Abstract
Despite substantial research on cancer therapeutics, systemic toxicity and drug-resistance limits the clinical application of many drugs like cisplatin. Therefore, new chemotherapeutic strategies against different malignancies are needed. Targeted cancer therapy is a new paradigm for cancer therapeutics which targets pathways or chemical entities specific to cancer cells than normal ones. Unlike normal cells, cancer cells contain elevated copper which plays an integral role in angiogenesis. Copper is an important metal ion associated with chromatin DNA, particularly with guanine. Thus, targeting copper via copper-specific chelators in cancer cells can serve as an effective anticancer strategy. New pharmacophore di(2-picolyl)amine (DPA)-3(bromoacetyl) coumarin (ligand-L) was synthesized and characterized by IR, ESI-MS, 1H- and 13C-NMR. Binding ability of ligand-L to DNA/Cu(II) was evaluated using a plethora of biophysical techniques which revealed ligand-L-DNA and ligand-L-Cu(II) interaction. Competitive displacement assay and docking confirmed non-intercalative binding mode of ligand-L with ctDNA. Cyclic voltammetry confirmed ligand-L causes quasi reversible Cu(II)/Cu(I) conversion. Further, acute toxicity studies revealed no toxic effects of ligand-L on mice. To evaluate the chemotherapeutic potential and anticancer mechanism of ligand-L, DNA damage via pBR322 cleavage assay and reactive oxygen species (ROS) generation were studied. Results demonstrate that ligand-L causes DNA cleavage involving ROS generation in the presence of Cu(II). In conclusion, ligand-L causes redox cycling of Cu(II) to generate ROS which leads to oxidative DNA damage and pro-oxidant cancer cell death. These findings will establish ligand-L as a lead molecule to synthesize new molecules with better copper chelating and pro-oxidant properties against different malignancies.
Introduction
DNA is the genetic material which regulates vital processes such as transcription, recombination, proliferation and cell survival. Targeting DNA to modify or inhibit its activity is a subject of extensive research for designing new anticancer agents against different malignancies [1]. Among various anticancer agents, DNA cleaving agents have attracted considerable interest in the field of molecular biology and drug development [2]. Metal-containing compounds have been developed with extensive applications in wide ranging fields such as material and biological sciences [3, 4]. The present treatment regiments for chemotherapy under the class of metal-containing compounds such as platinum-based drug (cisplatin) for different malignancies. However, the major disadvantages of cisplatin (heavy-metal based drug) include various side effects such as severe systemic toxicity (hepatotxicity and nephrotoxicity) and increased drug resistance [5, 6].
To minimize the toxicity and side effects of metal-containing drugs, new chemotherapeutic agents and therapies against different malignancies need to be developed. Targeted cancer therapy is a relatively new form of therapy which involves the use of drugs that block the growth of cancerous tissue by interfering with specific molecules/pathways and spare the normal cells [7]. The basic rationale of targeted cancer therapy is to target chemical entities/mutated proteins that are specific to cancer cells and absent in normal cells of the body [8].
One major difference between normal and cancer cells is the levels of copper which are elevated in malignant cells [9–11]. Copper is an important redox active metal ion attached to DNA bases, particularly guanine [12]. Elevated copper in cancer plays a role in angiogenesis by functioning as a co-factor of several pro-angiogenic molecules such as vascular endothelial growth factor (VEGF), angiogenin and basic fibroblast growth factor (bFGF) [13,14]. Copper chelating to decrease copper bioavailability has been investigated in many clinical studies to inhibit angiogenesis (e.g. clinicaltrials.gov id# NCT00003751, NCT00176800, NCT01837329, NCT02068079, NCT00405574) [15]. Many copper chelators have been synthesized to possess anticancer activities. For example, in murine model of hepatocellular carcinoma, trientine induced apoptosis through interaction with cellular copper and ROS generation [16]. Lowering of copper by tetrathiomolybdate has been observed to reduce tumor growth and impede angiogenesis in murine mesothelioma tumor model [17]. D-penicillamine also shows anticancer activity against leukemia and breast cancer by chelating intracellular bioavailable copper [18]. Thus, under targeted cancer therapy copper-specific chelators should to be synthesized to lower bio-available copper and provide selective approach to target tumor cells. Cytotoxicity of copper-specific chelators arises from high redox activity with copper ions to produce reactive oxygen species (ROS) which leads to DNA damage and oxidative cell death [19, 20].
Coumarins (2H-1-benzopyran-2-one; 1,2-benzopyrone; cis-o-coumarinic acid lactone; coumarinic anhydride) are a large group of naturally occurring organic compounds synthesized by plants, bacteria and fungi [21, 22]. Coumarin represents a promising scaffold and used for the synthesis of coumarin-based derivatives which exhibit various pharmacological properties such as antifungal, antioxidant, anticoagulant, antiviral, antiproliferative, antialzheimer, anticancer and anti-HIV [23–30]. The biochemical properties of coumarin and its derivatives have suggested their use in clinical medicine [31]. Various coumarin derivatives are under clinical trials in different malignancies such as prostate cancer, renal cell carcinoma, malignant melanoma, breast cancer and leukemia [32, 33]. Recently, several studies were conducted that described the addition of bioactive moieties such as pyran, pyridine, thiazole and pyrazole into coumarin nucleus to synthesize new anticancer agents [34–37]. However, no progress has been made towards efficient synthesis of coumarin-based dipic derivatives. Dipic ligands such as di(2-picolyl)amine (DPA) and N-substituted DPA derivatives have been investigated for anticancer activities against malignant cells [38, 39]. Also, it is important to note that DPA exhibits co-ordination properties in metal complexes [40].
In continuation of our pursuit to design and synthesize copper-specific chelators against cancer cells, we herein report the synthesis, characterization and biological activity of new coumarin nucleus-based DPA derivative (ligand-L). Binding studies of ligand-L to DNA/Cu(II) were explored to ascertain the interaction and binding mode. pBR322 DNA cleavage experiments were also conducted to assess the DNA damage caused by ligand-L induced ROS generation in the presence of Cu(II). Results indicate that ligand-L acts as pro-oxidant in the presence of Cu(II) leading to its cytotoxic action. Ligand-L also follows all the parameters under ‘Rule of five’ (no violations) showing tendency towards drug-likeness and drug score with no toxicity. The results of this investigation would be useful in establishing ligand-L as a lead molecule to synthesize new chemical molecules with better copper chelating and pro-oxidant properties against cancer cells.
Materials and methods
General experimental procedures
Chemicals and solvents used in this study were purchased from Merck, India and Sigma-Aldrich, St. Louis, MO, USA and used without further purification. IR spectrum was recorded using Perkin Elmer Spectrum Two IR spectrometer by potassium bromide (KBr) pellet method and values are given in cm-1. 1H and 13C NMR spectra were run in DMSO-d6 on a Bruker Avance II 400 NMR spectrometer at 400 and 100 MHz, respectively. Chemical shifts are reported in ppm (Δ) relative to internal standard tetramethylsilane (TMS). Mass spectrum was recorded on a JEOL SX 102/DA-6000 mass spectrometer. Melting point was recorded on Buchi melting point apparatus B-545. Elemental analysis (C, H, N) was recorded on Perkin Elmer 2400 Series II system. Thin layer chromatography (TLC) glass plates (20×5 cm) were coated with silica gel G and exposed to iodine vapours to check the homogeneity as well as progress of the reaction.
General method for the synthesis of coumarin-based di(2-picolyl)amine (DPA) derivative
An equimolar mixture of 3-(2-bromoacetyl)-2H-chromen-2-one and di(2-picolyl)amine (DPA) (2 mmol each), was stirred in dichloromethane (20 ml) in the presence of sodium carbonate. The reaction mixture was allowed to stir at room temperature for 6–8 h. After completion of the reaction, as evident from TLC, the solvent was removed under reduced pressure. The crude product obtained was washed with water, dried and crystallized from appropriate solvents.
3-(2-{bis[(pyridin-2-yl)methyl]amino}acetyl)-2H-chromen-2-one (ligand-L)
Ligand-L crystallized from CHCl3-MeOH as light reddish solid; Yield: 89%; mp 203°C; IR (KBr, cm-1): 1731.25 (lactone carbonyl-coumarin nucleus), 1688.87 (α, β-unsaturated carbonyl), 1613.28 (α, β- C = C). 1H NMR (400 MHz, DMSO-d6, Δ, ppm): 3.82 (s, 4H), 4.15 (s, 2H), 7.20–7.53 (m, 8H), 7.64–7.68 (m, 2H), 7.81 (d, 2H), 8.51 (s, 1H). 13C NMR (100 MHz, DMSO-d6, Δ, ppm): 61.34, 61.35, 64.20, 116.53, 121.80, 121.83, 124.20, 124.23, 125.81, 127.90, 128.62, 131.24, 136.31, 139.25, 139.29, 149.45, 149.48, 152.20, 153.09, 157.82, 157.85, 166.81, 181.79. Electrospray ionization mass spectrometry (ESI-MS): m/z 385 [M+]. Anal. Calcd. for C23H19N3O3: C, 71.67; H, 4.97; N, 10.90. Found: C, 71.64; H, 4.98; N, 10.91.
Calf thymus DNA (ctDNA) preparation: DNA binding experiments
Stock solution of ctDNA was prepared in 10 mM Tris-HCl buffer (pH 7.2) and later stored at 4°C. Purity of DNA solution was analyzed by recording the absorbance ratio i.e. A260nm / A280nm. Absorbance ratio was between 1.8 and 1.9, and therefore no further purification was required. DNA concentrations used in different experiments were determined using average molar extinction coefficient value of 6600 M-1 cm-1 of a single nucleotide at 260 nm [41]. Stock solution (3 mM) of ligand-L was prepared in DMSO.
UV-Vis spectroscopy of ligand-L with ct-DNA
UV-Vis spectral study of ligand-L was carried out using UV-VIS spectrophotometer (UV-1800, Shimadzu Corp., Tokyo, Japan). Absorbance spectra of ligand-L were recorded in the absence and presence of increasing concentrations of ct-DNA. Briefly, fixed concentration of ligand-L (5 μM) was titrated with increasing concentrations of ct-DNA (0–35 μM) in 10 mM Tris-HCl buffer (pH 7.2).
