was similar to HSCs (Fig. 7D and E), and in four occasions out of 42 HSCs and 77 MPPs analyzed, we found an HSC and an MPP in the vicinity of a single IL-7+ cell (Fig. 7D), suggesting that HSCs and MPPs can interact with the same IL-7+ cell. These findings led us to ask whether CXCL12 produced by IL-7+ cells was important for HSC and MPP maintenance, and B lymphocyte development. To address this question, we conditionally deleted Cxcl12 from IL-7+ cells by crossing Cxcl12fl/fl mice (Ding and Morrison, 2013) with Il7-cre transgenic mice, and quantified HSCs, MPPs, CLPs, and developing B cell subsets in BM and spleen by flow cytometry. We found significant reductions in the number of HSCs, MPPs, CLPs, and developing B cell subsets in BM (Fig. 7F), whereas in the spleen these cell populations were numerically equivalent (Fig. 7G). These data indicated that CXCL12 produced by IL-7+ cells played an important role in the generation and/or maintenance of normal numbers of HSCs, but not in HSC retention within BM. Besides CXCL12, HSC maintenance also requires cell-intrinsic cKit signaling triggered by membrane-bound SCF (mSCF). Of note, mSCF was readily detected in ~17% of IL-7+ cells (Fig. 7H and I). Importantly, conditional Scf deletion from IL-7+ cells resulted in a significant reduction of HSC and MPP numbers in BM (Fig. 7J). Control experiments showed that Il7-cre marks ~80% of mesenchymal LEPR+ cells in BM, whereas Lepr-cre acts on more than 95% of mesenchymal LEPR+ cells in BM (Fig. S7C–E). These data showed that HSC niche cells expressed factors essential for HSC and MPP maintenance, as well as for lymphoid-lineage commitment and differentiation.
Figure 7. HSCs and MPPs reside in, and are maintained by, niches formed by IL-7+ cells.
A, Femur whole-mount from Il7-ECFP mice stained with antibodies to detect CFP, lineage, nuclei (DAPI), and CD150. Scale bar is 10 μm. Orange arrow indicates HSC. B, Femur whole-mount from Il7-ECFP mice transplanted with Fgd5Zsgreen/+ BM cells stained with antibodies to detect CFP and lineage (blue). Zsgreen+ cells were directly visualized. Scale bar is 10 μm. Orange arrow indicates HSC. C, Quantification of HSCs and randomly selected cells in contact with IL-7+ (CFP+) BM cells (left), and average distances to nearest IL-7+ cell (right). Circles indicate average of at least 9 HSCs or > 100 randomly selected cells in individual mice analyzed. Green filled circle indicates chimera from Fgd5Zsgreen/+ BM; lines connect random cells and observed HSCs in individual mice. Statistical significance calculated with paired t test. D, Femur whole-mount section from Il7-ECFP mice stained with antibodies to detect CFP, lineage, cKit, FLT3, and CD150. Scale bar is 10 μm. Orange arrow indicates HSC; yellow arrow indicates MPP. E, Quantification of cell distance to nearest IL-7+ cell: HSCs (n = 52), MPPs (n = 25) and randomly selected cells. Circles indicate individual cells analyzed from 5 independent experiments for HSCs, and 2 independent experiments for MPPs. Statistical significance calculated with unpaired t test. F and G, Enumeration of hematopoietic cell subsets in BM (n=15–20) (F) and spleen (n=9) (G) of Il7-cre+; Cxcl12fl/fl and littermate controls. Bars indicate the mean±S.E.M. Statistical significance calculated with unpaired t test. H, Membrane-bound SCF expression in IL-7+ BM cells. Left panel indicates CD31+CD144 expression versus IL-7-GFP in gated live CD45+Ter119− BM cells. Right panel shows mSCF expression in gated IL-7+ cells; filled histogram is staining control. I, Frequency of mSCF+ cells within the IL-7+ BM cell gate. Bar indicates average, circles represent individual mice analyzed. J, Enumeration of HSCs and MPPs in BM of Il7-cre+; Scffl/fl mice and littermate controls. Bars indicate the mean, circles depict individual mice analyzed. Statistical significance calculated with unpaired t test. See also Figure S7.
