Abstract
Numerical approaches to high-density single nucleotide polymorphism (SNP) data are often employed independently to address individual questions. We linked independent approaches in a bioinformatics pipeline for further insight. The pipeline driven by heterozygosity and Hardy-Weinberg equilibrium (HWE) analyses was applied to characterize Bos taurus and Bos indicus ancestry. We infer a gene co-heterozygosity network that regulates bovine fertility, from data on 18,363 cattle with genotypes for 729,068 SNP. Hierarchical clustering separated populations according to Bos taurus and Bos indicus ancestry. The weights of the first principal component were subjected to Normal mixture modelling allowing the estimation of a gene’s contribution to the Bos taurus-Bos indicus axis. We used deviation from HWE, contribution to Bos indicus content and association to fertility traits to select 1,284 genes. With this set, we developed a co-heterozygosity network where the group of genes annotated as fertility-related had significantly higher Bos indicus content compared to other functional classes of genes, while the group of genes associated with milk production had significantly higher Bos taurus content. The network analysis resulted in capturing novel gene associations of relevance to bovine domestication events. We report transcription factors that are likely to regulate genes associated with cattle domestication and tropical adaptation. Our pipeline can be generalized to any scenarios where population structure requires scrutiny at the molecular level, particularly in the presence of a priori set of genes known to impact a phenotype of evolutionary interest such as fertility.
Introduction
Genotype data from high-density single nucleotide polymorphism (SNP) arrays serves as a starting point for many genomic analyses as they can reflect a wide range of processes [1–3]. SNP data have been used to characterize linkage disequilibrium and estimate effective population size [4,5], to perform genome-wide association studies [6–8], to compress genomes and highlight regions of evolutionary interest in humans and livestock species [9–11], to study the genetic variants of common diseases [12–14], and to identify population structure and signatures of selection [5,15–18]. These numerical approaches are employed independently to address specific questions. Formally linking them in a computational routine can drive discovery. Population assignment at the DNA level can inform genotype-phenotype associations, because phenotypes of each population (or lineage) are distinct. Herein, we propose a computational routine to maximize the use of SNP data in a comparative genomics framework.
A typical use of SNP data for population genetics involves computation of percentage of heterozygosity (HET), fixation index (FST), and principal component analysis (PCA) [3]. The HET values serve as a summary of genotype data and provide first-hand information about the genetic diversity within a population. Related measures such as extended haplotype homozygosity (EHH) [19] and its variants have been used to identify selective sweeps and signatures within cattle breeds [20–23]. A literature gap is the exploration of HET values across genetically diverse cattle breeds. Computation of HET from SNP data could facilitate the discrimination of breeds with divergent ancestry (that is, sub-species of cattle: Bos indicus and Bos taurus). We proposed that gene ancestry can be calculated by computing the average HET of its SNP.
First attempts to classify livestock breeds using genetic markers were originally based on microsatellites [24–27] and most analysis included a few hundred animals and a handful of breeds. The Bovine HapMap Consortium [28] interrogated 37,470 SNP in 497 animals and used PCA to elucidate the genetic structure of diverse breeds. PCA was used to measure genetic divergence in Bos indicus and Bos taurus cattle [29,30] and to inform machine learning methods to predict cattle ancestry [29,31]. We distinguish our current work by developing new methods and expanding the dataset to include hundreds of animals per breed. We used PCA as a starting point to identify genes that have discriminatory power to identify cattle population as Bos indicus or Bos taurus. Then clustering methods were applied to average HET values to prove that our measure of gene ancestry is able to segregate cattle breeds according to known lineages, similarly to PCA. As HET values differ across breeds, we noticed a striking contrast between the set of genes that have high/low HET values in each breed. Gene ancestry was linked to biological processes in Gene Ontology enrichment analyses followed by annotation of gene attributes (whether a gene is a transcription factor, expressed in tissue-specific manner, codes a secreted protein, or codes kinases). Finally, we investigated if genes relevant to breed differences could interact with genes associated to fertility or lactation by building gene network based on average HET correlations.
Results and discussion
Overview of the bioinformatics pipeline
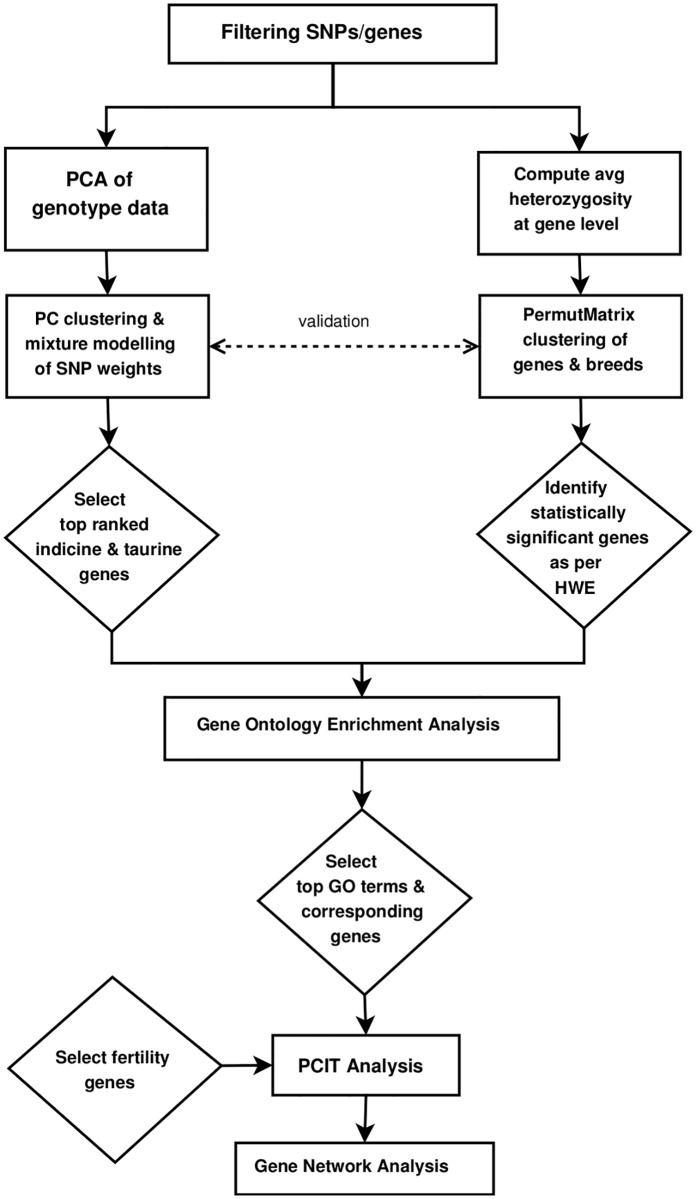
Our approach to analysing the genotype data of the various cattle breeds is schematically illustrated in the flowchart of Fig 1 and summarized in six steps: 1. Data pre-processing to select animal populations and genotypes for SNP in autosomal chromosomes within 1 kb of a known protein coding gene; 2. Principal component analysis (PCA) of the genotype data to characterize population structure; 3. Computation of the gene-level heterozygosity followed by clustering analysis to dissect population structure and selection of statistically significant genes based on deviation from Hardy-Weinberg equilibrium (HWE) values; 4. Perform Gene Ontology (GO) enrichment analysis on two gene lists–one derived from Bos indicus vs. Bos taurus content and another from HWE deviation; 5. Generation of a gene co-heterozygosity network using partial correlation and information theory [32] for candidate genes from the two lists, alongside fertility-related genes and milk-related genes from previous studies; 6. Analysis of the network structure and determination of the key genes. These six steps are detailed in S1 Text.
Fig 1. Flowchart of the pipeline for exploratory analysis of the effects of heterozygosity in the bovine genome.