UV-Vis spectroscopy of ligand-L with Cu(II)
Absorption spectra of ligand-L in absence and presence of Cu(II) were recorded in the wavelength range 235–400 nm using UV-VIS spectrophotometer (UV-1800, Shimadzu Corp., Tokyo, Japan). To the reaction mixture, fixed concentration of ligand-L (5 μM) was used and titrated against increasing concentrations of Cu(II) (0–35 μM) in 10 mM Tris-HCl (pH 7.2).
Steady state fluorescence studies of ligand-L with Cu(II) and ct-DNA
Fluorescence emission spectra were recorded on a RF-5301PC spectrofluorometer (Shimadzu Corp., Tokyo, Japan). Ligand-L was excited at 292 nm and emission spectra were recorded in the wavelength range 300–420 nm after setting the widths of excitation slit at 5 nm and emission slit at 10 nm. To a 1 ml reaction mixture, fixed concentration of ligand-L (5 μM) was used and titrated with increasing concentrations of ct-DNA/Cu(II) (0–40 μM). All fluorescence spectroscopy experiments were carried out in 10 mM Tris-HCl (pH 7.2). The fluorescence intensities were also corrected for inner filter effects using the equation [42]:
where, Fcorr and Fobs are the corrected and observed fluorescence intensities, respectively and A1 and A2 are the sum of absorbances of DNA and ligand-L at the excitation (292 nm) and emission (350 nm) wavelengths, respectively.
Competitive displacement assays
DNA binding dyes such as ethidium bromide (EtBr) [43] and Hoechst 33258 (HO) are used to decipher the binding mode of drug on interaction with DNA. In case of EtBr displacement assay, ctDNA (20 μM) and EtBr (2.5 μM) were dissolved in 10 mM Tris-HCl (pH 7.2). Later, increasing concentrations of ligand-L (0–50 μM) were added to EtBr-DNA solution. Solution was excited at 475 nm and emission spectra were recorded in the range 500–700 nm. Groove binding dye, HO (2 μg/ml) and ctDNA (20 μM) were dissolved in 10 mM Tris-HCl (pH 7.2) and titrated with increasing concentrations of ligand-L (0–50 μM). HO-DNA complex was excited at 343 nm and emission was recorded from 400–600 nm.
Isothermal titration calorimetry measurements (ITC)
ITC is an informative and sensitive method to study thermodynamic parameters of interactions between bio-macromolecules and ligands [44]. Thermodynamic parameters resulted from interaction between ligand-L and ctDNA were determined using VP-ITC titration microcalorimeter (MicroCal Inc., Northampton, MA, USA). All solutions used for ITC were properly degassed prior to use. Briefly, reference and calorimeter cell were loaded with 1X TE buffer (pH 8.0) and ctDNA (0.8 mM), respectively. ctDNA was titrated with 4.1 mM ligand-L up to 28 successive injections of 10 μl each. Titration cell was stirred continuously at 307 rpm and reference power was set at 16 μcal sec-1. Calorimetric data was fitted using independent binding model and analyzed using MicroCal Origin 7.0 software to calculate equilibrium binding constant (Kb), entropy change (ΔS0) and enthalpy change (ΔH0) of complex formation. Free energy change (ΔG0) of ligand-L-DNA complex was calculated using following equations:
| (1) |
| (2) |
where, T is the absolute temperature (298 K) and R is the gas constant with value 8.314 J mol-1 K-1.
Circular Dichroism (CD) studies
CD spectra of ctDNA and in the presence of increasing concentrations of ligand-L were recorded using JASCO-J-720 CD spectropolarimeter equipped with a Peltier-type temperature controller. CD experiments were carried out at 25°C. All the CD spectra were recorded in the range 228–300 nm with a scan speed 200 nm min-1 and spectral bandwidth of 1.0 nm. Each spectrum was the average of three scans. Background spectrum of buffer (10 mM Tris-HCl, pH 7.2) was subtracted from the spectra of DNA and ligand-L-DNA solution. The results were expressed as ellipticity (mdeg).
Potassium iodide (KI) quenching studies
Iodide quenching experiments were performed in the presence and absence of ctDNA. Briefly, ligand-L (30 μM) was dissolved in 10 mM Tris-HCl (pH 7.2) and titrated with increasing concentrations of KI (0–8 mM). Ligand-L was excited at 292 nm and emission spectra were recorded in the wavelength range 300–420 nm. In a different experiment, ligand-L (30 μM) and ctDNA (30 μM) were taken and then an increasing concentration of KI (0–8 mM) was added. Quenching constant (Ksv) values in the presence and absence of DNA was calculated via Stern-Volmer equation [45, 46].
Molecular docking studies
Binding mode of ligand-L in B-DNA was determined using AutoDock (v4.2) in Lamarckian Genetic Algorithm [47, 48]. Ligand-L chemical structure was prepared using ChemDraw 12.0 and saved in MOL format. Mol file was converted into PDB using Avogadro 1.0.1 [49]. Later, structure optimization was carried out using AM1 (Austin Model 1) in Arguslab 4.0.1 and the best conformer exhibiting lowest energy was saved in PDB format for docking. Target receptor (PDBID: 1BNA, sequence d(CGCGAATTCGCG)2) and ligand-L were prepared via docking protocol and saved into ‘PDBQT’ format. Blind docking was then performed to determine the most favourable binding mode of ligand-L in DNA. The input ‘grid parameter’ files were adjusted to X = 60, Y = 60 and Z = 110 with 0.375 nm grid spacing. Rest all docking parameters were set to default values. Energy-scoring function is used to determine the top ligand-DNA pose. The top pose conformation was visualised via PyMOL software (Molecular Graphics System, version 1.5.0.1, Schrodinger.LLC) [50].
Electrochemical measurements of Cu(II)/Cu(I) conversion in presence of ligand-L
Cyclic voltammetry studies were performed on Princeton Applied Research model 263A-1 potentiostat/galvanostat. Voltammetric experiments were carried out using a 3-electrode setup cell consisting of a glassy carbon disk as the working electrode, a platinum wire as the auxiliary electrode and Ag/AgCl electrode system saturated with potassium chloride (KCl) as the reference electrode. Experiments were performed with ligand-L solution in the absence and presence of Cu(II) ions. A solution of 10 mM Tris-HCl (pH 7.2) was used as a supporting electrolyte. Solutions were purged for 5 min before recording the voltammograms. Data was recorded at a scan rate of 150 mV sec-1 at 25°C.
Prediction of drug-likeness (Lipinski’s rule of five)
Lipinski’s rule of five [51] is a rule of thumb that evaluates the drug-likeness properties of chemical compounds. To determine the drug-likeness of ligand-L, physiochemical properties such as octanol-water partition coefficient (log P), molecular weight (MW), rotatable bonds, polar surface area, hydrogen bond donors and acceptors were calculated using molinspiration server (www.molinspiration.com/cgi-bin/properties) [52] and ChemAxon (www.chemicalize.org) [53].
In vitro toxicity test for synthesized compound
In vitro toxicity of ligand-L was checked using erythrocyte lysis test. Briefly, fresh heparinised blood was collected from a healthy non smoking volunteer (Author herself) in EDTA tubes and then centrifuged at 1500 × g for 15 min at 4°C. After centrifugation, buffy coat and plasma present at the top was discarded and erythrocytes at the bottom of tube were washed three times with phosphate buffer saline (PBS) (pH 7.4). The washed erythrocytes were diluted in isotonic solution and 5% hematocrit was prepared for testing toxicity. Red blood cells (RBCs) suspension was incubated with 0.5 ml of 2% DMSO solution (vehicle control for ligand-L) and increasing concentrations of ligand-L (25–100 μM) at 37°C for 1 hr. After complete incubation, the reaction mixture was centrifuged at 1500 × g and the supernatant was collected to measure released (hemoglobin) Hb at λmax = 576 nm.
Percent hemolysis was measured via the formula:
where, Asample is the absorbance of sample treated with ligand-L, Acontrol is the absorbance of samples incubated in PBS and Apositive control is the absorbance of samples treated with 1% Triton X-100.
In vivo acute toxicity study
Ligand-L toxicity was assessed using organisation for economic co-operation and development (OECD) protocol [54]. All animal experiments were approved by Institutional Animal Ethical Committee (IAEC) of Department of Biochemistry, Faculty of Life Sciences, Aligarh Muslim University, Aligarh, India (714/02/a/CPCSEA). Twelve adult swiss albino mice (43–45 g) were maintained in hygienic large cages at 25 ± 2°C on a 12 h light/dark cycles and acclimatized for 1 week before the treatment. Mice were divided into two groups: (1) Group A (n = 6) received only vehicle control (DMSO) intraperitoneally; Group B (n = 6) received 250 mg/kg ligand-L dissolved in DMSO intraperitoneally. Animals were observed for 4 hours post injection and three times a day thereafter injection to notice any change in behaviour and physiological activities or mortality. After 14 days, the animals were sacrificed by cervical dislocation. Blood samples were collected, allowed to clot and centrifuged at 1000 × g for 15 min at room temperature for serum biochemical examination (liver and kidney function test). Kidney and liver were excised and processed for histological analysis via H&E staining.
ROS measurement
ROS namely superoxide anion and hydroxyl radical production were assessed. Superoxide generation by ligand-L alone and in the presence of Cu(II) ions was determined using nitroblue tetrazolium (NBT) assay [55]. Briefly, the assay mixture contains 50 mM sodium phosphate buffer (pH 7.5), 0.3 mM NBT, 0.1 mM EDTA and 0.06% Triton X-100 in a total reaction volume of 3 ml. The reaction was started by adding ligand-L in the presence and absence of Cu(II) ions and absorbance was recorded at 560 nm against a blank solution after 1 h incubation. Hydroxyl radical production by ligand-L in the presence and absence of Cu(II) ions was detected by the method of Quinlan and Gutteridge [56]. ctDNA (300 μg) was used as a substrate and the generation of malondialdehyde from deoxyribose radicals was assayed by recording the absorbance at 532 nm.
Plasmid nicking assay
DNA damage by Cu(II)-ligand-L interaction was assessed by plasmid nicking assay. Reaction mixture (25 μl) contained 10 mM Tris-HCl (pH 7.2), 0.5 μg pBR322 plasmid DNA and other components as indicated in legends. Incubation was performed for 1 h at 37°C. After complete incubation, 5 μl of 5X tracking dye (40 mM EDTA, 0.05% bromophenol blue and 50% (v/v) glycerol) was added and the complete reaction mixture was subjected to electrophoresis in 1% agarose gel. DNA bands were stained with 0.5 μg/ml EtBr solution and visualized under UV illumination gel-doc system (Bio-Rad; Hercules, CA).