Discussion
Here we showed that the BM niche formed by IL-7+ cells represented a common niche for hematopoietic progenitors at multiple stages of differentiation. The majority of IL-7+ cells formed a subset of mesenchymal progenitor CAR cells capable of multilineage differentiation, and a minor fraction of IL-7+ cells were BM sinusoidal endothelial cells. CXCR4 was critically required in CLPs for positioning in the vicinity of IL-7+ cells in BM, and proximity to IL-7 enabled optimal IL-7R signaling and B cell developmental progression. Furthermore, a considerable fraction of HSCs and MPPs were positioned in direct contact with IL-7+ cells, which regulated HSC and MPP numbers through CXCL12 and membrane-bound SCF expression.
HSCs require short-range extrinsic factors, such as mSCF, produced by rare MSPCs and endothelial cells in BM for their long-term maintenance. SCF-producing cells overlap entirely with CAR cells (Ding et al., 2012), and IL-7 expression marked the mesenchymal progenitor subset that expresses the highest amounts of CXCL12. Furthermore, Nestin-GFP expressing mesenchymal cells are also critical components of HSC niches in BM, and these cells also express essential HSC maintenance and lymphoid differentiation factors, and largely overlap with IL-7+ and IL-7− CAR cells. As HSCs are highly dependent on CXCR4 signaling for quiescence and long-term maintenance (Nie et al., 2008; Sugiyama et al., 2006), our studies suggest that HSC quiescence and lymphopoiesis are distinct cellular decisions controlled by highly overlapping cellular niches.
Mesenchymal progenitor cells respond to long-range signals, such as cytokines and hormones, which regulate mesenchymal progenitor cell activity. For example, adrenergic signals controlled by circadian rhythms are sensed by beta 3 adrenergic receptors expressed on mesenchymal progenitor cells and reduce CXCL12 production (Mendez-Ferrer et al., 2008). Consequently, HSPCs are displaced from HSC niches and exit the BM in a circadian manner, although the physiological role for HSPC recirculation remains poorly understood. As our data showed that hematopoietic cell lineages are differentially sensitive to CXCR4 signaling (lymphoid lineages being more dependent on CXCR4 than myeloid and erythro-megakaryocyte lineages), we suggest that circadian fluctuations in CXCL12 production contribute to the regulation of both HSC and MPP quiescence as well as multilineage differentiation. Periodic fluctuations in CXCR4 signaling may allow HSCs and MPPs to uncouple from niche signals that promote cellular quiescence (e.g. mSCF) and signals that support lymphoid lineage commitment, thereby allowing for cell division, self-renewal, and commitment to the myeloid lineage. Similarly, fluctuations in CXCL12 production may regulate IL7Ra signaling and EBF1 and PAX5 expression in CLPs (Dias et al., 2005), and lead to consequent fluctuations in the activation of B cell fate gene programs and repression of T, NK and innate lymphoid cell fates (Nechanitzky et al., 2013). In this model, constitutive CXCR4 signaling would prevent HSC and MPP uncoupling from quiescence signals to divide and differentiate, which is highly consistent with evidence that CXCR4 signaling intensity inversely correlated with hematopoietic reconstitution in mice and in humans (McDermott et al., 2015).
B cells develop through multiple developmental stages in which entry and exit from cell cycle and RAG protein expression are tightly controlled. Some evidence suggests that distinct BM niches control RAG expression and cell cycle, particularly at the preB cell stage where cell proliferation driven by IL-7R signaling inhibits Rag gene re-expression and prevents Ig light chain gene recombination (Clark et al., 2014). Specifically, CXCR4 expression is increased in preB cells when compared to proB cells, and such an increase was proposed to attenuate IL7R signaling by positioning preB cells away from IL-7+ cells (Johnson et al., 2008). However, our studies using IL-7 and CXCL12 reporter mice demonstrated that Il7 is expressed by BM cells that express the highest amounts of CXCL12. Given the notorious difficulty we, and others (Mazzucchelli et al., 2009), had in visualizing IL-7+ cells in situ, it is possible that the previous studies that identified IL-7+ cells in BM (Tokoyoda et al., 2004) were able to visualize IL-7+ CXCL12− cells only. In our studies, we found that IL-7 produced by endothelial cells contributed primarily to proB and preB cell development, suggesting that perivascular niches might be important sites for proB and preB cell differentiation. Alternatively, as proB and preB cell proliferation is highly dependent on IL-7, and both subsets divide more rapidly than CLPs, it is possible that proB and preB cells are more sensitive than CLPs to small reductions in IL-7 in BM. Thus, studies designed to quantify the distribution of proB and preB cells in relationship to endothelial cells, IL-7+ and IL-7− CAR cells, are needed for a detailed understanding of the BM niches controlling B cell developmental transitions.