Principal component and heterozygosity analyses reveal population structure in accordance to Bos indicus and Bos taurus ancestry of cattle breeds
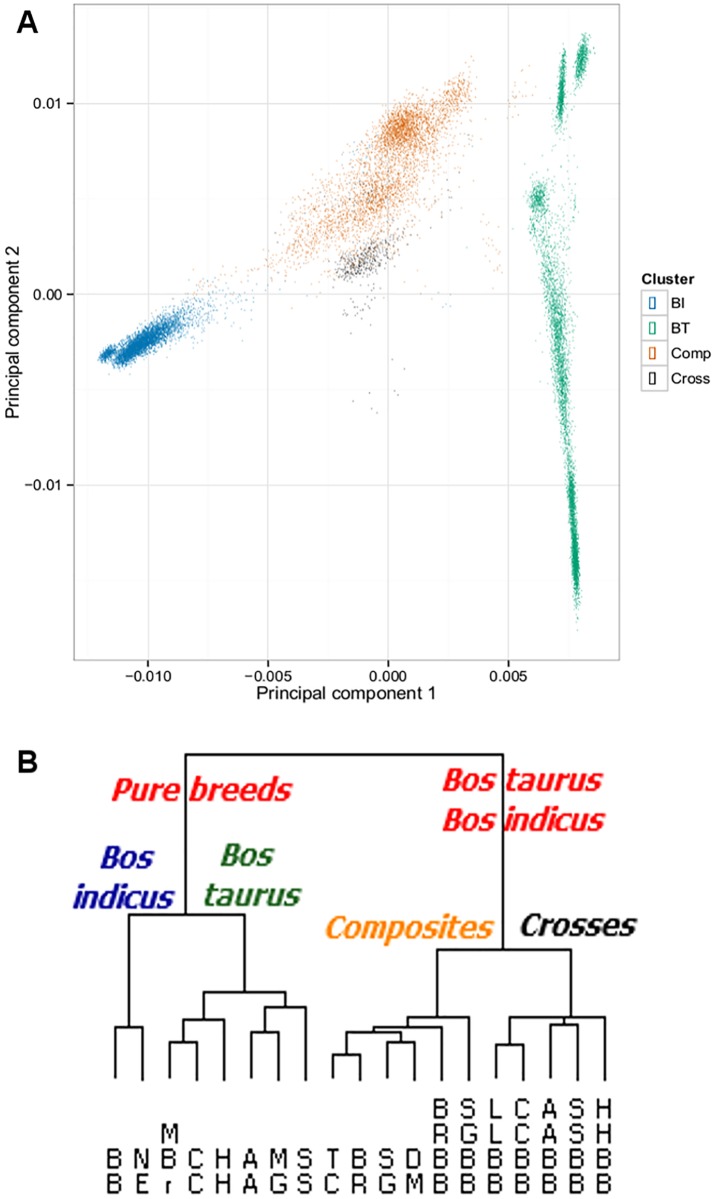
We performed a principal component analysis (PCA) of the genotype data (246,864 SNP) for 18,363 cattle of 19 breeds. A clear separation between the breeds based on their lineage is evident from PCA analyses (Fig 2A). The first two principal components explained 21.8% (PC1) and 2.3% (PC2) of the variation. We observed the pure Bos indicus breeds (BI) on the extreme left and the pure Bos taurus breeds (BT) on the extreme right of the PC1 spectrum. The middle region of the plot depicts the cattle corresponding to the Bos taurus-Bos indicus crossbreeds. These observations are consistent with documented knowledge of cattle history [28,33]. The crossbreeds LLBB, CCBB, AABB, SSBB, HHBB have similar genetics and clustered together (black cluster in Fig 2A). Similarly, the tropically-adapted breeds TC, BR, DM, and SG are clustered together (orange cluster in Fig 2A).
Fig 2.
(A) Principal Component Analysis of SNP genotypes corresponding to cattle breeds grouped based on their lineage. Left illustrates Bos indicus (BI), Middle shows Bos taurus—Bos indicus (cross-breeds and composite breeds), and Right corresponds to Bos taurus (BT); (B) Hierarchical clustering analysis of heterozygosity of 8,631 genes across the 19 cattle breeds produces a dendrogram showing the clustering of breeds consistent with their respective lineages.
Bos taurus breeds are genetically more conserved compared to the pure Bos indicus breeds–the Bos taurus breeds showed higher LD (r2 = 0.45) than their indicine (r2 = 0.25) and composite (r2 = 0.32) counterparts. This higher LD in taurine breeds was attributed to a smaller effective population size and a stronger bottleneck during breed formation [5].
A relative smaller variation within the Bos taurus breeds was observed, largely scattered along PC2 (see Fig 2A). In contrast, the Bos indicus breeds have larger variation along PC1 and we observe a gradual transition into the Bos taurus-Bos indicus breeds, consistent with previous findings [33]. The link between PC1 and Bos indicus content has motivated us to formally ascertain this relationship by computing the contribution of individual SNP to the Bos indicus content of the cattle. Recall that each principal component is a weighted linear combination of the features (SNP) in the data set. As part of PCA, we obtained the SNP weights for each of the principal components and as such, the importance of the SNP to each principal component.
We considered PC1 and analysed the SNP weights along this vector with an expectation, based on Fig 2A, that the pure Bos indicus breeds would have a negative value, the pure Bos taurus breeds would have positive values, and the Bos taurus-Bos indicus breeds would have a combination of positive and negative values. The empirical distribution of the SNP weights followed two distinct modes that required a mixture model with two normal distributions to quantify the contribution of the SNP to the Bos indicus content in cattle (S2 Text). Membership of 31% of SNP to Bos indicus and 69% to Bos taurus components was estimated (S1 Fig). We provided our entire list of 8,631 genes and their contributions to the indicine and taurine components of the bovine genome in S3 Text.
Hierarchical cluster analysis with respect to HET values was carried and also revealed the separation of cattle into distinct groups based on their ancestry and breed type (Fig 2B). The first partition in the hierarchy corresponds to purebreds and crossbreds towards the left and right, respectively. Within each pure versus cross-bred partition, we observed a remarkable separation based on the lineage of breeds. BB and NE (pure Bos indicus) have their own cluster while the breeds MBr, CC, AA, HH, MG, and SS are clustered together (pure Bos taurus). Among the cross-breeds with Bos taurus- Bos indicus lineage, we observed that the cross-breeds LLBB, CCBB, AABB, SSBB, and HHBB are clustered together. Similarly, the composite breeds TC, BR, DM, SG, BRBB, and SGBB are clustered together. These results align with those reported above for the PCA method [33]. The clustering method was able to detect this hidden population structure based only on the heterozygosity values at the gene level.
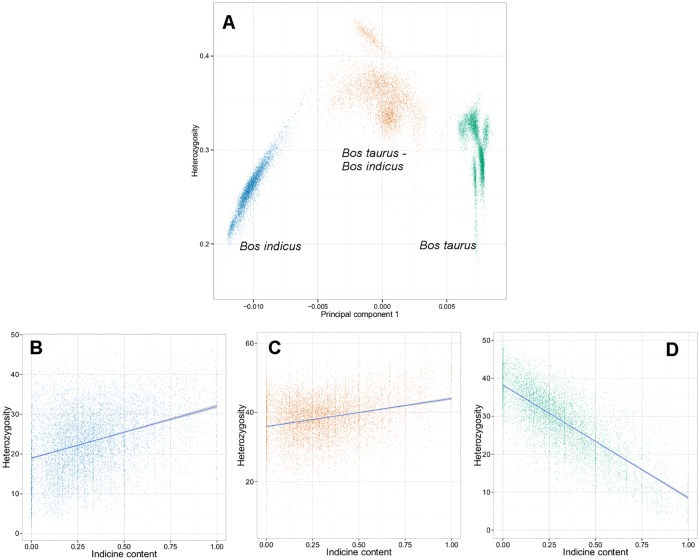
Heterozygosity and Bos indicus content were correlated metrics at the animal level and at the gene level within lineages (Fig 3). At the animal level, we found a strong non-linear relationship between the PC1 values and heterozygosity. This inverted V pattern has been recently reported by Samuels et al. [34] with various human populations. Its recapitulation here (Fig 3A) with beef cattle suggests some universal law by which heterozygosity alone governs the principal population structure in a genetically diverse sample. For the 8,631 genes under consideration, we observed a strong linear relationship between a gene’s heterozygosity and its contribution to Bos indicus content within the Bos taurus lineage (Fig 3D; Pearson correlation, r = –0.74), while the correlation strengths with the Bos indicus (Fig 3B) and Bos taurus-Bos indicus (Fig 3C) lineages are 0.35 and 0.28, respectively. The negative sign indicates that genes with low heterozygosity contribute significantly to the Bos indicus content in Bos taurus breeds. On the other hand, the positive correlations observed for the Bos indicus and Bos taurus- Bos indicus lineages, indicate that an increase in the heterozygosity of genes relates to an increase in the net Bos indicus content.
Fig 3. Relationship between heterozygosity and Bos indicus content at the animal and gene-level derived from PC1.