Statistical analysis
Experimental values were expressed as mean ± SEM of three independent experiments. Data was analysed by one way-analysis of variance (ANOVA) using GraphPad Prism 5.01 (California, USA) to examine statistically significant differences. p-values < 0.05 were considered statistically significant.
Results
Chemistry
Ligand-L was typically synthesized via a condensation reaction between 3-(2-bromoacetyl)-2H-chromen-2-one and di(2-picolyl)amine (DPA) with the elimination of HBr molecule under stirring at room temperature (Fig 1). The compound was obtained in excellent yield (89%) with high degree of purity. The structure of ligand-L was characterized by IR, 1H NMR, 13C NMR, ESI-MS and elemental analysis. The characterisation studies have been in good corroboration with the expected structural framework of ligand-L. IR spectrum of ligand-L displayed characteristic signals for lactone carbonyl (coumarin nucleus) (1731.25 cm-1), α, β-unsaturated carbonyl (1688.87 cm-1) and α, β- C = C (1613.28 cm-1) (S1 Fig). In 1H NMR spectral analysis, ligand-L exhibited a sharp downfield singlet resonating at around Δ 8.51 ppm assigned to olefinic proton (H-4) (= C-H) (S2 Fig). This appreciable downfield shift of H-4 olefinic proton may be due to possible H-bonding with the adjacent carbonyl group. A pair of singlet peaks resonating at Δ 3.82 [-N-(CH2)2] and Δ 4.15 (-CO-CH2-N-) correspond to four and two methylene protons (-CH2), respectively. A doublet at Δ 7.81 ppm for two protons has been assigned to aromatic H-5 and H-8 protons of coumarin nucleus. A pair of multiplet resonating at Δ 7.64–7.68 and Δ 7.20–7.53 have been allocated to H-6, H-7 (coumarin nucleus) (2 protons) and pyridine nucleus (8 protons), respectively. In 13C NMR spectral study, the absorption bands resonating at Δ 181.79 and Δ 166.81 have been assigned to α, β-unsaturated carbonyl and lactone carbonyl groups, respectively (S3 Fig). The spectrum also displayed the presence of three methylene carbons (-CH2) resonating at Δ 64.20, 61.35 and 61.34, confirming the condensation of two substrate moieties to form ligand-L. The absorption bands Δ 116.53–157.85 have been assigned to other aromatic ring carbons of coumarin and pyridine nucleus. The mass spectral analysis was found to be in good conformity with the proposed structure.
Fig 1. Scheme 1: Synthetic route for the synthesis of ligand-L.
Absorption studies of ligand-L with ctDNA and Cu(II)
To understand the interaction of ligand-L with ctDNA and Cu(II) ions, UV-Vis spectroscopy was performed. Results showed that ligand-L exhibits maximum absorbance at ~292 nm (Fig 2). Addition of increasing concentration of ctDNA resulted in hyperchromism with no shift in the position of maximum absorption peak (Fig 2A). In general, reports suggest that hyperchromism is due to interaction of molecules outside the DNA helix, whereas hypochromism is suggestive of intercalatory mode of binding between ligand and DNA [57, 58]. Hence, absorbance spectral studies provide an evidence of non-intercalative binding mode of ligand-L with DNA. Further, no clear isosbestic point suggests more than one type of binding between ligand-L and DNA. Absorption studies also indicate that with increasing concentration of Cu(II), hyperchromism with a blue shift was observed suggesting interaction between ligand-L and Cu(II) ions (Fig 2B). Also, absence of any isosbestic point in ligand-L-Cu(II) spectra suggests more than one type of complex formation between ligand-L and Cu(II). These results confirm the binding of ligand-L to ct-DNA and Cu(II) ions.
Fig 2. Absorption spectra of ligand-L (5 μM) in the absence and presence of ct-DNA (0–35 μM) and Cu(II) (0–35 μM).
(A) Increasing concentration of ct-DNA showed hyperchromic shift suggesting ct-DNA-Ligand-L interaction. (B) Increasing concentration of Cu(II) showed hyperchromic shift suggesting Cu(II)- Ligand-L interaction.
Interaction studies via steady state fluorescence
Fluorescence spectroscopy has been widely used to determine the interaction and binding mode of drugs/molecules to DNA due to high sensitivity and accuracy [59]. In the absence of ctDNA or Cu(II) ions, ligand-L exhibited an emission maximum at 350 nm after excitation at 292 nm (Fig 3). On addition of increasing concentrations of DNA, quenching with no shift in emission maxima peak position of ligand-L was observed (Fig 3A). Similarly, addition of increasing concentrations of Cu(II) to ligand-L solution resulted in quenching of fluorescence emission intensity with no significant shift in λmax emission of ligand-L (Fig 3B). This quenching (hypochromism) confirms the interaction of ligand-L with ctDNA and Cu(II) ions.
Fig 3.
Fluorescence emission spectra of ligand-L (5 μM) in the absence and presence of ct-DNA (0–40 μM) (A) and Cu(II) (0–40 μM) (B). Increasing concentrations of DNA and Cu(II) lead to quenching in fluorescence intensity of ligand-L.
To further understand the interaction of ligand-L with DNA and Cu(II) ions, quenching constant (Ksv) was obtained from Stern-Volmer equation [45, 46]:
where, F0 and F are the fluorescence intensities in the absence and presence of ctDNA or Cu(II) ions, respectively and [Q] is the concentration of ctDNA or Cu(II) ions in the solution. Ksv was determined by plotting the ratio of fluorescence intensity (F0/F) in the absence and presence of ctDNA or Cu(II) ions as a function of increasing concentrations of DNA or Cu(II) ions. Ksv values were calculated from the slopes of Fig 4 and found to be (7.41 ± 0.01) × 103 M-1 and (8.53 ± 0.01) × 103 M-1 for ctDNA and Cu(II) ions, respectively. Ligand-L-DNA complex exhibits a quenching constant value lower than the classical intercalators [60, 61], and therefore indicating non-intercalative mode of binding with DNA.
Fig 4.
Stern-Volmer plots for interaction of ligand-L with ct-DNA (A) and Cu(II) (B).
Later, steady state fluorescence data was also used to calculate binding stoichiometry (n) and binding constant (K) of ligand-L-DNA and ligand-L-Cu(II) complexes using the equation:
where, F0 and F are the fluorescence intensities in the absence and presence of DNA or Cu(II) ions, respectively. [Q] is the concentration of DNA or Cu(II) used in the experiments. The values of binding stoichiometry and binding constant were determined by the slope and intercept of the plot log [(F0 –F)/F] vs log [Q], respectively. The results so obtained are summarized in Table 1. It was found that both the ligand-L-DNA and ligand-L-Cu(II) systems exhibit an almost identical stoichiometry or binding sites (n value). This result suggests that the molecular population of two different systems contribute equally in molecular interactions in ligand-L complex formation with DNA and Cu(II).
Table 1. Parameters obtained using fluorescence studies.
| Complex | Ksv (×103) (M-1) |
K (×103) (M-1) |
n | R2 |
|---|---|---|---|---|
| Ligand-L + DNA | 7.41 ± 0.01 | 4.80 ± 0.03 | 0.958 | 0.990 |
| Ligand-L + Cu(II) | 8.53 ± 0.01 | 5.23 ± 0.04 | 0.957 | 0.984 |
Thermodynamic parameters were also calculated to determine the nature of binding forces between ligand-L and DNA/Cu(II). There are several kinds of binding forces that exist in interactions between drug and biomolecules such as hydrophobic forces, van der Waals forces, electrostatic interactions and hydrogen bonds [62, 63]. Enthalpy change (ΔH0) and entropy change (ΔS0) are the main quantities that determine the interaction forces: (1) ΔH0 > 0 and ΔS0 > 0, hydrophobic forces; (2) ΔH0 < 0 and ΔS0 < 0, van der Waals force’s and hydrogen bonds; (3) ΔH0 << 0 and ΔS0 > 0, electrostatic forces. The values of entropy change and enthalpy change were calculated at different temperatures using the van’t Hoff equation:
where, K represents the binding constant at absolute temperature and R is the gas constant (1.985 × 10−3 kcal mol-1 K-1). The value of ΔH0 and ΔS0 were obtained from the slope and intercept of the plot ln k vs 1/T, respectively (S4 and S5 Figs). ΔG was determined at three different temperatures (298K, 303 K and 310 K) using the equation: ΔG = ΔH0 − TΔS0. Results of such calculations at different temperatures for the interaction of ligand-L to DNA/Cu(II) are given in S1 and S2 Tables. As evident from the results, negative ΔH0 and ΔS0 values indicate that the major interaction forces in ligand-L-DNA complexation are hydrogen bonding and van der Waals forces. Negative ΔH0 and positive ΔS0 confirm the presence of electrostatic interactions between ligand-L and Cu(II). The negative values of ΔG0 confirmed that the interaction of ligand-L to DNA/Cu(II) is spontaneous in nature.
Competitive displacement assay studies
Competitive binding experiments are extensively used to reveal the binding mode of small molecules to DNA. In order to decipher the binding mode, various DNA binding dyes such as EtBr (intercalator) or Hoechst 33258 (groove binder) are used. This assay is based on the principle that any small molecule/drug that displaces the bound dye from DNA is expected to bind in a similar fashion as bound dye [64–67]. Therefore, changes in the fluorescence of dye-DNA complex upon addition of small molecule/drug help to determine the binding mode of molecule in DNA. As evident from the results of EtBr displacement assay, addition of increasing concentrations of ligand-L did not change the fluorescence intensity of EtBr-DNA complex (Fig 5A). This suggests non-intercalative mode of binding of ligand-L with ctDNA.
Fig 5. Competitive displacement assays.
(A) Fluorescence titration of EtBr-DNA complex with ligand-L. EtBr-DNA complex was excited at 471 nm and emission spectra were recorded from 520–700 nm. No change in fluorescence intensity was recorded on addition of increasing concentration of ligand-L. (B) Fluorescence titration of Hoechst-DNA complex with ligand-L. Hoechst-DNA complex was excited at 343 nm and emission spectra were recorded from 400–600 nm. Fluorescence intensity decreases on addition of increasing concentration of ligand-L.