Osteolineage cells, and specifically osteoblasts, have been implicated as essential regulators of BM niches supporting the development of lymphoid lineages. Using in vivo cell ablation strategies, previous studies demonstrated significant reductions in CLPs and downstream lymphoid progenitors when osteolineage cells and osteoblasts were induced to undergo apoptosis (Terashima et al., 2016; Visnjic et al., 2004; Yu et al., 2016; Zhu et al., 2007). However, the use of conditional approaches for inducing osteoblast-lineage cell death result in synchronized cell apoptosis within BM that may not be efficiently cleared by phagocytic macrophages, which may drive an inflammatory response. In turn, inflammatory signals not only reduce B lymphopoiesis (Ueda et al., 2004), but may also affect HSC numbers (Glatman Zaretsky et al., 2014), and these changes were evident in some of these studies (Visnjic et al., 2004; Zhu et al., 2007). Adding support to this possibility, a recent study showed an inflammation-dependent increase in myeloid cell populations in BM presumably caused by osteolineage cell ablation toxicity (Yu et al., 2016).
Other studies lending support to the model that endosteal niches control lymphopoiesis focused on the effects of conditional gene deletion in osteolineage cells and osteoblasts. For example, Gs alpha subunit (Gnas) and Il7 deletion in osteolineage cells and osteoblasts revealed moderate to profound reductions in lymphopoiesis (Terashima et al., 2016; Wu et al., 2008; Yu et al., 2016). However, these studies used conditional gene deletion strategies that are highly efficient in mesenchymal progenitor stages prior to osteolineage commitment. Specifically, constitutive Sp7-driven (Sp7 encodes the transcription factor Osterix) cre activity has been shown to mark mesenchymal-lineage progenitors, including osteolineage cells (Greenbaum et al., 2013). Additionally, an inducible Sp7-cre strain revealed that Sp7-cre activity is active in distinct waves of mesenchymal multipotent progenitors during embryonic and postnatal life until 8 weeks of age, after which inducible Sp7-cre activity becomes restricted to osteoblasts (Mizoguchi et al., 2014). Importantly, the large majority of mesenchymal-lineage cells marked by inducible Sp7-cre activity at 6 weeks of age contained very few osteoblasts (Yu et al., 2016). Thus, these data strongly suggest that conditional Il7 deletion using the inducible Sp7-cre at 6 weeks of age (Terashima et al., 2016) favored IL-7 deletion from MSPCs, and very little from osteoblasts. Likewise, the constitutive osteocalcin promoter-driven cre expression used to delete Il7 in osteoblasts (Terashima et al., 2016) targets ~70% of CAR cells (Zhang and Link, 2016), which include osteolineage progenitors (Omatsu et al., 2010). Finally, Cxcl12 deletion from osteoblasts using the Col2.3-cre transgene led to a small but significant reduction in BM CLPs that did not cause B-lymphopenia (Ding and Morrison, 2013). However, it should be noted that Col1a1 expression is detectable in the transcriptome of Nestin+ MSPCs and SCF+ BM stromal cells (Ding et al., 2012; Kunisaki et al., 2013; Mendez-Ferrer et al., 2010) and thus it is possible that the Col2.3-cre transgene is already active in a small fraction of MSPCs.
In summary, our studies revealed that IL-7 was produced in BM by a rare and heterogeneous cell subset composed of mesenchymal cells with multilineage differentiation potential, by sinusoidal endothelial cells, and by periarteriolar pericytes. The majority of IL-7+ cells expressed the highest amount of CXCL12 in BM. Consequently, CLPs rely on CXCR4 signaling for positioning in proximity to IL-7+ cells, which was essential for accessing limiting IL-7 and for receiving sufficient IL-7R signaling. Although IL-7 is essential for B and T cell development in adult mice (Carvalho et al., 2001), and possibly in adult humans (Parrish et al., 2009), NK cell development is mostly dependent on IL-15. NK cells also developed poorly from CXCR4-deficient MPPs and CLPs, and CAR cells express IL-15 (Noda et al., 2011). Combined, these data demonstrate that the control of cell positioning in BM niches is an essential checkpoint in lymphopoiesis as it enables hematopoietic precursor cells to access limiting lymphoid-instructive cytokines. The findings that HSCs and MPPs were located near or in contact with IL-7+ cells and that CXCL12 and SCF expressed by IL-7+ cells are important for HSC and MPP maintenance favor a model in which HSC maintenance and multilineage differentiation are distinct cell lineage decisions controlled by overlapping BM niches. The functional and evolutionary advantage(s) for the local control of HSC maintenance and multilineage differentiation remain(s) to be determined.