(A) Heterozygosity against PC1 in each animal results in an inverted-V pattern; (B) Heterozygosity at gene-level based on lineage for Bos indicus; (C) Bos taurus–Bos indicus; and (D) Bos taurus.
Heterozygosity clustering detection was possible even when only 86 fertility-related (FE) genes were used in the analyses (S2 Fig). For FE genes that were common across at least three publications [35–37],we present heterozygosity results and Bos indicus content in Table 1.
Table 1. Fertility-related genes common across the literature sources and their heterozygosity and Bos indicus/Bos taurus contributions.
| Gene | Number of SNP | Functional AttributesA | Heterozygosity (Lineage) | Posterior Probability (gene’s contribution) | |||
|---|---|---|---|---|---|---|---|
| Bos indicus | Bos taurus | Bos taurus–Bos indicus | Bos indicus | Bos taurus | |||
| ADH6 | 13 | TS | 23.81 | 40.56 | 45.42 | 27.02 | 72.98 |
| E2F3 | 26 | TF | 13.81 | 30.44 | 30.66 | 8.62 | 91.38 |
| ELF5 | 17 | TF, TS | 14.38 | 37.20 | 44.55 | 17.64 | 82.36 |
| ETS1 | 24 | TF | 17.61 | 36.80 | 41.89 | 20.78 | 79.22 |
| ETV6 | 50 | TF | 17.23 | 33.85 | 37.88 | 19.16 | 80.84 |
| LHX4 | 12 | TF | 17.31 | 21.74 | 28.60 | 25.17 | 74.83 |
| OVGP1 | 7 | SE | 32.28 | 40.23 | 43.98 | 0.55 | 99.45 |
| PPARG | 18 | TF | 19.70 | 34.74 | 42.04 | 16.97 | 83.03 |
| PPP3CA | 94 | TS | 25.43 | 29.59 | 36.96 | 26.34 | 73.66 |
| SOX5 | 164 | TF | 20.23 | 25.81 | 37.03 | 30.79 | 69.21 |
| TSHR | 44 | TS, SE | 26.47 | 29.91 | 37.05 | 33.12 | 66.88 |
ATF = transcription factor; TS = tissue specific; SE = secreted.
Analysis of indicine and taurine content in fertility and milk related genes
We collected 86 fertility genes as detailed in S1 Text. The milk related genes were sourced from the Cattle component (http://www.animalgenome.org/cgi-bin/QTLdb/BT/genesrch?gwords=milk) of the Animal QTL database [38] and from literature [39,40]. We collected 231 milk related genes and 125 of these were represented in our entire list of 8,631 genes.
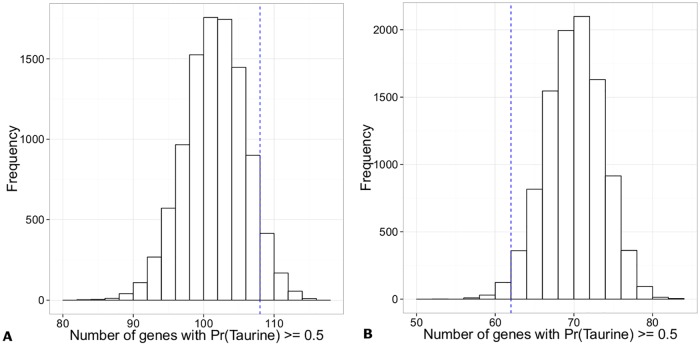
For both the fertility and milk related genes, we computed their memberships to the indicine and taurine components. For the milk related genes, we observed that 108 (out of 125) genes have a posterior probability of at least 0.5 of having a taurine origin. To prove that this has not occurred by chance alone, we conducted a permutation test with 10,000 experiment trials. In each experiment, we randomly sampled 125 genes from the 8,631 genes and checked how many of them have at least 0.5 posterior probability of belonging to the taurine component. The corresponding histogram is shown in Fig 4A. From the distribution, we notice that 108 belongs to the 93.5th percentile, which suggests that there is only about 6% chance that the 108 genes belong to the taurine component by chance alone. This suggests that the milk related genes are strongly associated with the taurine axis and this has been previously discussed in the literature [41–44]. Thus, we provided a proof-of-concept where our designed methodology of dissecting the bovine genome is able to identify genes that contribute to a phenotype of interest (milk related).
Fig 4. Distribution of the number of genes that have a Pr(Taurine) > = 0.5 after conducting 10,000 permutation tests.
(A) Milk related genes; and (B) Fertility related genes. The vertical blue line indicates the observation of 108 milk related genes and 62 fertility related genes that showed a Pr(Taurine) > = 0.5 in our selected list of 8,631 genes.
For the list of fertility genes, 62 out of 86 genes had a posterior probability of at least 0.5 of belonging to the taurine component. However, the permutation test indicated that 62 corresponds to the 16th percentile as shown in Fig 4B. This suggests that the fertility genes are not associated with the taurine axis but are strongly associated with the indicine component of the bovine genome. This novel discovery could yield new insights into the evolution of fertility traits in the bovine genome.
Analysis of Bos indicus content by chromosome
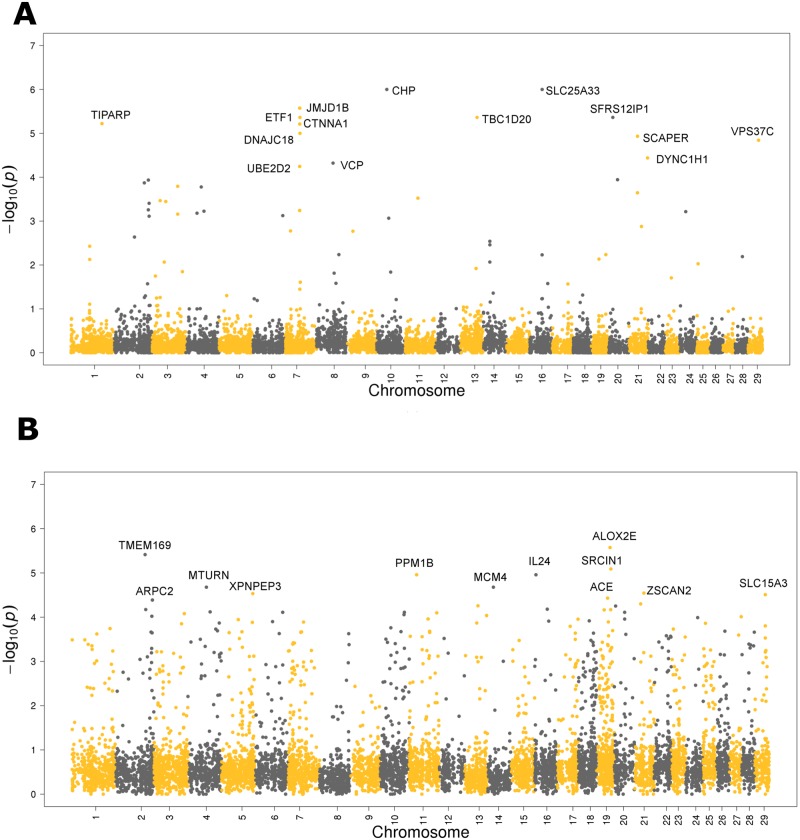
The contribution of each of the genes to the Bos indicus and Bos taurus components allowed us to compute a chromosome’s contribution by averaging the posterior probabilities across the genes within a chromosome. The resulting genome-wide distribution plots are shown in Fig 5, where each point corresponds to a gene from our list of 8,631 genes sorted on the x-axis by genome map position. The y-axis indicates the–log(p), where p is the posterior probability, m1 for Bos indicus and m2 for Bos taurus in Equation 1 (S1 Text). We observed there are fewer genes that stand out with respect to their contribution to the Bos indicus content (Fig 5A), while there are a greater number of genes contributing to the Bos taurus content (Fig 5B). A more detailed analysis, revealed 14 genes with a significant contribution (–log(p) > 4) to the Bos indicus content (TIPARP, JMJD1B, ETF1, CTNNA1, DNAJC18, UBE2D2, VCP, CHP, TBC1D20, SLC25A33, SFRS12IP1, LOC100140107, LOC537748, and VPS37C). One enriched GO term from this list is GO:0071822 (Protein complex subunit organization) with a FDR p-value = 0.00505. Worth mentioning is DNAJC18 (DnaJ heat shock protein family member C18) due to its recently reported association with heat stress in contrasting Bos taurus and Bos indicus cattle [45,46]. Also noteworthy is SLC25A33 estimated to have a contribution to Bos indicus of 100% and encoded at 44.8 Mb of BTA16 in a hard-sweep region recently reported to be shared among four Bos taurus breeds [23] and possibly related to the initial cattle domestication events.