To further confirm the non-intercalative mode of binding, Hoechst dye based displacement assay was performed. Hoechst dye holds an ability to bind to the minor groove region of DNA and increases fluorescence intensity on binding to DNA [66, 67]. Results suggest that increase in concentration of ligand-L decreases the fluorescence intensity of Hoechst-DNA system (Fig 5B). This confirms that binding mode of ligand-L is in the groove rather than intercalation.
Binding studies using ITC method
The representative thermodynamic parameters of ligand-L interaction with ctDNA were evaluated using ITC titration method. Table 2 and Fig 6 show the thermodynamic parameters of ligand-L-DNA complex calculated via independent binding model. Ligand-L interaction with DNA resulted in negative value of enthalpy change indicating that the interaction is exothermic in nature. Sequential titrations of ligand-L to DNA solution lead to large negative entropy term (TΔS0 = -5.03 × 102 kcal/mol), which suggests that the binding of ligand-L with ctDNA is enthalpy driven. It can also be seen from results in Table 2 that both enthalpy and entropy changes have negative values indicating that the binding of ligand-L with ctDNA is predominately governed by hydrogen bonding and van der Waals interactions. Further, ITC analysis revealed a negative Gibb’s free energy change (∆G0) for ligand-L-DNA complex which indicates that the interaction process proceeds spontaneously. It is also important to note that intercalative binding interactions are entropy driven whereas groove binding mode is enthalpy driven [68, 69]. Thermodynamic results obtained from ITC experiment support the competitive binding experiment results (Fig 5), further supporting the non-intercalative binding mode of ligand-L with DNA. Regarding the binding parameters (Kb and Ksv), it should be noted that ITC and fluorescence spectroscopy experiments provide different values (Tables 1 and 2). This difference may be due the fact that ITC measures global or bulk changes in binding and thermodynamic parameters, while fluorescence spectroscopy measures only local changes around the fluorophore [70, 71]. As a result, ITC encounters large molecular interactions among entire molecular population, whereas fluorescence spectroscopy determines only limited or localized molecular interactions in the test sample.
Table 2. The binding constant, binding stoichiometry and thermodynamic parameters for the binding of ligand-L to ctDNA determined with ITC at 25°C.
| Complex | Kb (M-1) | n | ∆G° (kcal mol-1) | ∆H° (kcal mol-1) | ∆S° (kcal mol-1 K-1) |
|---|---|---|---|---|---|
| Ligand-L + DNA | (1.45 ± 0.18) × 104 | 0.037 ± 0.008 | -6.38 | (-5.10 ± 0.43) × 102 | -1.69 |
Fig 6. ITC curve (upper panel) and the binding isotherm (lower panel) for ligand-L interaction with ct-DNA at 25°C.
Circular Dichroism (CD) studies
CD spectroscopy technique is a sensitive method to detect changes in the secondary structure of DNA [72]. Changes in the intrinsic CD spectra of DNA backbone depends on the non-covalent interaction of molecules with DNA [73]. As seen from Fig 7, CD spectrum of DNA alone exhibits a positive peak at ~ 276 nm and a negative peak at ~ 244 nm. This is consistent with CD spectra of double helical DNA in B conformation [74, 75]. The positive band at ~ 276 nm is due to base stacking and negative band at ~ 244 nm corresponds to helicity of right-handed B-form of DNA [76, 77]. Further, it is important to note that both these bands (peaks) are very sensitive to the interaction of small molecules with DNA [77, 78]. Groove binding molecules cause less or no perturbation on the positive and negative bands of CD spectra of ctDNA, whereas intercalating molecules are known to produce significant effect on intensities of both bands [79, 80]. Binding mode of ligand-L with ctDNA was studied using CD spectroscopy. Fig 7 shows that no detectable change in CD spectrum was recorded upon addition of ligand-L to ctDNA solution. These results confirmed the non-intercalative binding mode of ligand-L with ctDNA.
Fig 7. Effect of ligand-L on CD spectra of ctDNA.
CD spectra of ctDNA (50 μM) with varying concentrations of ligand-L (0–100 μM).
KI quenching studies
Iodide quenching studies help to determine the binding mode of drug with DNA. Such an assay depends on the fact that when small molecules are intercalated in DNA, iodide ions are repelled by negatively charged phosphate groups of DNA and fluorescence of such molecules remains unaffected in the presence of DNA. However, molecules which are present as groove binders (exposed to external surface) are easily approachable to quenchers even in the presence of DNA [81].
The quenching constant of anionic quenchers in the absence and presence of DNA is calculated via Stern-Volmer equation [45, 46]:
where, F0 and F are the fluorescence intensities in the absence and presence of anionic quencher, KI and [Q] is the concentration of KI in the solution. Ksv is the quenching constant obtained from the slope of F0/F vs [Q] plot. Ksv values determine the type of binding of molecules with DNA. Decrease in Ksv occurs in intercalation and it remains unchanged when interaction is electrostatic or when molecules bind to the groove. As evident from results, no significant difference in Ksv values is observed in the absence and presence of DNA for ligand-L (Fig 8). Therefore, it can be inferred that ligand-L exhibits non-intercalative binding mode with ctDNA.
Fig 8. KI quenching experiment.
Stern-Volmer plot for fluorescence quenching of ligand-L (30 μM) by KI in the absence and presence of ctDNA (30 μM). Difference in Ksv value (quenching constant) was used to investigate the binding mode of ligand-L with ctDNA. R is the correlation coefficient.
Molecular docking studies
In order to determine the exact interaction of ligand-L with DNA, docking studies were performed (Table 3 and Fig 9). Surface view interaction of the docked structure in Fig 9A clearly indicates that ligand-L preferentially binds in the groove region of DNA. In total, ligand-L forms 5 hydrogen bonds with adenine (A-3, 4, 7) and thymine (T-5, 8) residues of DNA with binding energy -6.36 kcal/mol and inhibition constant 21.69 μM. Thus, hydrogen bonding plays a major role in the interaction of ligand-L with DNA. More negative binding energy (-6.36 kcal mol-1) and lower inhibition constant (21.69 μM) confirm strong binding of ligand-L with B-DNA. Further, higher hydrogen bonding interaction number (5 H-bonds) confirms the greater stability of ligand-L with B-DNA. Finally, it is interesting to note that both the experimental binding free energy (ITC, Table 2) and theoretical binding energy (docking, Table 3) have negative values, indicating that ligand-L-DNA interaction is spontaneous at room temperature. This shows that molecular docking results are in accordance with the results of ITC experiments.
Table 3. AutoDock results (binding energy, inhibition constant and no. of hydrogen bonds) of ligand-L in B-DNA.
| Ligand | AutoDock binding energy (kcal/mol) | AutoDock inhibition constant (μM) | No. of Hydrogen bonds |
|---|---|---|---|
| Ligand-L | -6.36 | 21.69 | 5 |
Fig 9. Molecular docked structure of ligand-L complexed with B-DNA.
(A) Surface view interaction of ligand-L with B-DNA. (B) Hydrogen bonding interactions (5) of ligand-L with B-DNA (PDB ID: 1BNA).
Cyclic voltammetric studies of Cu(II)-ligand-L interaction
Cyclic voltammetry is an important technique for characterizing the redox properties of metal complexes [82]. The cyclic voltammograms (CV) recorded at a scan rate 150 mV sec-1 for ligand-L and Cu(II)-ligand-L solution are shown in Fig 10. CV of ligand-L shows flattened cathodic and anodic peaks. However, addition of Cu(II) ions to ligand-L solution leads to the formation of pronounced cathodic and anodic peaks at = 0.03 V and = -0.45 V, respectively. These two peaks couple together to form a well-defined Cu(II)/Cu(I) qausi-reversible redox couple [83, 84] with E0 ½ = 0.48 V (half-wave potential). The separation between the cathodic and anodic peak current (Ipa/Ipc = 0.84) clearly confirms the formation of redox couple Cu(II)/Cu(I) via one electron transfer redox process.
Fig 10. Cyclic voltammogram of ligand-L (25 μM) in absence and presence of Cu(II) (25 μM).
In silico study of ADME/T prediction
For a compound to be orally active (drug-likeness), it should follow Lipinski ‘Rule of five’ which includes the following parameters: (1) Mass < 500; (2) Octanol-water partition coefficient (logP) ≤ 5; (3) Rotatable bonds ≤ 10; (4) Polar surface area ≤ 150 Å2; (5) Hydrogen bond acceptors ≤ 10; (6) Hydrogen bond donors ≤ 5. Ligand-L under investigation follows all the parameters under ‘Rule of five’ (no violations) calculated via molinspiration and chemicalize.org servers, and revealed higher tendency of ligand-L towards drug-likeness (S3 Table).
Ligand-L exerts no toxic effect on RBCs in vitro
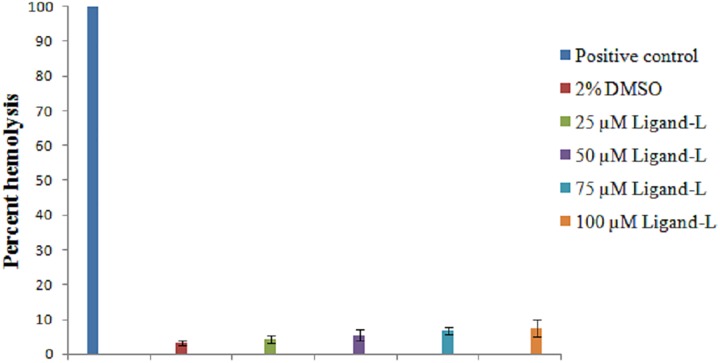
Toxicity is the major side effect of synthesized cancer chemotherapeutic agents. Therefore, we investigated the toxic effect of ligand-L by in vitro RBC lysis testing (Fig 11). As evident from our results, ligand-L did not induce significant hemolysis and such an effect was also not observed even at high concentrations 100 μM. These results clearly indicate ligand-L is safe, free of any toxicity constraints and exhibits blood compatibility.
Fig 11. Erythrocyte lysis test.
In vitro toxicity measured on treatment with vehicle control (2% DMSO) and increasing concentrations of ligand-L (25–100 μM). Values expressed as mean ± SEM of three independent experiments.
Ligand-L exerts no toxic effect on liver and kidney
Acute toxicity of ligand-L was evaluated using albino mice. Mice well tolerated the drug and no sign of pharmacotoxicity was observed at 250 mg/kg body weight dose. Liver and renal function test biomarkers in the serum of control and treated-mice are reported in Tables 4 and 5 and no significant difference was observed in control and treated liver/kidney of mice. Further, no obvious histopathological changes were observed in treated liver and kidney structures as compared to normal control group (Fig 12). These results clearly suggest that ligand-L is devoid of hepatotoxicity and nephrotoxicity effects and could be used as anticancer agent.