Experimental Procedures
Mice
Adult C57BL/6 (CD45.2+) and Boy/J (CD45.1+) were purchased from The Jackson Laboratories or National Cancer Institute. Cxcl12fl/fl, Scffl/fl, Lepr-cre, Rosa26YFP, Rosa26tdtomato/+ (stock 007909), Tie2-cre (stock 004128), Fgd5ZsGreen/+, and Prx1-cre (stock 005584) mice were also obtained from The Jackson Laboratories. Il7-cre, Il7racre/+, Cxcr4fl/fl and Il7fl/fl mice were from internal colonies. Flk2-cre mice were a gift from Dr. E. Camilla Forsberg (University of California, Santa Cruz). We noted that the Flk2-cre transgene is inherited from the Y-chromosome, but hematopoietic cell composition in BM and secondary lymphoid organs of Flk2-cre and littermate controls is indistinguishable. Il7-ECFP mice were provided by Dr. Scott Durum (National Institute of Health). Some Il7GFP/+ mice (Hara et al. 2012) were from our internal colony (Kyoto University); some Il7GFP/+ mice were obtained from Dr. Susan Kaech (Yale University). Cxcl12DsRed/+ mice were a kind gift from Dr. Sean Morrison (University of Texas Southwestern Medical Center). Col2.3-cre mice were kindly provided by T. Nakamura (Kyoto University). Vav-cre mice were kindly provided by Dr. Thomas Graf (Albert Einstein College of Medicine). All mice were maintained under specific pathogen-free conditions at Yale Animal Resources Center and the Experimental Research Center for Infectious Diseases in the Institute for Virus Research, Kyoto University, and used according to the protocol approved by the Yale University Institutional Animal Care and Use Committee and by the Animal Research Committee at the Institute for Virus Research, Kyoto University.
Immunostaining of bone marrow
BM Whole Mounts. Whole mounts were stained with primary antibodies for 2–3 days at 4°C and secondary antibodies for 1 day at 4°C. For random positioning analysis, spots were placed on all DAPI+ or LIN+ cells, and each spot was given a unique identification number ranging from 0 to N. The randbetween() function in Microsoft Excel was then used to generate random numbers between 0 and N (each of which corresponded to a DAPI+ or LIN+ spot). The distance between the selected spot and the closest IL-7+ cell was then measured using the measurement function in Imaris. Images were acquired on a Leica SP8 confocal microscope.
CLP single cell differentiation in vitro assays
Single live Ly6D+ CLP (Lin− CD127+ cKITint SCA1+ FLT3+ Ly6D+) were sorted to 96-well seeded with OP9 stromal cells in RPMI supplemented with Flt3L, SCF, IL-7 and IL-2 (final concentration of 50 ng/mL). On day 5, culture medium was replaced, and on day 10 scores were assigned for B cells (CD19+), myeloid (CD11b+), NK cells (NK1.1+) and dendritic cells (CD11c+) by flow cytometry.
Mesenchymal progenitor cell sorting, and mSCF analyses
Fresh unfixed collagenase-digested BM cells were stained with rat anti-mouse SCF antibody (R&D) for 1 hour. SCF staining was revealed with an anti-rat AlexaFluor 555 antibody. Cells were acquired and sorted on a BD FACS Aria II equipped with UV (355nm), Violet (405nm), Blue (488nm), Green (532nm) and Red (640nm) lasers, and with FACSDiva 7.
Supplementary Material
Acknowledgments
We thank Peter Cresswell, Michael McCune and Susan Kaech for mice and equipment. ACG was funded by FCT (SFRH/BD/73782/2010); VYL was supported by A*STAR, Singapore. EN was sponsored by NIH (1F32AR063574-01A1). DHB was supported by the Austrian Science Fund (J3220-B19). This work was supported by the NIH (R56AI098996 and RO1AI113040), by the Ministry of Education, Culture, Sports, Science, and Technology of Japan ([C] 25460589, 25111504 and 15H01153 to K.I.; [B] 24790468 and 26860322 to T.H.), by Fujiwara Memorial Foundation, by BioLegend/Tomy Digital Biology, and by the Shimizu Foundation for Immunology and Neuroscience.