Fig 5. Genome-wide distribution plots depicting the highly contributing genes to the Bos indicus and Bos taurus contents in the bovine genome.
Each point corresponds to a gene from our list of 8,631 genes along the genome. The likelihood of a gene being of Bos indicus or Bos taurus origin is plotted along the Y-axis. (A) Genes with high Bos indicus (low Bos taurus) content (B) Genes with low Bos indicus (high Bos taurus) content.
Similarly, we found 29 genes with statistically significant contribution to Bos taurus content (–log(p) > 4): TMEM169, ARPC2, SRRM1, EPHA8, UTP11L, LOC615685, INHBA, XPNPEP3, HNRNPD, ACOT2, EIF2B2, PPM1B, CIZ1, CHGB, ADNP, MCM4, IL24, GPR157, ALOX12E, LOC535629, ACE, LOC506185, FASN, BNIP1, LOC782185, ZSCAN2, CLK3, NR1D2, and SLC15A3. From this list, we highlight FASN (fatty acid synthase) and INHBA (Inhibin, beta activin beta-A chain). Ample evidence from the Animal QTL database [38] (http://www.animalgenome.org/cgi-bin/QTLdb/index) suggests the presence of QTL in the coding region of FASN associated with body weight, marbling and milk fat yield in cattle. The same source documents INHBA as harbouring QTL for semen volume, sperm counts and motility. Fortes et al. [47] propose SNP associated with serum levels of Inhibin in Brahman bulls as an early biomarker of sexual development. This same QTL was absent when Tropical Composite bulls were subject to GWAS for the same phenotypes [48]. These contrasting GWAS results reinforce the idea that INHBA polymorphism segregation and association with reproduction differs according to Bos indicus content of each breed.
Analysis of heterozygosity at the chromosome level
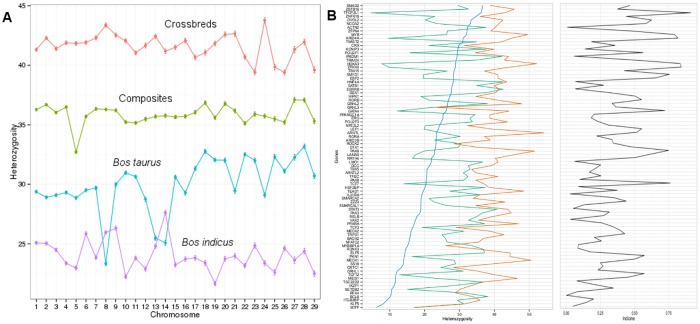
In addition to the analysis of heterozygosity at the gene level, the average heterozygosity across the 29 autosomal chromosomes in the bovine genome reveals a striking contrast in the heterozygosity across the four different lineages (Fig 6A). The Bos indicus lineage has the least heterozygosity while the cross-breeds have the highest heterozygosity across the genome. Within each lineage, there appeared to be some chromosomes with dramatic changes in heterozygosity relative to the other chromosomes. For instance, BTA14 in Bos indicus has a greater heterozygosity and has a relatively large value when compared to its immediate neighbours which indicates that the BTA14 may be an important locus for introgression of Bos taurus genes. In fact, the importance of BTA14 and its role in milk production and ovulation rate has been well documented in the literature [49–51]. Furthermore, there is the age at first calving QTL on BTA14 that was detected in Nelore cattle [52–54].
Fig 6.
(A) Average chromosome heterozygosity across the four cattle lineages. (B) Variation of heterozygosity across the cattle lineages (blue, orange and green for Bos indicus, Bos taurus–Bos indicus and Bos taurus, respectively) for the list of 84 network genes (out of 1,284) that are TF and fertility related as well as at least one of the other functional attributes (TS, SE, KI). The right panel shows the contribution to the indicine content for the same set of genes.
Similarly, BTA8, BTA13, and BTA14 in Bos taurus contain the lowest average heterozygosity. Further, BTA5 within the composite breeds contains the lowest heterozygosity, while BTA27 and BTA28 contain the highest heterozygosity. BTA13, BTA14 and BTA28 have been reported to harbour QTL for carcass traits [55] while QTL on BTA5 are known to have a pronounced effect on reproductive efficiency in cattle [56–58]. The heterozygosity analysis brings to light some of the important regions in the cattle genome for breed discrimination.
Gene Ontology (GO) enrichment analysis
The exploration of possible biological functions inherent in candidate gene lists is often done by a GO enrichment analysis [59]. The objective is to identify the set of genes which are significantly overrepresented in a target set of genes relative to a background set of genes. For each cattle breed, we have a target list of genes which deviate significantly from HWE. These gene lists are important as they could potentially be the variants which give cattle sub-species their distinctive Bos indicus or Bos taurus phenotypes. As a result, we have 19 target lists corresponding to each cattle breed. We conducted 19 separate analyses and collected the enriched GO terms statistically overrepresented up to a p-value level of 0.1%. This resulted in 142 GO terms, each occurring a maximum of three times with 7 of them (about 5%) occurring exactly thrice. These 7 GO terms together correspond to 1,193 genes in our list of 8,631 genes (Table 2). According to this analysis, genes involved with the regulation of developmental processes are overrepresented in genome regions that deviate from HWE. It is possible to extrapolate that the formation of cattle subspecies by phenotypic selection, has afforded particular importance to genome regions involved with fine-tuning the development of tissues and organs during development.
Table 2. Overrepresented GO terms in cattle genome regions which deviate from HWE corresponding to a total of 1,193 genes.
Each GO term is enriched thrice.
| GO Term | Description | p-value | Genes |
|---|---|---|---|
| GO:0007155 | cell adhesion | 1.40e-04 | 388 |
| GO:0022610 | biological adhesion | 1.51e-04 | 389 |
| GO:0031344 | regulation of cell projection organization | 1.94e-05 | 258 |
| GO:0043547 | positive regulation of GTPase activity | 8.94e-05 | 222 |
| GO:0050793 | regulation of developmental process | 3.62e-04 | 746 |
| GO:0051960 | regulation of nervous system development | 3.27e-04 | 302 |
| GO:2000026 | regulation of multicellular organismal development | 2.93e-04 | 576 |
We also considered the list of top ranked genes based on their contribution to the Bos indicus and Bos taurus content We selected those genes which have a membership of at least 95% to the Bos indicus and Bos taurus clusters (see Methods section and S1 Text for details). This resulted in 64 and 718 genes with high contribution to the Bos indicus and Bos taurus clusters, respectively that were targeted in two separate GO enrichment analyses. A striking enrichment of gene annotation terms associated with RNA splicing and mRNA processing was observed (Table 3). It is tempting to consider the possibility that the post-transcriptional processing machinery is overrepresented among the genes which potentially discriminate between the Bos taurus and Bos indicus subspecies. Post-transcriptional processing is an important element of gene regulation and could well contribute to sub-species differences.
Table 3. Overrepresented GO terms among a list of cattle genes which significantly contribute to the Bos indicus/Bos taurus content of cattle genomes, corresponding to a total of 52 genes.
Each GO term occurs once.
| GO Term | Description | p-value | Genes |
|---|---|---|---|
| GO:0000398 | mRNA splicing, via spliceosome | 4.85e-05 | 18 |
| GO:0002082 | regulation of oxidative phosphorylation | 5.95e-04 | 2 |
| GO:0002467 | germinal center formation | 2.06e-04 | 4 |
| GO:0002544 | chronic inflammatory response | 6.53e-05 | 5 |
| GO:0006397 | mRNA processing | 8.55e-04 | 25 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 7.05e-04 | 10 |
| GO:0008380 | RNA splicing | 4.43e-05 | 24 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 4.91e-04 | 15 |
Selection of genes for network analysis
Given that fertility phenotypes are an important consideration in the formation of domestic breeds, we constructed a co-heterozygosity network in order to further scrutinise the potential role of fertility-related genes in regions of high heterozygosity. The genes included in the co-heterozygosity network were selected from three possibly overlapping lists: 1) Genes based on their deviation from HWE; 2) Genes based on their significant contribution to the Bos indicus/Bos taurus content; and 3) Fertility (FE) related genes.