Table 4. Effect of ligand-L treatment on liver function test of mice.
Values expressed as mean ± SEM.
| Group | Total bilirubin (μmol/L) | Albumin (g/L) |
ALP (IU/L) |
ALT (IU/L) |
AST (IU/L) |
GGT (IU/L) |
|---|---|---|---|---|---|---|
| A: Vehicle | 1.01 ± 0.41 | 20.55 ± 1.55 | 40.72 ± 1.27 | 52.10 ± 2.54 | 187.15 ± 3.18 | 3.28 ± 0.28 |
| B: 250 mg per kg dose | 0.83 ± 0.62 | 22.32 ± 2.21 | 43.17 ± 0.73 | 50.73 ± 3.21 | 193.2 ± 4.3 | 3.0 ± 0.11 |
Table 5. Effect of ligand-L treatment on renal function test of mice.
Values expressed as mean ± SEM.
| Group | Na+ (mmol/L) | K+ (mmol/L) |
Cl- (mmol/L) |
Urea (mmol/L) |
Creatinine (μmol/L) |
|---|---|---|---|---|---|
| A: Vehicle | 142 ± 1.35 | 7.7 ± 0.47 | 111.83 ± 1.55 | 8.3 ± 2.03 | 18.23 ± 0.51 |
| B: 250 mg per kg dose | 145.8 ± 0.34 | 8.2 ± 0.18 | 109.1 ± 0.91 | 9.4 ± 1.11 | 18.07 ± 0.78 |
Fig 12. Histological analysis of ligand-L treated groups (H&E staining).
Histological sections of liver (first row) and kidney (second and third row). Untreated (control group) liver (Panel 1) and kidney (Panels 3 and 5) of mice. Liver (Panel 2) and kidney (Panels 4 and 6) from ligand-L (250 mg/kg body weight) treated mice. No significant differences in structures were observed of liver and kidney of control and treated groups. Panels 3 and 4 represent tubular region of control and treated kidney respectively, whereas panels 5 and 6 represent glomerulus region of control and treated kidney respectively.
Cu(II)-ligand-L interaction induces ROS generation
Increased oxidative stress beyond the threshold level serves as an effective therapeutic strategy to target cancer cells [85]. ROS mainly include superoxide anion (O2.-) and hydroxyl radical (OH.) which are harmful to bio-macromolecules of cells. Superoxide anion and hydroxyl radicals were estimated using NBT assay and ctDNA substrate assay (Figs 13 and 14). As evident from results of Fig 13A, ligand-L alone did not induce significant superoxide anion generation. Addition of Cu(II) ions in ligand-L solution leads to significant superoxide anion generation (Fig 13B). Such superoxide anion generation was quenched by copper chelator, neocuproine (Fig 13B). Further, the presence of SOD (enzymatic ROS scavenger) largely abrogated the superoxide generation by Cu(II)-ligand-L interaction (Fig 13B). Fig 14 demonstrates the results of hydroxyl radical generation by ligand-L in the absence and presence of Cu(II) ions. Ligand-L did not induce significant hydroxyl radical generation (Fig 14A). However, presence of Cu(II) in ligand-L solution causes significant hydroxyl radical production (Fig 14B). Also, hydroxyl radical generation was abrogated on addition of neocuproine and thiourea (Fig 14B). These results confirm that ligand-L causes redox cycling of Cu(II) ions leading to ROS generation.
Fig 13. NBT reduction assay.
(A) Estimation of superoxide anion generation by increasing concentrations of ligand-L (25–100 μM). (B) Superoxide anion generation by ligand-L in the presence of Cu(II) ions and effect of copper chelator (neocuproine) and superoxide dismutase (ROS scavenger) on ROS generation by ligand-L-Cu(II) system. (1) Ligand-L (25 μM) (2) Cu(II) (25 μM) (3) Ligand-L (25 μM) + Cu(II) (25 μM) (4) Ligand-L (25 μM) + Cu(II) (25 μM) + Neocuproine (50 μM) (5) Ligand-L (25 μM) + Cu(II) (25 μM) + SOD (20 μg/ml). *P < 0.05 with respect to ligand-L (25 μM) set and #P < 0.05 with respect to ligand-L (25 μM) + Cu(II) (25 μM) set.
Fig 14.
(A) Estimation of hydroxyl radical generation by increasing concentrations of ligand-L (25–100 μM). (B) Hydroxyl radical generation by ligand-L in the presence of Cu(II) ions and effect of copper chelator (neocuproine) and thiourea (ROS scavenger) on ROS generation by ligand-L-Cu(II) system. (1) Control-No treatment (2) Ligand-L (25 μM) (3) Cu(II) (25 μM) (4) Ligand-L (25 μM) + Cu(II) (25 μM) (5) Ligand-L (25 μM) + Cu(II) (25 μM) + Neocuproine (50 μM) (6) Ligand-L (25 μM) + Cu(II) (25 μM) + thiourea (0.2 mM). *P < 0.05 with respect to control and #P < 0.05 with respect to Ligand-L (25 μM) + Cu(II) (25 μM) set.
Treatment of supercoiled plasmid pBR322 with Cu(II)-ligand-L system
Cytotoxicity of Cu(II)-ligand-L system was assessed on supercoiled pBR322 plasmid DNA. As seen from the agarose gel pattern, increasing concentrations of ligand-L alone (25–75 μM) and Cu(II) alone (25–75 μM) did not cause any DNA cleavage (S6 Fig). On the other hand, increasing concentration of Cu(II) (25–100 μM) in the presence of 25 μM ligand-L leads to nicking of plasmid from its supercoiled form to open circular topological structure of DNA (Fig 15A). At molar ratio 1:4 of ligand-L:Cu(II) system, the cleavage becomes more pronounced with the appearance of linear form of plasmid DNA (Fig 15B). Further, addition of neocuproine and ROS scavengers abrogated plasmid nicking via ligand-L:Cu(II) system [Ligand-L (25 μM) + Cu(II) (25 μM)] (Fig 15B). These results suggest that ligand-L is capable of DNA damage in the presence of copper via redox cycling of copper ions (Cu(I) acts as intermediate in DNA damage) and ROS generation.
Fig 15. Cu(II)-Ligand-L system induced plasmid DNA damage.
(A) Treatment of plasmid pBR322 DNA with ligand-L alone (25 μM) (Lane 1) and Cu(II)-Ligand-L system i.e. Ligand-L (25 μM) + Cu(II) (25 μM) (Lane 2), Ligand-L (25 μM) + Cu(II) (50 μM) (Lane 3), Ligand-L (25 μM) + Cu(II) (75 μM) (Lane 4), Ligand-L (25 μM) + Cu(II) (100 μM) (Lane 5). (B) Effect of copper chelator (neocuproine) and ROS scavengers (thiourea, catalase and SOD) on plasmid pBR322 DNA damage induced by ligand-L-Cu(II) system. (Lane1 1): Ligand-L (25 μM) + Cu(II) (25 μM). (Lane 2): Ligand-L (25 μM) + Cu(II) (25 μM) + Neocuproine (50 μM). (Lane 3): Ligand-L (25 μM) + Cu(II) (25 μM) + thiourea (0.2 mM). (Lane 4): Ligand-L (25 μM) + Cu(II) (25 μM) + catalase (20 μg/ml). (Lane 5): Ligand-L (25 μM) + Cu(II) (25 μM) + SOD (20 μg/ml). Lane ‘C’ depicts the ‘Control’ untreated plasmid DNA.
Discussion
Copper uptake is markedly increased in all types of tumor cells as it is required for tumor growth and angiogenesis [86, 87]. Tumor cells express high levels of CTR1, primary copper transporter which aids in the uptake and accumulation of copper required to maintain internal homeostasis [88]. Redox active copper complexes mediate direct DNA stand scission via ROS generation and such complexes have been screened for anticancer activity in vitro and in vivo [89–91]. The basic rationale behind targeted cancer therapy using redox active copper chelating agents is to generate high levels of ROS to surpass threshold limit leading to oxidative stress mediated irreversible damage and cancer cell death.
Here, we synthesized 3-(2-{bis[(pyridin-2-yl)methyl]amino}acetyl)-2H-chromen-2-one (ligand-L) which binds to DNA and Cu(II). In vitro fluorescence quenching study reveals that the binding of ligand-L to DNA/Cu(II) is strong because the binding constant, K (Table 1) is high. Therefore, it is very likely that ligand-L will also bind with DNA/Cu(II) in in vivo conditions. Our results of high binding constant (~ 103 M-1) are supported by the studies present in literature where it was shown that curcumin (binding constant with DNA: 2.97 × 103 M-1) causes DNA damage in cancer cells and also inhibits tumor growth in in vitro and in vivo xenograft model [92–94].
ROS generation and DNA cleavage assays are suggestive of a role of Cu(II)-ligand-L interaction in ROS generation that causes DNA breakage. Further, experiments using neocuproine (copper chelator) and ROS scavengers have shown inhibition of DNA cleavage and ROS generation by Cu(II)-ligand-L complex, suggesting that ligand-L toxicity requires interaction with copper. Thus, copper ions act as a molecular target of ligand-L in mediating redox cycling that leads to ROS generation (hydroxyl radicals) and selective cytotoxicity against cancer cells.
Ligand-L in the presence of transition metals like copper generates ROS via Fenton-like reaction. Such a quasi reversible Cu(II)/Cu(I) redox couple decomposes hydrogen peroxide to generate hydroxyl radicals, which act as proximal DNA cleaving agent [95].
We believe that copper-dependent ROS generation by ligand-L is the primary step in inducing oxidative stress and DNA damage. Further, up-regulation of p53 mitochondrial gene, ATM/ATR pathway, down-regulation of anti-apoptotic protein (Bcl-2) and cell cycle related proteins (cyclin, cyclin dependent kinases-CDKs) might be the secondary downstream effects of ROS generation induced by Cu(II)-ligand-L interaction [96]. These effects may be ultimately involved in inducing apoptosis of cancer cells.