Footnotes
Author contributions
ACG and VYL designed and performed experiments, and wrote the paper. S T-I, TS, EN and DHB performed some experiments and revised the paper. SS, HRR, and ER generated Il7raCre/+ and Il7-cre mice, respectively, and revised the paper. TN, KI and RAF supervised some studies and revised the paper. TH designed, performed, and supervised some experiments, and wrote the manuscript. JPP conceptualized and supervised studies, performed some experiments, and wrote the paper.
References
- Ara T, Tokoyoda K, Sugiyama T, Egawa T, Kawabata K, Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19:257–267. doi: 10.1016/s1074-7613(03)00201-2. [DOI] [PubMed] [Google Scholar]
- Avecilla ST, Hattori K, Heissig B, Tejada R, Liao F, Shido K, Jin DK, Dias S, Zhang F, Hartman TE, et al. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat Med. 2004;10:64–71. doi: 10.1038/nm973. [DOI] [PubMed] [Google Scholar]
- Beck TC, Gomes AC, Cyster JG, Pereira JP. CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. The Journal of experimental medicine. 2014;211:2567–2581. doi: 10.1084/jem.20140457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer SW, Schroeder AV, Smith-Berdan S, Forsberg EC. All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell. 2011;9:64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryon PA, Gentilhomme O, Fiere D. Histomorphometric analysis of bone-marrow adipose density and heterogeneity in myeloid aplasia and dysplasia (author’s transl) Pathol Biol (Paris) 1979;27:209–213. [PubMed] [Google Scholar]
- Carvalho TL, Mota-Santos T, Cumano A, Demengeot J, Vieira P. Arrested B lymphopoiesis and persistence of activated B cells in adult interleukin 7(-/)- mice. The Journal of experimental medicine. 2001;194:1141–1150. doi: 10.1084/jem.194.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol. 2014;14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Silva H, Jr, Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. The Journal of experimental medicine. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R, Mandal PK, Ebina W, Ben-Zvi A, Nombela-Arrieta C, Silberstein LE, Rossi DJ. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. The Journal of experimental medicine. 2014;211:1315–1331. doi: 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatman Zaretsky A, Engiles JB, Hunter CA. Infection-induced changes in hematopoiesis. J Immunol. 2014;192:27–33. doi: 10.4049/jimmunol.1302061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Shitara S, Imai K, Miyachi H, Kitano S, Yao H, Tani-ichi S, Ikuta K. Identification of IL-7-producing cells in primary and secondary lymphoid organs using IL-7-GFP knock-in mice. J Immunol. 2012;189:1577–1584. doi: 10.4049/jimmunol.1200586. [DOI] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T, Kollet O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2m(null) mice. Leukemia. 2002;16:1992–2003. doi: 10.1038/sj.leu.2402684. [DOI] [PubMed] [Google Scholar]
- Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, Kream BE. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. The International journal of developmental biology. 2004;48:645–653. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, Scadden DT. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci U S A. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Jones D, Springer TA. The chemokine receptor CXCR4 is required for the retention of B lineage and granulocytic precursors within the bone marrow microenvironment. Immunity. 1999;10:463–471. doi: 10.1016/s1074-7613(00)80046-1. [DOI] [PubMed] [Google Scholar]
- Mazzucchelli RI, Warming S, Lawrence SM, Ishii M, Abshari M, Washington AV, Feigenbaum L, Warner AC, Sims DJ, Li WQ, et al. Visualization and identification of IL-7 producing cells in reporter mice. PLoS One. 2009;4:e7637. doi: 10.1371/journal.pone.0007637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott DH, Gao JL, Liu Q, Siwicki M, Martens C, Jacobs P, Velez D, Yim E, Bryke CR, Hsu N, et al. Chromothriptic cure of WHIM syndrome. Cell. 2015;160:686–699. doi: 10.1016/j.cell.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CN, Hartigan-O’Connor DJ, Lee MS, Laidlaw G, Cornelissen IP, Matloubian M, Coughlin SR, McDonald DM, McCune JM. IL-7 production in murine lymphatic endothelial cells and induction in the setting of peripheral lymphopenia. Int Immunol. 