We obtained 1,193 genes not in HWE and 52 genes which predominantly contributed to the Bos indicus and Bos taurus content. These two lists were combined with the 86 FE genes sourced from the literature. The three lists contained 1,284 unique genes that were further categorized based on their functional attributes: transcription factors (TF), tissue-specific (TS), secreted (SE) and kinases (KI). We identified 84 out of the 1,284 genes that are TF and were also classified as either TS, SE, KI or FE. The variation of heterozygosity in these set of 84 TF and across the Bos taurus (BT), Bos indicus (BI) and Bos taurus—Bos indicus (BTI) lineages is shown in Fig 6B. The number of network genes that overlap with TF include 73 genes that are expressed in a tissue-specific manner, 10 genes that code for proteins that could potentially be secreted outside the cytoplasm (ARNTL, GRHL3, IL31RA, KCNIP3, LAMA5, MEIS1, SATB1, SMARCA2, TCF12, TRIM24), 3 genes that code for kinases (HIPK1, PKN1, ROCK2), and 61 genes that are fertility related.
Gene co-heterozygosity network
We generated a gene co-heterozygosity network using the PCIT algorithm to identify significant connections based on correlated heterozygosity values for the 1,284 genes. These correlations were used to establish gene to gene edges in the network inference. This approach is able to point to genes for which the Bos taurus or Bos indicus origin is particularly crucial for animal performance. Imposing a correlation threshold of 0.95, we obtained a sub-network of 328 genes with 1,098 significant connections. Other thresholds were explored and further details are provided in S2 Text. We observed that the degree distribution on a logarithm scale follows a scale-free network, shown in S6 Fig (correlation of 0.85 and p-value of 2.2 x 10−16). The maximum degree is 47 and corresponds to the SPEN gene, which is a known transcriptional regulator [60–62]. The contribution to Bos indicus for SPEN was estimated at 64.80% placing it in the top 9% of all 8,631 genes.
We observed a significantly higher Bos indicus content for the 86 FE genes as compared to the remaining 1,198 network genes. This was attributed to fertility genes being under strong selection among the various cattle lineages [63]. Further exploitation of FE and their roles in the predicted co-heterozygosity network are offered in the S2 Text.
Some of the interesting genes that are present in this network include BRCA1 which is involved in bovine mastitis [64,65], MCF2L which is known to play a critical part in joint tissue development in humans [66], FOXP2 which is a TF required for proper development of speech and language regions of the brain during embryogenesis in humans [67], CREBBP which acts as a binding protein that is important in embryonic development, growth control, and has been implicated in the embryo-placenta signalling in bovine embryos [68,69].
Conclusion
Our pipeline can be generalized to any scenarios where population structure requires scrutiny at the molecular level, particularly in the presence of a priori set of genes known to impact a phenotype of evolutionary interest such as fertility.
Methods
Animal Care and Use Committee approval was not required for this study because the data were obtained from existing phenotypic and genotype databases from the Cooperative Research Centre for Beef Genetic Technologies (“Beef CRC”; http://www.beefcrc.com).
Data collection and pre-processing
Genotypes from 17,867 cattle representing 18 breeds were extracted from data previously reported [70]. Genotypes from 496 Nelore (NE), a pure Bos indicus breed were also included [71]. In total, 18,363 cattle of 19 breeds were studied (S3 Fig), of which Brahman and Nelore are Bos indicus (BI), six breeds are Bos taurus (BT), and eleven breeds are Bos taurus-Bos indicus composites (BTI). Cattle were genotyped with high-density chip (over 700,000 SNP). SNP mapped to sex chromosomes were removed from analyses as these behave differently with respect to HWE and genotypes were from both female and male cattle. We targeted a 1kb region surrounding known genes in order to capture SNP associated with protein-coding regions. Only genes that have at least the median number of corresponding SNP (six) were included in subsequent analyses. The final set comprised 246,864 SNP located in 8,631 genes.
Principal component analysis, mixture modelling and gene ancestry
Principal component analysis (PCA) was performed with PLINK [72]. We extracted the weights of the first principal component (PC1) as it explains the maximum variability. Like others, we found that PC1 captured the Bos indicus component of cattle breeds [29,31,73]. It is conceivable that some SNP, mapped to certain genes, contribute more than others to the Bos indicus components. Bolormaa et al. [74] assigned chromosome segments to be of Bos indicus or Bos taurus ancestry using a weighted regression model of SNP allele frequencies. However, their method required pre-defined segment length and was not informed by PCA analyses. We used PCA output as a first step to project the data on to the maximum variable direction and used statistical machine learning and two-component mixture modelling to quantify the Bos indicus and Bos taurus content of a gene. Our method identifies gene ancestry and lists genes that contribute significantly to Bos indicus or Bos taurus ancestry. These genes harbour informative SNP for determination of cattle lineage. The two-component mixture used is detailed in S1 Text. Mixture parameters were estimated via maximum likelihood using EMMIX software [75]. After estimating mixture parameters, the contribution of each SNP to each component is given by its posterior probability of belonging to Bos indicus or Bos taurus components of the mixture model. SNP in coding region were collapsed to estimate gene contribution to Bos indicus ancestry, which implies that a gene has higher/lower probability of membership to Bos indicus or Bos taurus components.
Heterozygosity, Hardy-Weinberg equilibrium and clustering of breeds
Percentage heterozygosity (HET) was computed for each SNP as the proportion of animals with a heterozygous genotype. HET was computed for each SNP and averaged over all the animals in a given breed. HET values of SNP were averaged to obtain gene level HET, in a cumulative text statistic [76] (S1 Text). Gene HET was used to cluster cattle breeds using PermutMatrix [77] software. Allelic frequencies were used determine deviation from HWE. A nominal P-value of 1% served as threshold to select genes with significant deviation from HWE.
Gene Ontology (GO) enrichment analyses
We performed GO enrichment analysis using GOrilla [78,79] to aid biological interpretation of genes deems significant for each breed, based on SNP deviation from HWE. Genes with significant deviation were contrasted to background (all 8,631 genes studied). Ranked gene lists based on PC1 estimated contribution to Bos indicus and Bos taurus ancestry were also analysed with GOrilla.
Functional attributes and bovine fertility related genes
Genes were catalogued as transcription factors (TF), genes which are expressed in a tissue-specific (TS) manner, genes encoding secreted proteins (SE), and kinases (KI) as shown in S1A Fig. TF were defined according to the Animal Transcription Factor Database (http://www.bioguo.org/AnimalTFDB/) [80]. TS were identified from the Tissue-specific Gene Expression and Regulation [81] in humans. SE were identified with the Human Protein Atlas [82], and KI with the Human Kinome database [83]. Human databases were used in the absence of similar cattle resources.
Genes associated with heifer puberty and other cattle fertility traits were retrieved from previous studies [35–37,84], shown in S4B Fig. The fertility-related genes were catalogued as per above criteria (TF, TS, SE, KI) and checked for overlapping with our list of 8,631 genes. In total, 1,157 genes related to fertility were in our dataset, shown in S4C Fig.
Gene co-heterozygosity network
We inferred a co-heterozygosity gene network using the partial correlation and information theory (PCIT) algorithm [32] to identify significant edges. Genes that deviated significantly from HWE, genes that contributed strongly to Bos indicus or Bos taurus ancestry, and fertility-related genes were included in the network prediction. Cytoscape [85] was used to visualise and analyse the resulting network. A search algorithm was employed to locate the minimal trio of fertility-related genes that span the majority of the network topology [86].
Supporting information
(DOCX)
(DOCX)
(TXT)
Left and Right modes describe the Bos indicus and Bos taurus components, respectively. Red indicates the actual distribution of SNP weights, grey curves are the individual Normal distributions, and black curve is the mixture model obtained by combining the two Normal distributions.
(TIFF)
The gradient from green to black to red correspond to low, medium and high heterozygosity.
(TIFF)
(TIFF)
(A) The list of 2,891 genes from the entire list of 8,631 genes that belong to the four main functional categories of transcription factor (TF), secreted hormones (SE), kinases (KI) and genes expressed in tissue-specific (TS) manner; (B) The subset of 1,157 fertility genes collected from the literature where Canovas, MThomas, Fortes_Rev and Fortes correspond to [35], [37], [84] and [36]respectively; (C) The same list of 1,157 fertility genes across the four functional attributes (excluding 10 genes that are not TF, TS, SE, KI).