Among oxygen radicals generated by Cu(II)-ligand-L system, hydroxyl radicals are the most electrophilic with high reactivity and also possess small diffusion radius. For hydroxyl radicals to cause strand scission, ROS should be generated in close proximity of cellular DNA [97–99]. For the pro-oxidant activity of ligand-L, it is important that ligand-L should be associated with DNA prior to generation of hydroxyl radicals at that site. Thus, ligand-L should bind to DNA where it can interact with chromatin bound Cu(II) to form Cu(II)-DNA-ligand-L ternary complex to mediate ROS generation.
To establish the role of ROS in cancer treatment, it is important to note that tumor cells have higher basal levels of intra-cellular ROS as compared to normal cells [100]. Studies also suggest that cancer cells exhibit up-regulated antioxidant system attributed to evade altered redox status [85]. Thus, pro-oxidant approaches (via anticancer agents) should be implemented to further increase the oxidative stress above the toxicity threshold level where the antioxidant system of these cells is also not sufficient to contain toxic ROS levels, representing an effective mechanism for cancer treatment [85, 101].
In summary, we synthesized, characterized and examined the biological relevance of coumarin nucleus-based DPA derivative (ligand-L). A series of biophysical and docking studies were implemented to predict the binding characteristics and interaction of ligand-L with ctDNA and Cu(II). Results clearly confirmed the formation of ligand-L-DNA and ligand-L-Cu(II) complex. Ligand-L engages in Cu(II) redox cycling to generate ROS that leads to DNA damage (nicking). Thus, ligand-L in all probabilities acts as pro-oxidant in the presence of Cu(II), leading to its cytotoxic action. Since tumor cells have elevated copper than normal cells, this provides a window of opportunity for ligand-L to be used as robust therapeutic agent for selective cytotoxic action against different malignancies with copper as the molecular target. Further studies are in progress in our laboratory to provide a deep insight into the molecular mechanism of anticancer activity of ligand-L in malignant cells. We expect this study to establish ligand-L as a lead molecule to identify or synthesize new anti-cancer drugs with better copper chelating and pro-oxidant properties.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Treatment of plasmid pBR322 DNA with increasing concentrations of ligand-L alone (25–100 μM) (Lanes 1–3) and Cu(II) ions alone (25–100 μM) (Lanes 4–6). Lane C represents untreated (control) plasmid. Ligand-L and Cu(II) treatment alone were ineffective in plasmid DNA cleavage.
(PDF)
(PDF)
(DOCX)
Virtual screening of ligand-L showing drug-likeliness by (A) Molinspiration (B) chemicalize.org servers.
(PDF)
Acknowledgments
The authors are thankful to Central Instrumentation Facility (CIF) of Chemistry Department, AMU, Aligarh for FTIR, CV and CD facilities. We thank SAIF, Punjab University, Chandigarh for NMR spectral studies. We would also like to thank the Chairperson, Biochemistry Department, AMU, Aligarh, for providing the necessary laboratory facilities. The UGC-MANF JR fellowship to SK is acknowledged.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
SK received financial support from the UGC-MANF JR Fellowship. No additional funding was received for this study. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002; 2:188–200. doi: 10.1038/nrc749 [DOI] [PubMed] [Google Scholar]
- 2.Burger RM. Cleavage of nucleic acids by bleomycin. Chem Rev. 1998; 98:1153–1170. [DOI] [PubMed] [Google Scholar]
- 3.Boerner LJ, Zaleski JM. Metal complex-DNA interactions: from transcription inhibition to photoactivated cleavage. Curr Opin Chem Biol. 2005; 9:135–144. doi: 10.1016/j.cbpa.2005.02.010 [DOI] [PubMed] [Google Scholar]
- 4.Zhang CX, Lippard SJ. New metal complexes as potential therapeutics. Curr Opin Chem Biol. 2003; 7:481–489. [DOI] [PubMed] [Google Scholar]
- 5.Boulikas T, Vougiouka M. Cisplatin and platinum drugs at the molecular level. (Review). Oncol Rep. 2003; 10:1663–1682. [PubMed] [Google Scholar]
- 6.Hassan I, Chibber S, Naseem I. Ameliorative effect of riboflavin on cisplatin induced nephrotoxicity and hepatotoxicity under photoillumination. Food Chem Toxicol. 2010; 48:2052–2058. doi: 10.1016/j.fct.2010.05.004 [DOI] [PubMed] [Google Scholar]
- 7.Blagosklonny MV. Matching targets for selective cancer therapy. Drug Discov Today. 2003; 8:1104–1107. [DOI] [PubMed] [Google Scholar]
- 8.Dobbelstein M, Moll U. Targeting tumour-supportive cellular machineries in anticancer drug development. Nat Rev Drug Discov. 2014; 13:179–196. doi: 10.1038/nrd4201 [DOI] [PubMed] [Google Scholar]
- 9.Yoshida D, Ikeda Y, Nakazama S. Quantitative analysis of copper, zinc and copper/zinc ratio in selective human brain tumors. J Neurooncol. 1993; 16:109–115. [DOI] [PubMed] [Google Scholar]
- 10.Ebadi M, Swanson S. The status of zinc, copper and metallothionein in cancer patients. Prog Clin Biol Res. 1988; 259:167–175. [PubMed] [Google Scholar]
- 11.Nasulewicz A, Mazur A, Opolski A. Role of copper in tumor angiogenesis—clinical implications. J Trace Elem Med Biol. 2004; 18:1–8. doi: 10.1016/j.jtemb.2004.02.004 [DOI] [PubMed] [Google Scholar]
- 12.Kagawa TF, Geierstanger BH, Wang AH, Ho PS. Covalent modification of guanine bases in double stranded DNA: the 1:2-AZDNA structure of dc(CACACG) in the presence of CuCl2. J Biol Chem. 1991; 266:20175–20184. [DOI] [PubMed] [Google Scholar]
- 13.Brewer GJ. Copper lowering therapy with tetrathiomolybdate as an antiangiogenic strategy in cancer. Curr Cancer Drug Targets. 2005; 5:195–202. [DOI] [PubMed] [Google Scholar]
- 14.Pan Q, Kleer CG, van Golen KL, Irani J, Bottema KM, Bias C, et al. Copper deficiency induced by tetrathiomolybdate suppresses tumor growth and angiogenesis. Cancer Res. 2002; 62:4854–4859. [PubMed] [Google Scholar]
- 15.Denoyer D, Masaldan S, La Fontaine S, Cater MA. Targeting copper in cancer therapy: 'Copper That Cancer'. Metallomics. 2015; 7:1459–1476. doi: 10.1039/c5mt00149h [DOI] [PubMed] [Google Scholar]
- 16.Yoshii J, Yoshiji H, Kuriyama S, Ikenaka Y, Noguchi R, Okuda H, et al. The copper-chelating agent, trientine, suppresses tumor development and angiogenesis in the murine hepatocellular carcinoma cells. Int J Cancer. 2001; 94:768–773. [DOI] [PubMed] [Google Scholar]
- 17.Crowe A, Jackaman C, Beddoes KM, Ricciardo B, Nelson DJ. Rapid copper acquisition by developing murine mesothelioma: decreasing bioavailable copper slows tumor growth, normalizes vessels and promotes T cell infiltration. PLoS One. 2013; 8:e73684 doi: 10.1371/journal.pone.0073684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupte A, Mumper RJ. Copper chelation by D-penicillamine generates reactive oxygen species that are cytotoxic to human leukemia and breast cancer cells. Free Radic Biol Med. 2007; 43:1271–1278. doi: 10.1016/j.freeradbiomed.2007.07.003 [DOI] [PubMed] [Google Scholar]
- 19.Trejo-Solis C, Jimenez-Farfan D, Rodriguez-Enriquez S, Fernandez-Valverde F, Cruz-Salgado A, Ruiz-Azuara L, et al. Copper compound induces autophagy and apoptosis of glioma cells by reactive oxygen species and JNK activation. BMC Cancer. 2012; 12:156 doi: 10.1186/1471-2407-12-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prisecaru A, McKee V, Howe O, Rochford G, McCann M, Colleran J, et al. Regulating bioactivity of Cu2+ bis-1,10-phenanthroline artificial metallonucleases with sterically functionalized pendant carboxylates. J Med Chem. 2013; 56:8599–8615. doi: 10.1021/jm401465m [DOI] [PubMed] [Google Scholar]
- 21.Raistrick H, Stickings CE, Thomas R. Studies in the biochemistry of micro-organisms. 90. Alternariol and alternariol monomethyl ether, metabolic products of Alternaria tenuis. Biochem J. 1953; 55:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinman JW, Caron EL, Hoeksema H. The Structure of Novobiocin. J Am Chem Soc. 1957; 79:3789–3800. [Google Scholar]
- 23.Curir P, Galeotti F, Dolci M, Barile E, Lanzotti V. Pavietin, a coumarin from Aesculus pavia with antifungal activity. J Nat Prod. 2007; 70:1668–1671. doi: 10.1021/np070295v [DOI] [PubMed] [Google Scholar]
- 24.Beillerot A, Domínguez JC, Kirsch G, Bagrel D. Synthesis and protective effects of coumarin derivatives against oxidative stress induced by doxorubicin. Bioorg Med Chem Lett. 2008; 18:1102–1105. doi: 10.1016/j.bmcl.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Wang X, Xu W, Farzaneh F, Xu R. The structure and pharmacological functions of coumarins and their derivatives. Curr Med Chem. 2009; 16:4236–4260. [DOI] [PubMed] [Google Scholar]
- 26.Hwu JR, Singha R, Hong SC, Chang YH, Das AR, Vliegen I, et al. Synthesis of new benzimidazole-coumarin conjugates as anti-hepatitis C virus agents. Antiviral Res. 2008; 77:157–162. doi: 10.1016/j.antiviral.2007.09.003 [DOI] [PubMed] [Google Scholar]