2013;25:471–483. doi: 10.1093/intimm/dxt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Pinho S, Ahmed J, Kunisaki Y, Hanoun M, Mendelson A, Ono N, Kronenberg HM, Frenette PS. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nature immunology. 2013;14:867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- Nie Y, Han YC, Zou YR. CXCR4 is required for the quiescence of primitive hematopoietic cells. The Journal of experimental medicine. 2008;205:777–783. doi: 10.1084/jem.20072513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M, Omatsu Y, Sugiyama T, Oishi S, Fujii N, Nagasawa T. CXCL12-CXCR4 chemokine signaling is essential for NK-cell development in adult mice. Blood. 2011;117:451–458. doi: 10.1182/blood-2010-04-277897. [DOI] [PubMed] [Google Scholar]
- Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, Nagasawa T. The Essential Functions of Adipo-osteogenic Progenitors as the Hematopoietic Stem and Progenitor Cell Niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Parrish YK, Baez I, Milford TA, Benitez A, Galloway N, Rogerio JW, Sahakian E, Kagoda M, Huang G, Hao QL, et al. IL-7 Dependence in human B lymphopoiesis increases during progression of ontogeny from cord blood to bone marrow. J Immunol. 2009;182:4255–4266. doi: 10.4049/jimmunol.0800489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, Nagler A, Ben-Hur H, Many A, Shultz L, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science (New York, NY) 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- Pereira JP, An J, Xu Y, Huang Y, Cyster JG. Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids. Nature immunology. 2009;10:403–411. doi: 10.1038/ni.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repass JF, Laurent MN, Carter C, Reizis B, Bedford MT, Cardenas K, Narang P, Coles M, Richie ER. IL7-hCD25 and IL7-Cre BAC transgenic mouse lines: new tools for analysis of IL-7 expressing cells. Genesis. 2009;47:281–287. doi: 10.1002/dvg.20497. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Terashima A, Okamoto K, Nakashima T, Akira S, Ikuta K, Takayanagi H. Sepsis-Induced Osteoblast Ablation Causes Immunodeficiency. Immunity. 2016;44:1434–1443. doi: 10.1016/j.immuni.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Tsapogas P, Zandi S, Ahsberg J, Zetterblad J, Welinder E, Jonsson JI, Mansson R, Qian H, Sigvardsson M. IL-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors. Blood. 2011;118:1283–1290. doi: 10.1182/blood-2011-01-332189. [DOI] [PubMed] [Google Scholar]
- Tzeng YS, Li H, Kang YL, Chen WC, Cheng WC, Lai DM. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117:429–439. doi: 10.1182/blood-2010-01-266833. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Yang K, Foster SJ, Kondo M, Kelsoe G. Inflammation controls B lymphopoiesis by regulating chemokine CXCL12 expression. The Journal of experimental medicine. 2004;199:47–58. doi: 10.1084/jem.20031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnjic D, Kalajzic Z, Rowe DW, Katavic V, Lorenzo J, Aguila HL. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258–3264. doi: 10.1182/blood-2003-11-4011. [DOI] [PubMed] [Google Scholar]
- Wu JY, Purton LE, Rodda SJ, Chen M, Weinstein LS, McMahon AP, Scadden DT, Kronenberg HM. Osteoblastic regulation of B lymphopoiesis is mediated by Gs{alpha}-dependent signaling pathways. Proc Natl Acad Sci U S A. 2008;105:16976–16981. doi: 10.1073/pnas.0802898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VW, Lymperi S, Oki T, Jones A, Swiatek P, Vasic R, Ferraro F, Scadden DT. Distinctive Mesenchymal-Parenchymal Cell Pairings Govern B Cell Differentiation in the Bone Marrow. Stem cell reports. 2016;7:220–235. doi: 10.1016/j.stemcr.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Link DC. Targeting of Mesenchymal Stromal Cells by Cre-Recombinase Transgenes Commonly Used to Target Osteoblast Lineage Cells. J Bone Miner Res. 2016 doi: 10.1002/jbmr.2877. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Garrett R, Jung Y, Zhang Y, Kim N, Wang J, Joe GJ, Hexner E, Choi Y, Taichman RS, Emerson SG. Osteoblasts support B-lymphocyte commitment and differentiation from hematopoietic stem cells. Blood. 2007;109:3706–3712. doi: 10.1182/blood-2006-08-041384. [DOI] [PubMed] [Google Scholar]
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.