(TIFF)
(TIFF)
(A) At a correlation cut-off of 0.90 comprising of 858 genes and 12,958 significant connections (B) At a correlation cut-off of 0.95 comprising of 328 genes and 1,098 connections.
(TIFF)
All corresponds to the 8,631 genes in our analysis and PCIT corresponds to the 1,284 network genes. The only category for which significant differences exists (p-value < 0.01) in the indicine percentage is for fertility-related genes.
(TIFF)
The size of the node corresponds to the indicine content. The nodes in green are transcription factors and remaining nodes in the network are purple-coloured. Nodes that are triangle-shaped are fertility-related genes and others are denoted by circles: (A) PCIT network after applying a threshold of 0.90; (B) The network spanned by the trio of fertility related genes GATA4, NR1H4, VAX2; (C) The network spanned by the trio of fertility related genes ELF5, ROCK2, POU2F1.
(TIFF)
Acknowledgments
The Northern Pastoral Group of Companies, Department of Employment, Economic Development and Innovation (DEEDI) and CSIRO collaborated to collect the Beef CRC phenotypes and genotypes data. The authors wish to thank the many colleagues from the Beef CRC who contributed towards data collection for this project.
Data Availability
The minimal raw data is the SNP genotype data - 18,363 cattle and 729,068 SNP genotypes. The raw SNP genotype data are part of the Beef CRC project (http://www.beefcrc.com/) and are co-owned with Meat and Livestock Australia, and can be made available subject to the agreement of the owners. Any parties seeking access to the raw SNP genotype data should contact: Dr. Toni Reverter-Gomez (toni.reverter-gomez@csiro.au or +61732142392). Further, we have provided data on the 8,631 genes collated and their indicine/taurine proportions as part of the paper and its Supporting Information files.
Funding Statement
Embrapa Southeast Livestock has provided support in the form of salaries for authors [MM and LR]. They provided the Nelore data and contributed to the drafting of the manuscript. The specific roles of these authors are articulated in the 'author contributions' section.
References
- 1.Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, et al. (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 4: e5350 doi: 10.1371/journal.pone.0005350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vignal A, Milan D, SanCristobal M, Eggen A (2002) A review on SNP and other types of molecular markers and their use in animal genetics. Genetics Selection Evolution 34: 275–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4: 981–994. doi: 10.1038/nrg1226 [DOI] [PubMed] [Google Scholar]
- 4.Canas-Alvarez JJ, Mouresan EF, Varona L, Diaz C, Molina A, Baro JA, et al. (2016) Linkage disequilibrium, persistence of phase, and effective population size in Spanish local beef cattle breeds assessed through a high-density single nucleotide polymorphism chip. J Anim Sci 94: 2779–2788. doi: 10.2527/jas.2016-0425 [DOI] [PubMed] [Google Scholar]
- 5.Porto-Neto LR, Kijas JW, Reverter A (2014) The extent of linkage disequilibrium in beef cattle breeds using high-density SNP genotypes. Genet Sel Evol 46: 22 doi: 10.1186/1297-9686-46-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlier C, Coppieters W, Rollin F, Desmecht D, Agerholm JS, Cambisano N, et al. (2008) Highly effective SNP-based association mapping and management of recessive defects in livestock. Nat Genet 40: 449–454. doi: 10.1038/ng.96 [DOI] [PubMed] [Google Scholar]
- 7.Page BT, Casas E, Heaton MP, Cullen NG, Hyndman DL, Morris CA, et al. (2002) Evaluation of single-nucleotide polymorphisms in CAPN1 for association with meat tenderness in cattle. J Anim Sci 80: 3077–3085. [DOI] [PubMed] [Google Scholar]
- 8.Zhang K, Qin ZS, Liu JS, Chen T, Waterman MS, Sun F (2004) Haplotype block partitioning and tag SNP selection using genotype data and their applications to association studies. Genome Res 14: 908–916. doi: 10.1101/gr.1837404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson NJ, Porto-Neto L, Kijas JW, Reverter A (2015) Compression distance can discriminate animals by genetic profile, build relationship matrices and estimate breeding values. Genetics Selection Evolution 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson NJ, Porto-Neto LR, Kijas J, McWilliam S, Taft RJ, Reverter A (2014) Information compression exploits patterns of genome composition to discriminate populations and highlight regions of evolutionary interest. BMC Bioinformatics 15: 66 doi: 10.1186/1471-2105-15-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kijas JW, Lenstra JA, Hayes B, Boitard S, Porto Neto LR, San Cristobal M, et al. (2012) Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol 10: e1001258 doi: 10.1371/journal.pbio.1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- 13.Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22: 139–144. doi: 10.1038/9642 [DOI] [PubMed] [Google Scholar]
- 14.Weiss KM, Clark AG (2002) Linkage disequilibrium and the mapping of complex human traits. Trends in Genetics 18: 19–24. [DOI] [PubMed] [Google Scholar]
- 15.Koufariotis L, Chen YPP, Bolormaa S, Hayes BJ (2014) Regulatory and coding genome regions are enriched for trait associated variants in dairy and beef cattle. Bmc Genomics 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacEachern S, Hayes B, McEwan J, Goddard M (2009) An examination of positive selection and changing effective population size in Angus and Holstein cattle populations (Bos taurus) using a high density SNP genotyping platform and the contribution of ancient polymorphism to genomic diversity in Domestic cattle. BMC Genomics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purfield DC, Berry DP, McParland S, Bradley DG (2012) Runs of homozygosity and population history in cattle. Bmc Genetics 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randhawa IA, Khatkar MS, Thomson PC, Raadsma HW (2016) A Meta-Assembly of Selection Signatures in Cattle. PLoS One 11: e0153013 doi: 10.1371/journal.pone.0153013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, et al. (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. doi: 10.1038/nature01140 [DOI] [PubMed] [Google Scholar]
- 20.Bomba L, Nicolazzi EL, Milanesi M, Negrini R, Mancini G, Biscarini F, et al. (2015) Relative extended haplotype homozygosity signals across breeds reveal dairy and beef specific signatures of selection. Genetics Selection Evolution 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan DF, Zhang SL, Jiang JC, Jiang L, Zhang Q, Liu JF (2013) Genome-Wide Detection of Selective Signature in Chinese Holstein. Plos One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qanbari S, Pimentel ECG, Tetens J, Thaller G, Lichtner P, Sharifi AR, et al. (2010) A genome-wide scan for signatures of recent selection in Holstein cattle. Animal Genetics 41: 377–389. doi: 10.1111/j.1365-2052.2009.02016.x [DOI] [PubMed] [Google Scholar]
- 23.Boitard S, Boussaha M, Capitan A, Rocha D, Servin B (2016) Uncovering Adaptation from Sequence Data: Lessons from Genome Resequencing of Four Cattle Breeds. Genetics 203: 433–450. doi: 10.1534/genetics.115.181594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blott SC, Williams JL, Haley CS (1999) Discriminating among cattle breeds using genetic markers. Heredity 82: 613–619. [DOI] [PubMed] [Google Scholar]
- 25.Machugh DE, Loftus RT, Bradley DG, Sharp PM, Cunningham P (1994) Microsatellite DNA Variation within and among European Cattle Breeds. Proceedings of the Royal Society B-Biological Sciences 256: 25–31. [DOI] [PubMed] [Google Scholar]
- 26.Wiener P, Burton D, Williams JL (2004) Breed relationships and definition in British cattle: a genetic analysis. Heredity 93: 597–602. doi: 10.1038/sj.hdy.6800566 [DOI] [PubMed] [Google Scholar]
- 27.Tambasco-Talhari D, de Alencar MM, de Paz CCP, da Cruz GM, Rodrigues ADA, Packer IU, et al. (2005) Molecular marker heterozygosities and genetic distances as correlates of production traits in F-1 bovine crosses. Genetics and Molecular Biology 28: 218–224. [Google Scholar]