- 27.Donnelly AC, Mays JR, Burlison JA, Nelson JT, Vielhauer G, Holzbeierlein J, et al. The design, synthesis, and evaluation of coumarin ring derivatives of the novobiocin scaffold that exhibit antiproliferative activity. J Org Chem. 2008; 73:8901–8920. doi: 10.1021/jo801312r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piazzi L, Cavalli A, Colizzi F, Belluti F, Bartolini M, Mancini F, et al. Multi-target-directed coumarin derivatives: hAChE and BACE1 inhibitors as potential anti-Alzheimer compounds. Bioorg Med Chem Lett. 2008; 18:423–426. doi: 10.1016/j.bmcl.2007.09.100 [DOI] [PubMed] [Google Scholar]
- 29.Kashman Y, Gustafson KR, Fuller RW, Cardellina JH, McMahon JB, Currens MJ, et al. The calanolides, a novel HIV-inhibitory class of coumarin derivatives from the tropical rainforest tree, Calophyllum lanigerum. J Med Chem. 1992; 35:2735–2743. [DOI] [PubMed] [Google Scholar]
- 30.Mohareb RM, MegallyAbdo NY. Uses of 3-(2-Bromoacetyl)-2H-chromen-2-one in the Synthesis of Heterocyclic Compounds Incorporating Coumarin: Synthesis, Characterization and Cytotoxicity. Molecules 2015; 20:11535–11553. doi: 10.3390/molecules200611535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacy A, O'Kennedy R. Studies on coumarins and coumarin-relatedcompounds to determine their therapeuticrole in the treatment of cancer. Curr Pharm Des. 2004; 10:3797–3811. [DOI] [PubMed] [Google Scholar]
- 32.Salem MA, Marzouk MI, El-Kazak AM. Synthesis and Characterization of Some New Coumarins with in Vitro Antitumor and Antioxidant Activity and High Protective Effects against DNA Damage. Molecules. 2016; 21:249 doi: 10.3390/molecules21020249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanway SJ, Purohit A, Woo LW, Sufi S, Vigushin D, Ward R, et al. Phase I study of STX 64 (667 Coumate) in breast cancer patients: the first study of a steroid sulfatase inhibitor. Clin Cancer Res. 2006; 12:1585–1592. doi: 10.1158/1078-0432.CCR-05-1996 [DOI] [PubMed] [Google Scholar]
- 34.Mohareb RM, Megally Abdo NY. Synthesis and Cytotoxic Evaluation of Pyran, Dihydropyridine and Thiophene Derivatives of 3-Acetylcoumarin. Chem Pharm Bull (Tokyo). 2015; 63:678–687. [DOI] [PubMed] [Google Scholar]
- 35.Elshemy HA, Zaki MA. Design and synthesis of new coumarin hybrids and insight into their mode of antiproliferative action. Bioorg Med Chem. 2017; 25:1066–1075. doi: 10.1016/j.bmc.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 36.Jashari A, Imeri F, Ballazhi L, Shabani A, Mikhova B, Dräger G, et al. Synthesis and cellular characterization of novel isoxazolo- and thiazolohydrazinylidene-chroman-2,4-diones on cancer and non-cancer cell growth and death. Bioorg Med Chem. 2014; 22:2655–2661. doi: 10.1016/j.bmc.2014.03.026 [DOI] [PubMed] [Google Scholar]
- 37.Hafez OM, Nassar MI, El-Kousy SM, Abdel-Razik AF, Sherien MM, El-Ghonemy MM. Synthesis of some new carbonitriles and pyrazolecoumarin derivatives with potent antitumor and antimicrobial activities. Acta Pol Pharm. 2014; 71:594–601. [PubMed] [Google Scholar]
- 38.Ibrahim MM, Ramadan AM, Mersal GAM, El-Shazly SA. Synthesis, superoxide dismutase, nuclease, and anticancer activities of copper(II) complexes incorporating bis(2-picolyl)amine with different counter anions. J Mol Struct. 2011; 998:1–10. [Google Scholar]
- 39.Subasinghe A, Perera IC, Pakhomova S, Perera T. Synthesis, Characterization, and Biological Studies of a Piperidinyl Appended Dipicolylamine Ligand and Its Rhenium Tricarbonyl Complex as Potential Therapeutic Agents for Human Breast Cancer. Bioinorg Chem Appl. 2016; 2016:2675937 doi: 10.1155/2016/2675937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonioli B, Büchner B, Clegg JK, Gloe K, Gloe K, Götzke L, et al. Interaction of an extended series of N-substituted di(2-picolyl)amine derivatives with copper(II). Synthetic, structural, magnetic and solution studies. Dalton Trans. 2009; 24:4795–4805. [DOI] [PubMed] [Google Scholar]
- 41.Kumar CV, Asuncion EH. DNA binding studies and site selective fluorescence sensitization of an anthryl probe. J Am Chem Soc. 1993; 115:8547–8553. [Google Scholar]
- 42.Omidvar Z, Asoodeh A, Chamani J. Studies on the Antagonistic Behavior Between Cyclophosphamide Hydrochloride and Aspirin with Human Serum Albumin: Time-Resolved Fluorescence Spectroscopy and Isothermal Titration Calorimetry. J Solution Chem. 2013; 42:1005–1017. [Google Scholar]
- 43.Moosavi-Movahedi AA, Golchin AR, Nazari K, Chamani J, Saboury AA, Bathaie SZ, et al. Microcalorimetry, energetics and binding studies of DNA–dimethyltin dichloride complexes. Thermochim Acta. 2004; 414:233–241. [Google Scholar]
- 44.Chamani J, Moosavi-Movahedi AA, Saboury AA, Gharanfoli M, Hakimelahi GH. Calorimetric indication of the molten globule-like state of cytochrome c induced by n-alkyl sulfates at low concentrations. J Chem Thermodynamics. 2003; 35:199–207. [Google Scholar]
- 45.Stern O, Volmer M. The extinction period of fluorescence. Phys Z. 1919; 20:183–193. [Google Scholar]
- 46.Lackowic Z J. Principle of Fluorescence Spectroscopy, 2006. Springer Science and Business Media, LLC, New York ISBN. 13:978–0 [Google Scholar]
- 47.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009; 30:2785–2791. doi: 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998; 19:1639–1662. [Google Scholar]
- 49.Peng C, Ayali PW, Schlegel HB, Frisch MJ. Using redundant internal coordinates to optimize equilibrium geometries and transition states. J Comput Chem. 1996; 17:49–56. [Google Scholar]
- 50.DeLano WL. The pymol molecular graphic system. DeLano Scientific, Palo Alto, 1997. [Google Scholar]
- 51.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001; 46:3–26. [DOI] [PubMed] [Google Scholar]
- 52.Karthick V, Ramanathan K, Shanthi V, Rajasekaran R. Identification of potential inhibitors of H5N1 influenza A virus neuraminidase by ligand-based virtual screening approach. Cell Biochem Biophys. 2013; 66:657–669. doi: 10.1007/s12013-012-9510-7 [DOI] [PubMed] [Google Scholar]
- 53.Southan C, Stracz A. Extracting and connecting chemical structures from text sources using chemicalize.org. J Cheminform. 2013; 5:20 doi: 10.1186/1758-2946-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chemicals dofo. OECD Guideline for testing of chemicals. guidelines for the testing of chemicals and related documents. http://www.oecd.org/chemicalsafety/testing/oecdguidelinesforthetestingofchemicals.htm; Accessed: 2012 Nov 13 (2005).
- 55.Nakayama T, Kimura T, Kodama M, Nagata C. Generation of hydrogen peroxide and superoxide anion from active metabolites of naphthylamines and aminoazo dyes: its possible role in carcinogenesis. Carcinogenesis. 1983; 4:765–769. [DOI] [PubMed] [Google Scholar]
- 56.Quinlan GJ, Gutteridge JM. Oxygen radical damage to DNA by rifamycin SV and copper ions. Biochem Pharmacol. 1987; 36:3629–3633. [DOI] [PubMed] [Google Scholar]
- 57.Shahabadi N, Maghsudi M. Multi-spectroscopic and molecular modeling studies on the interaction of antihypertensive drug; methyldopa with calf thymus DNA. Mol Biosyst. 2014; 10:338–347. doi: 10.1039/c3mb70340a [DOI] [PubMed] [Google Scholar]
- 58.Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK. Mixed Ligand Complexes of Ruthenium(II): Factors Governing Binding to DNA. J Am Chem Soc. 1989; 111:3051–3058. [Google Scholar]
- 59.Zaki M, Afzal M, Ahmad M, Tabassum S. Synthesis and crystal structure elucidation of new copper(II)-based chemotherapeutic agent coupled with 1,2-DACH and orthovanillin: Validated by in vitro DNA/HSA binding profile and pBR322 cleavage pathway. J Photochem Photobiol B. 2016; 161:318–327. doi: 10.1016/j.jphotobiol.2016.05.030 [DOI] [PubMed] [Google Scholar]
- 60.Chen QY, Li DH, Zhao Y, Yang HH, Zhu QZ, Xu JG. Interaction of a novel red-region fluorescent probe, Nile blue, with DNA and its application to nucleic acids assay. Analyst. 1999; 124:901–906. [DOI] [PubMed] [Google Scholar]
- 61.Kashanian S, Dolatabadi JEN. In vitro studies on calf thymus DNA interaction and 2-tert-butyl-4-methylphenol food additive. Eur Food Res Technol. 2010; 230:821–825. [Google Scholar]
- 62.Moghaddam MM, Pirouzi M, Saberi MR, Chamani J. Comparison of the binding behavior of FCCP with HSA and HTF as determined by spectroscopic and molecular modeling techniques. Luminescence. 2014; 29:314–331. doi: 10.1002/bio.2546 [DOI] [PubMed] [Google Scholar]
- 63.Omidvar Z, Parivar K, Sanee H, Amiri-Tehranizadeh Z, Baratian A, Saberi MR, et al. Investigations with Spectroscopy, Zeta Potential and Molecular Modeling of the Non-Cooperative Behaviour Between Cyclophosphamide Hydrochloride and Aspirin upon Interaction with Human Serum Albumin: Binary and Ternary Systems from the View Point of Multi-Drug Therapy. J Biomol Struct Dyn. 2011; 29:181–206. doi: 10.1080/07391102.2011.10507382 [DOI] [PubMed] [Google Scholar]
- 64.Song Y, Kang J, Zhou J, Wang Z, Lu X, Wang L, et al. Study on the fluorescence spectra and electrochemical behavior of ZnL2 and Morin with DNA. Spectrochim Acta A Mol Biomol Spectrosc. 2000; 56A:2491–2497. [DOI] [PubMed] [Google Scholar]