- 28.Consortium TBH (2009) Genome-Wide Survey of SNP Variation Uncovers the Genetic Structure of Cattle Breeds. Science 324: 528–532. doi: 10.1126/science.1167936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertolini F, Galimberti G, Calo DG, Schiavo G, Matassino D, Fontanesi L (2015) Combined use of principal component analysis and random forests identify population-informative single nucleotide polymorphisms: application in cattle breeds. Journal of Animal Breeding and Genetics 132: 346–356. doi: 10.1111/jbg.12155 [DOI] [PubMed] [Google Scholar]
- 30.Porto-Neto LR, Sonstegard TS, Liu GE, Bickhart DM, Da Silva MVB, Machado MA, et al. (2013) Genomic divergence of zebu and taurine cattle identified through high-density SNP genotyping. Bmc Genomics 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis J, Abas Z, Dadousis C, Lykidis D, Paschou P, Drineas P (2011) Tracing Cattle Breeds with Principal Components Analysis Ancestry Informative SNPs. Plos One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reverter A, Chan EKF (2008) Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics 24: 2491–2497. doi: 10.1093/bioinformatics/btn482 [DOI] [PubMed] [Google Scholar]
- 33.Porto-Neto LR, Reverter A, Prayaga KC, Chan EK, Johnston DJ, Hawken RJ, et al. (2014) The genetic architecture of climatic adaptation of tropical cattle. PLoS One 9: e113284 doi: 10.1371/journal.pone.0113284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuels DC, Wang J, Ye F, He J, Levinson RT, Sheng Q, et al. (2016) Heterozygosity Ratio, a Robust Global Genomic Measure of Autozygosity and Its Association with Height and Disease Risk. Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canovas A, Reverter A, DeAtley KL, Ashley RL, Colgrave ML, Fortes MR, et al. (2014) Multi-tissue omics analyses reveal molecular regulatory networks for puberty in composite beef cattle. PLoS One 9: e102551 doi: 10.1371/journal.pone.0102551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortes MR, Reverter A, Zhang Y, Collis E, Nagaraj SH, Jonsson NN, et al. (2010) Association weight matrix for the genetic dissection of puberty in beef cattle. Proc Natl Acad Sci U S A 107: 13642–13647. doi: 10.1073/pnas.1002044107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas M. SNP Discovery in RNA-Seq Across Breeds of Cattle in Puberty-Related Candidate Genes (ie, Network Hubs). Plant and Animal Genome. [Google Scholar]
- 38.Hu ZL, Park CA, Reecy JM (2016) Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res 44: D827–833. doi: 10.1093/nar/gkv1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole JB, VanRaden PM, O'Connell JR, Van Tassell CP, Sonstegard TS, Schnabel RD, et al. (2009) Distribution and location of genetic effects for dairy traits. J Dairy Sci 92: 2931–2946. doi: 10.3168/jds.2008-1762 [DOI] [PubMed] [Google Scholar]
- 40.Pegolo S, Cecchinato A, Mele M, Conte G, Schiavon S, Bittante G (2016) Effects of candidate gene polymorphisms on the detailed fatty acids profile determined by gas chromatography in bovine milk. J Dairy Sci 99: 4558–4573. doi: 10.3168/jds.2015-10420 [DOI] [PubMed] [Google Scholar]
- 41.Cohen-Zinder M, Seroussi E, Larkin DM, Loor JJ, Everts-van der Wind A, Lee JH, et al. (2005) Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res 15: 936–944. doi: 10.1101/gr.3806705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daley DR, McCuskey A, Bailey CM (1987) Composition and yield of milk from beef-type Bos taurus and Bos indicus X Bos taurus dams. J Anim Sci 64: 373–384. [DOI] [PubMed] [Google Scholar]
- 43.Kaupe B, Winter A, Fries R, Erhardt G (2004) DGAT1 polymorphism in Bos indicus and Bos taurus cattle breeds. J Dairy Res 71: 182–187. [DOI] [PubMed] [Google Scholar]
- 44.Tantia MS, Vijh RK, Mishra BP, Mishra B, Kumar ST, Sodhi M (2006) DGAT1 and ABCG2 polymorphism in Indian cattle (Bos indicus) and buffalo (Bubalus bubalis) breeds. BMC Vet Res 2: 32 doi: 10.1186/1746-6148-2-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapila N, Sharma A, Kishore A, Sodhi M, Tripathi PK, Mohanty AK, et al. (2016) Impact of Heat Stress on Cellular and Transcriptional Adaptation of Mammary Epithelial Cells in Riverine Buffalo (Bubalus Bubalis). PLoS One 11: e0157237 doi: 10.1371/journal.pone.0157237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kishore A, Sodhi M, Kumari P, Mohanty AK, Sadana DK, Kapila N, et al. (2014) Peripheral blood mononuclear cells: a potential cellular system to understand differential heat shock response across native cattle (Bos indicus), exotic cattle (Bos taurus), and riverine buffaloes (Bubalus bubalis) of India. Cell Stress Chaperones 19: 613–621. doi: 10.1007/s12192-013-0486-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fortes MR, Reverter A, Hawken RJ, Bolormaa S, Lehnert SA (2012) Candidate genes associated with testicular development, sperm quality, and hormone levels of inhibin, luteinizing hormone, and insulin-like growth factor 1 in Brahman bulls. Biol Reprod 87: 58 doi: 10.1095/biolreprod.112.101089 [DOI] [PubMed] [Google Scholar]
- 48.Fortes MR, Reverter A, Kelly M, McCulloch R, Lehnert SA (2013) Genome-wide association study for inhibin, luteinizing hormone, insulin-like growth factor 1, testicular size and semen traits in bovine species. Andrology 1: 644–650. doi: 10.1111/j.2047-2927.2013.00101.x [DOI] [PubMed] [Google Scholar]
- 49.Looft C, Reinsch N, Karall-Albrecht C, Paul S, Brink M, Thomsen H, et al. (2001) A mammary gland EST showing linkage disequilibrium to a milk production QTL on bovine Chromosome 14. Mamm Genome 12: 646–650. doi: 10.1007/s003350020003 [DOI] [PubMed] [Google Scholar]
- 50.Marques E, Grant JR, Wang Z, Kolbehdari D, Stothard P, Plastow G, et al. (2011) Identification of candidate markers on bovine chromosome 14 (BTA14) under milk production trait quantitative trait loci in Holstein. J Anim Breed Genet 128: 305–313. doi: 10.1111/j.1439-0388.2010.00910.x [DOI] [PubMed] [Google Scholar]
- 51.Wibowo TA, Gaskins CT, Newberry RC, Thorgaard GH, Michal JJ, Jiang Z (2008) Genome assembly anchored QTL map of bovine chromosome 14. Int J Biol Sci 4: 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costa RB, Camargo GM, Diaz ID, Irano N, Dias MM, Carvalheiro R, et al. (2015) Genome-wide association study of reproductive traits in Nellore heifers using Bayesian inference. Genet Sel Evol 47: 67 doi: 10.1186/s12711-015-0146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa RB, Camargo GM, Diaz ID, Irano N, Dias MM, Carvalheiro R, et al. (2015) Erratum to: Genome-wide association study of reproductive traits in Nellore heifers using Bayesian inference. Genet Sel Evol 47: 72 doi: 10.1186/s12711-015-0150-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyeong KE, Iqbal A, Kim JJ (2014) A Genome Wide Association Study on Age at First Calving Using High Density Single Nucleotide Polymorphism Chips in Hanwoo (Bos taurus coreanae). Asian-Australasian Journal of Animal Sciences 27: 1406–1410. doi: 10.5713/ajas.2014.14273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espigolan R, Baldi F, Boligon AA, Souza FR, Fernandes GA Junior, Gordo DG, et al. (2015) Associations between single nucleotide polymorphisms and carcass traits in Nellore cattle using high-density panels. Genet Mol Res 14: 11133–11144. doi: 10.4238/2015.September.22.7 [DOI] [PubMed] [Google Scholar]
- 56.Adams HA, Sonstegard TS, VanRaden PM, Null DJ, Van Tassell CP, Larkin DM, et al. (2016) Identification of a nonsense mutation in APAF1 that is likely causal for a decrease in reproductive efficiency in Holstein dairy cattle. J Dairy Sci 99: 6693–6701. doi: 10.3168/jds.2015-10517 [DOI] [PubMed] [Google Scholar]
- 57.McDaneld TG, Kuehn LA, Thomas MG, Pollak EJ, Keele JW (2014) Deletion on chromosome 5 associated with decreased reproductive efficiency in female cattle. J Anim Sci 92: 1378–1384. doi: 10.2527/jas.2013-6821 [DOI] [PubMed] [Google Scholar]
- 58.Psaros KM, McDaneld TG, Kuehn LA, Snelling WM, Keele JW (2015) Evaluation of single nucleotide polymorphisms in chromosomal regions impacting pregnancy status in cattle. J Anim Sci 93: 978–987. doi: 10.2527/jas.2014-8509 [DOI] [PubMed] [Google Scholar]