- 65.Liu HK, Sadler PJ. Metal complexes as DNA intercalators. Acc Chem Res. 2011; 44:349–359. doi: 10.1021/ar100140e [DOI] [PubMed] [Google Scholar]
- 66.Kakkar R, Garg R, Suruchi. Theoretical study of tautomeric structures and fluorescence spectra of Hoechst 33258. J Mol Struct: THEOCHEM. 2002; 579:109–113. [Google Scholar]
- 67.Guan Y, Zhou W, Yao X, Zhao M, Li Y. Determination of nucleic acids based on the fluorescence quenching of Hoechst 33258 at pH 4.5. Anal Chim Acta. 2006; 570:21–28. [Google Scholar]
- 68.Haq I. Thermodynamics of drug-DNA interactions. Arch Biochem Biophys. 2002; 403:1–15. doi: 10.1016/S0003-9861(02)00202-3 [DOI] [PubMed] [Google Scholar]
- 69.Mati SS, Roy SS, Chall S, Bhattacharya S, Bhattacharya SC. Unveiling the groove binding mechanism of a biocompatible naphthalimide-based organoselenocyanate with calf thymus DNA: an "ex vivo" fluorescence imaging application appended by biophysical experiments and molecular docking simulations. J Phys Chem B. 2013; 117:14655–14665. doi: 10.1021/jp4090553 [DOI] [PubMed] [Google Scholar]
- 70.Shamsi A, Ahmed A, Bano B. Probing the interaction of anticancer drug temsirolimus with human serum albumin: Molecular docking and spectroscopic insight. J Biomol Struct Dyn. 2017; 18:1–11. [DOI] [PubMed] [Google Scholar]
- 71.Rehman MT, Shamsi H, Khan AU. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol Pharm. 2014; 11:1785–1797. doi: 10.1021/mp500116c [DOI] [PubMed] [Google Scholar]
- 72.Holm AI, Nielsen LM, Hoffmann SV, Nielsen SB. Vacuum-ultraviolet circular dichroism spectroscopy of DNA: a valuable tool to elucidate topology and electronic coupling in DNA. Phys Chem Chem Phys. 2010; 12:9581–9596. doi: 10.1039/c003446k [DOI] [PubMed] [Google Scholar]
- 73.Bonincontro A, Falivene M, Mesa CL, Risuleo G, Pena MR. Dynamics of DNA adsorption on and release from SDS-DDAB cat-anionic vesicles: a multitechnique study. Langmuir. 2008; 24:1973–1978. doi: 10.1021/la701730h [DOI] [PubMed] [Google Scholar]
- 74.Chanphai P, Agudelo D, Vesper AR, Bérubé G, Tajmir-Riahi HA. Effect of testosterone and its aliphatic and aromatic dimers on DNA morphology. Int J Biol Macromol. 2017; 95:850–855. doi: 10.1016/j.ijbiomac.2016.09.090 [DOI] [PubMed] [Google Scholar]
- 75.Agudelo D, Bourassa P, Bérubé G, Tajmir-Riahi HA. Review on the binding of anticancer drug doxorubicin with DNA and tRNA: Structural models and antitumor activity. J Photochem Photobiol B. 2016; 158:274–279. doi: 10.1016/j.jphotobiol.2016.02.032 [DOI] [PubMed] [Google Scholar]
- 76.Uma Maheswari P, Palaniandavar M. DNA binding and cleavage properties of certain tetrammine ruthenium(II) complexes of modified 1,10-phenanthrolines—effect of hydrogen-bonding on DNA-binding affinity. J Inorg Biochem. 2004; 98:219–230. [DOI] [PubMed] [Google Scholar]
- 77.Li Y, Zhang G, Pan J, Zhang Y. Determination of metolcarb binding to DNA by spectroscopic and chemometrics methods with the use of acridine orange as a probe. Sensors and Actuators B: Chemical. 2014; 191:464–472. [Google Scholar]
- 78.Ivanov IV, Minchenkova LE, Schyolkina AK, Poletayev AI. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers.1973; 12:89–110. doi: 10.1002/bip.1973.360120109 [DOI] [PubMed] [Google Scholar]
- 79.Jain SS, Polak M, Hud NV. Controlling nucleic acid secondary structure by intercalation: effects of DNA strand length on coralyne-driven duplex disproportionation. Nucleic Acids Res. 2003; 31:4608–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mergny JL, Duval-Valentin G, Nguyen CH, Perrouault L, Faucon B, Rougée M, et al. Triple helix-specific ligands. Science. 1992; 256:1681–1684. [DOI] [PubMed] [Google Scholar]
- 81.Moosavi-Movahedi AA, Chamani J, Ghourchian H, Shafiey H, Sorenson CM, Sheibani N. Electrochemical evidence for the molten globule states of cytochrome c induced by N-alkyl sulfates at low concentrations. J Protein Chem. 2003; 22:23–30. [DOI] [PubMed] [Google Scholar]
- 82.Lerman LS. Structural considerations in the interaction of DNA and acridines. J Mol Biol. 1961; 3:18–30. [DOI] [PubMed] [Google Scholar]
- 83.Siddiqi ZA, Sharma PK, Shahid M, Khalid M, Anjuli, Siddique A, Kumar S. Superoxide scavenging and antimicrobial activities of novel transition metal complexes of oxydiacetate dianion as primary ligand: spectral characterization, cyclic voltammetric investigations and crystal structure. Eur J Med Chem. 2012; 57:102–111. doi: 10.1016/j.ejmech.2012.08.043 [DOI] [PubMed] [Google Scholar]
- 84.Addison AW, Hendriks HMJ, Reedijk J, Thompson LK. Copper complexes of the "tripod" ligand tris(2-benzimidazolylmethyl)amine: five-and six-coordinate copper(II) derivatives and some copper(I) derivatives. Inorganic Chemistry. 1981; 20:103–110. [Google Scholar]
- 85.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009; 8:579–591. doi: 10.1038/nrd2803 [DOI] [PubMed] [Google Scholar]
- 86.Zowczak M, Iskra M, Torliński L, Cofta S. Analysis of serum copper and zinc concentrations in cancer patients. Biol Trace Elem Res. 2001; 82:1–8. doi: 10.1385/BTER:82:1-3:001 [DOI] [PubMed] [Google Scholar]
- 87.Lowndes SA, Harris AL. The role of copper in tumour angiogenesis. J Mammary Gland Biol Neoplasia. 2005; 10:299–310. doi: 10.1007/s10911-006-9003-7 [DOI] [PubMed] [Google Scholar]
- 88.Peng F, Lu X, Janisse J, Muzik O, Shields AF. PET of human prostate cancer xenografts in mice with increased uptake of 64CuCl2. J Nucl Med. 2006; 47:1649–1652. [PubMed] [Google Scholar]
- 89.Acilan C, Cevatemre B, Adiguzel Z, Karakas D, Ulukaya E, Ribeiro N, et al. Synthesis, biological characterization and evaluation of molecular mechanisms of novel copper complexes as anticancer agents. Biochim Biophys Acta. 2017; 1861:218–234. doi: 10.1016/j.bbagen.2016.10.014 [DOI] [PubMed] [Google Scholar]
- 90.Santini C, Pellei M, Gandin V, Porchia M, Tisato F, Marzano C. Advances in copper complexes as anticancer agents. Chem Rev. 2014; 114:815–862. doi: 10.1021/cr400135x [DOI] [PubMed] [Google Scholar]
- 91.Trejo-Solís C, Palencia G, Zúñiga S, Rodríguez-Ropon A, Osorio-Rico L, Luvia ST, et al. Cas IIgly induces apoptosis in glioma C6 cells in vitro and in vivo through caspase-dependent and caspase-independent mechanisms. Neoplasia. 2005; 7:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haris P, Mary V, Aparna P, Dileep KV, Sudarsanakumar C. A comprehensive approach to ascertain the binding mode of curcumin with DNA. Spectrochim Acta A Mol Biomol Spectrosc. 2017; 175:155–163. doi: 10.1016/j.saa.2016.11.049 [DOI] [PubMed] [Google Scholar]
- 93.Li ZX, Ouyang KQ, Jiang X, Wang D, Hu Y. Curcumin induces apoptosis and inhibits growth of human Burkitt's lymphoma in xenograft mouse model. Mol Cells. 2009; 27:283–289. doi: 10.1007/s10059-009-0036-9 [DOI] [PubMed] [Google Scholar]
- 94.Lee HP, Li TM, Tsao JY, Fong YC, Tang CH. Curcumin induces cell apoptosis in human chondrosarcoma through extrinsic death receptor pathway. Int Immunopharmacol. 2012; 13:163–169. doi: 10.1016/j.intimp.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 95.Shakir M, Azam M, Ullah MF, Hadi SM. Synthesis, spectroscopic and electrochemical studies of N,N-bis[(E)-2-thienylmethylidene]-1,8-naphthalenediamine and its Cu(II) complex: DNA cleavage and generation of superoxide anion. J Photochem Photobiol B. 2011; 104:449–456. doi: 10.1016/j.jphotobiol.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 96.Zafar A, Singh S, Naseem I. Cytotoxic activity of soy phytoestrogen coumestrol against human breast cancer MCF-7 cells: Insights into the molecular mechanism. Food Chem Toxicol. 2017; 99:149–161. doi: 10.1016/j.fct.2016.11.034 [DOI] [PubMed] [Google Scholar]
- 97.Rahman A, Shahabuddin, Hadi SM, Parish JH. Complexes involving quercetin, DNA and Cu(II). Carcinogenesis.1990; 11:2001–2003. [DOI] [PubMed] [Google Scholar]
- 98.Bhat R, Hadi SM. DNA breakage by tannic acid and Cu(II): sequence specificity of the reaction and involvement of active oxygen species. Mutat Res. 1994; 313:39–48. [DOI] [PubMed] [Google Scholar]
- 99.Ahsan H, Hadi SM. Strand scission in DNA induced by curcumin in the presence of Cu(II). Cancer Lett. 1998; 124:23–30. [DOI] [PubMed] [Google Scholar]
- 100.Nogueira V, Hay N. Molecular pathways: reactive oxygen species homeostasis in cancer cells and implications for cancer therapy. Clin Cancer Res. 2013; 19:4309–4314. doi: 10.1158/1078-0432.CCR-12-1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell 2006; 10:175–176. doi: 10.1016/j.ccr.2006.08.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Treatment of plasmid pBR322 DNA with increasing concentrations of ligand-L alone (25–100 μM) (Lanes 1–3) and Cu(II) ions alone (25–100 μM) (Lanes 4–6). Lane C represents untreated (control) plasmid. Ligand-L and Cu(II) treatment alone were ineffective in plasmid DNA cleavage.
(PDF)
(PDF)
(DOCX)
Virtual screening of ligand-L showing drug-likeliness by (A) Molinspiration (B) chemicalize.org servers.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.