- 59.Edwards SM, Sorensen IF, Sarup P, Mackay TF, Sorensen P (2016) Genomic Prediction for Quantitative Traits Is Improved by Mapping Variants to Gene Ontology Categories in Drosophila melanogaster. Genetics 203: 1871–1883. doi: 10.1534/genetics.116.187161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ariyoshi M, Schwabe JW (2003) A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev 17: 1909–1920. doi: 10.1101/gad.266203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.St-Pierre B, Cooper M, Jiang Z, Zacksenhaus E, Egan SE (2004) Dynamic regulation of the Stra13/Sharp/Dec bHLH repressors in mammary epithelium. Dev Dyn 230: 124–130. doi: 10.1002/dvdy.20013 [DOI] [PubMed] [Google Scholar]
- 62.VanderWielen BD, Yuan Z, Friedmann DR, Kovall RA (2011) Transcriptional repression in the Notch pathway: thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J Biol Chem 286: 14892–14902. doi: 10.1074/jbc.M110.181156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flori L, Fritz S, Jaffrezic F, Boussaha M, Gut I, Heath S, et al. (2009) The Genome Response to Artificial Selection: A Case Study in Dairy Cattle. PLoS ONE 4: e6595 doi: 10.1371/journal.pone.0006595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan Z, Chu G, Dan Y, Li J, Zhang L, Gao X, et al. (2012) BRCA1: a new candidate gene for bovine mastitis and its association analysis between single nucleotide polymorphisms and milk somatic cell score. Mol Biol Rep 39: 6625–6631. doi: 10.1007/s11033-012-1467-5 [DOI] [PubMed] [Google Scholar]
- 65.Yuan Z, Li J, Li J, Zhang L, Gao X, Gao HJ, et al. (2012) Investigation on BRCA1 SNPs and its effects on mastitis in Chinese commercial cattle. Gene 505: 190–194. doi: 10.1016/j.gene.2012.05.010 [DOI] [PubMed] [Google Scholar]
- 66.Shepherd C, Skelton AJ, Rushton MD, Reynard LN, Loughlin J (2015) Expression analysis of the osteoarthritis genetic susceptibility locus mapping to an intron of the MCF2L gene and marked by the polymorphism rs11842874. BMC Med Genet 16: 108 doi: 10.1186/s12881-015-0254-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morgan A, Fisher SE, Scheffer I, Hildebrand M (1993) FOXP2-Related Speech and Language Disorders. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A et al. editors. GeneReviews(R) Seattle (WA). [Google Scholar]
- 68.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859. doi: 10.1038/365855a0 [DOI] [PubMed] [Google Scholar]
- 69.Reverter A, Porto-Neto LR, Fortes MRS, McCulloch R, Lyons RE, Moore S, et al. (2016) Genomic analyses of tropical beef cattle fertility based on genotyping pools of Brahman cows with unknown pedigree1. Journal of Animal Science 94: 4096–4108. doi: 10.2527/jas.2016-0675 [DOI] [PubMed] [Google Scholar]
- 70.Bolormaa S, Pryce JE, Kemper K, Savin K, Hayes BJ, Barendse W, et al. (2013) Accuracy of prediction of genomic breeding values for residual feed intake and carcass and meat quality traits in Bos taurus, Bos indicus, and composite beef cattle. J Anim Sci 91: 3088–3104. doi: 10.2527/jas.2012-5827 [DOI] [PubMed] [Google Scholar]
- 71.Mudadu MA, Porto-Neto LR, Mokry FB, Tizioto PC, Oliveira PSN, Tullio RR, et al. (2016) Genomic structure and marker-derived gene networks for growth and meat quality traits of Brazilian Nelore beef cattle (vol 17, 235, 2016). Bmc Genomics 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7 doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gibbs RA, Taylor JF, Van Tassell CP, Barendse W, Eversoie KA, Gill CA, et al. (2009) Genome-Wide Survey of SNP Variation Uncovers the Genetic Structure of Cattle Breeds. Science 324: 528–532. doi: 10.1126/science.1167936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolormaa S, Hayes BJ, Hawken RJ, Zhang Y, Reverter A, Goddard ME (2011) Detection of chromosome segments of zebu and taurine origin and their effect on beef production and growth. Journal of Animal Science 89: 2050–2060. doi: 10.2527/jas.2010-3363 [DOI] [PubMed] [Google Scholar]
- 75.McLachlan GJ, Peel D, Basford KE, Adams P (1999) The EMMIX software for the fitting of mixtures of normal and t-components. Journal of Statistical Software 4: 1–14. [Google Scholar]
- 76.Davis CS (1982) The Distribution of a Linear Combination of Chi-Square Variables. Biometrics 38: 279–279. [Google Scholar]
- 77.Caraux G, Pinloche S (2005) PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21: 1280–1281. doi: 10.1093/bioinformatics/bti141 [DOI] [PubMed] [Google Scholar]
- 78.Eden E, Lipson D, Yogev S, Yakhini Z (2007) Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 3: e39 doi: 10.1371/journal.pcbi.0030039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 doi: 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang HM, Chen H, Liu W, Liu H, Gong J, Wang HL, et al. (2012) AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Research 40: D144–D149. doi: 10.1093/nar/gkr965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu X, Yu XP, Zack DJ, Zhu H, Qian J (2008) TiGER: A database for tissue-specific gene expression and regulation. Bmc Bioinformatics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. (2015) Tissue-based map of the human proteome. Science 347. [DOI] [PubMed] [Google Scholar]
- 83.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912-+. doi: 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 84.Fortes MR, Nguyen LT, Porto Neto LR, Reverter A, Moore SS, Lehnert SA, et al. (2016) Polymorphisms and genes associated with puberty in heifers. Theriogenology 86: 333–339. doi: 10.1016/j.theriogenology.2016.04.046 [DOI] [PubMed] [Google Scholar]
- 85.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research 13: 2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reverter A, Fortes MR (2013) Breeding and Genetics Symposium: building single nucleotide polymorphism-derived gene regulatory networks: Towards functional genomewide association studies. J Anim Sci 91: 530–536. doi: 10.2527/jas.2012-5780 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(TXT)
Left and Right modes describe the Bos indicus and Bos taurus components, respectively. Red indicates the actual distribution of SNP weights, grey curves are the individual Normal distributions, and black curve is the mixture model obtained by combining the two Normal distributions.
(TIFF)
The gradient from green to black to red correspond to low, medium and high heterozygosity.
(TIFF)
(TIFF)
(A) The list of 2,891 genes from the entire list of 8,631 genes that belong to the four main functional categories of transcription factor (TF), secreted hormones (SE), kinases (KI) and genes expressed in tissue-specific (TS) manner; (B) The subset of 1,157 fertility genes collected from the literature where Canovas, MThomas, Fortes_Rev and Fortes correspond to [35], [37], [84] and [36]respectively; (C) The same list of 1,157 fertility genes across the four functional attributes (excluding 10 genes that are not TF, TS, SE, KI).
(TIFF)
(TIFF)
(A) At a correlation cut-off of 0.90 comprising of 858 genes and 12,958 significant connections (B) At a correlation cut-off of 0.95 comprising of 328 genes and 1,098 connections.
(TIFF)
All corresponds to the 8,631 genes in our analysis and PCIT corresponds to the 1,284 network genes. The only category for which significant differences exists (p-value < 0.01) in the indicine percentage is for fertility-related genes.
(TIFF)
The size of the node corresponds to the indicine content. The nodes in green are transcription factors and remaining nodes in the network are purple-coloured. Nodes that are triangle-shaped are fertility-related genes and others are denoted by circles: (A) PCIT network after applying a threshold of 0.90; (B) The network spanned by the trio of fertility related genes GATA4, NR1H4, VAX2; (C) The network spanned by the trio of fertility related genes ELF5, ROCK2, POU2F1.
(TIFF)
Data Availability Statement
The minimal raw data is the SNP genotype data - 18,363 cattle and 729,068 SNP genotypes. The raw SNP genotype data are part of the Beef CRC project (http://www.beefcrc.com/) and are co-owned with Meat and Livestock Australia, and can be made available subject to the agreement of the owners. Any parties seeking access to the raw SNP genotype data should contact: Dr. Toni Reverter-Gomez (toni.reverter-gomez@csiro.au or +61732142392). Further, we have provided data on the 8,631 genes collated and their indicine/taurine proportions as part of the paper and its Supporting Information files.