Abstract
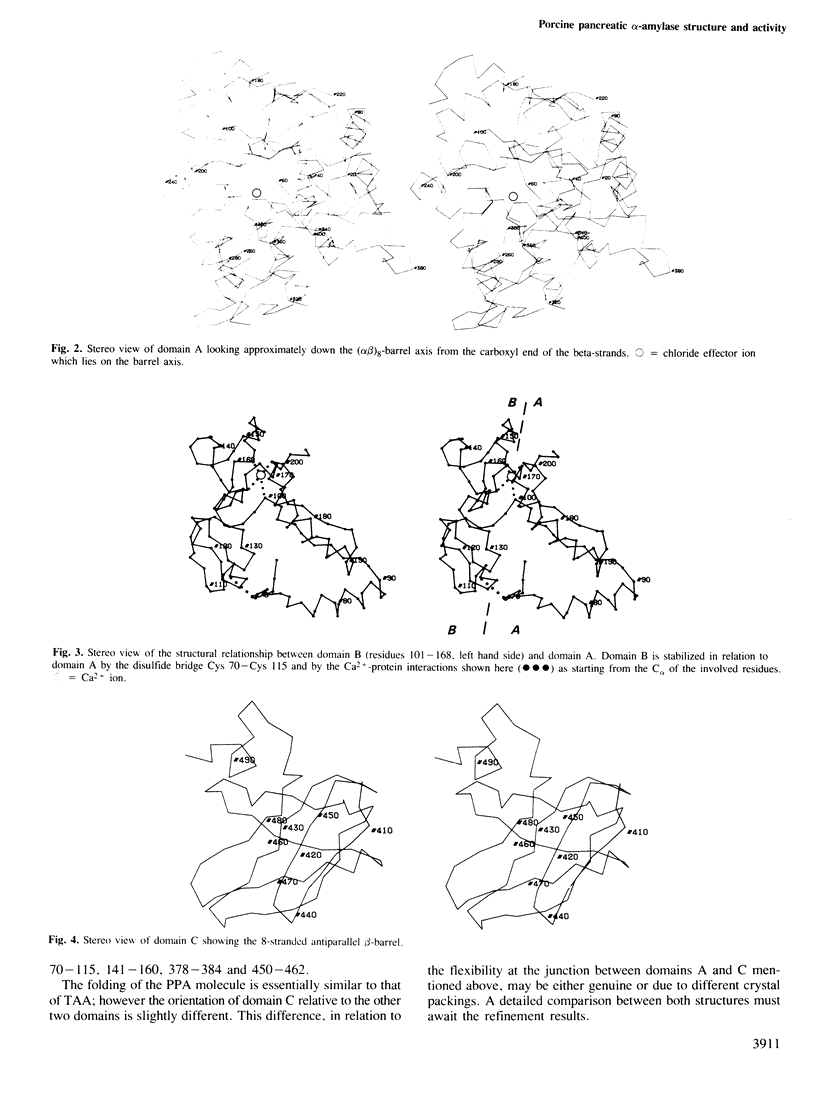
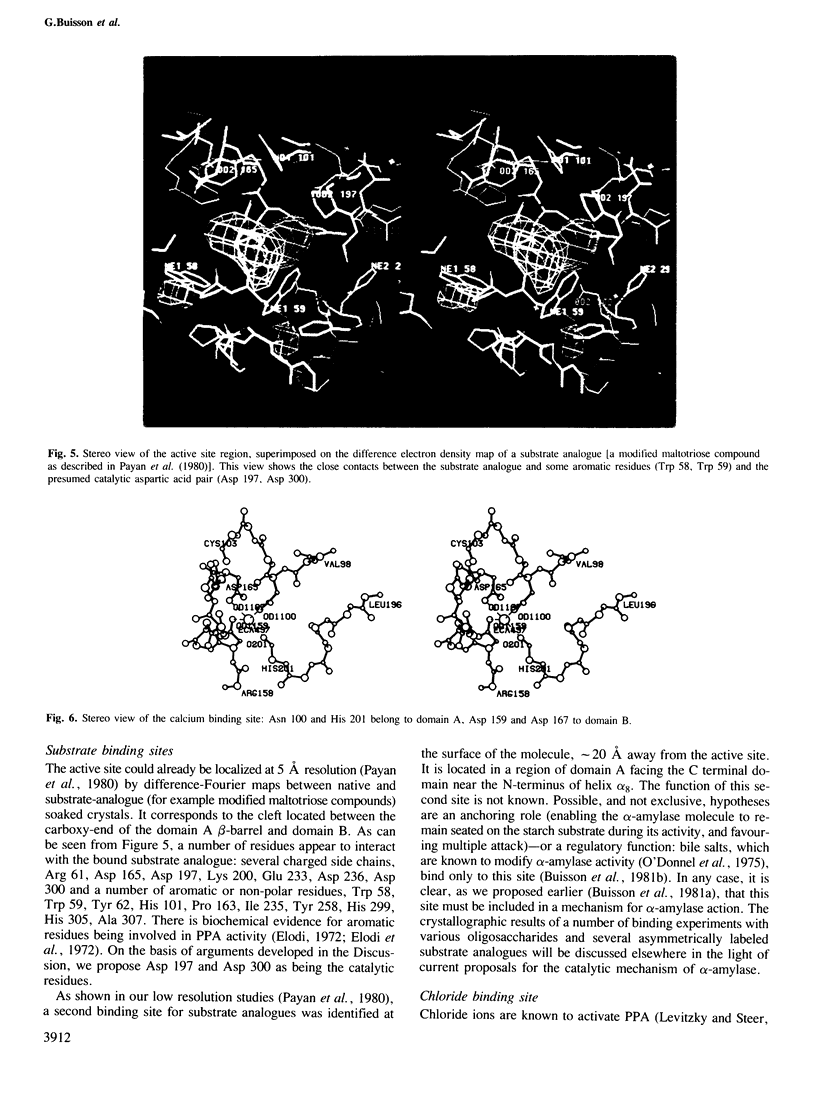
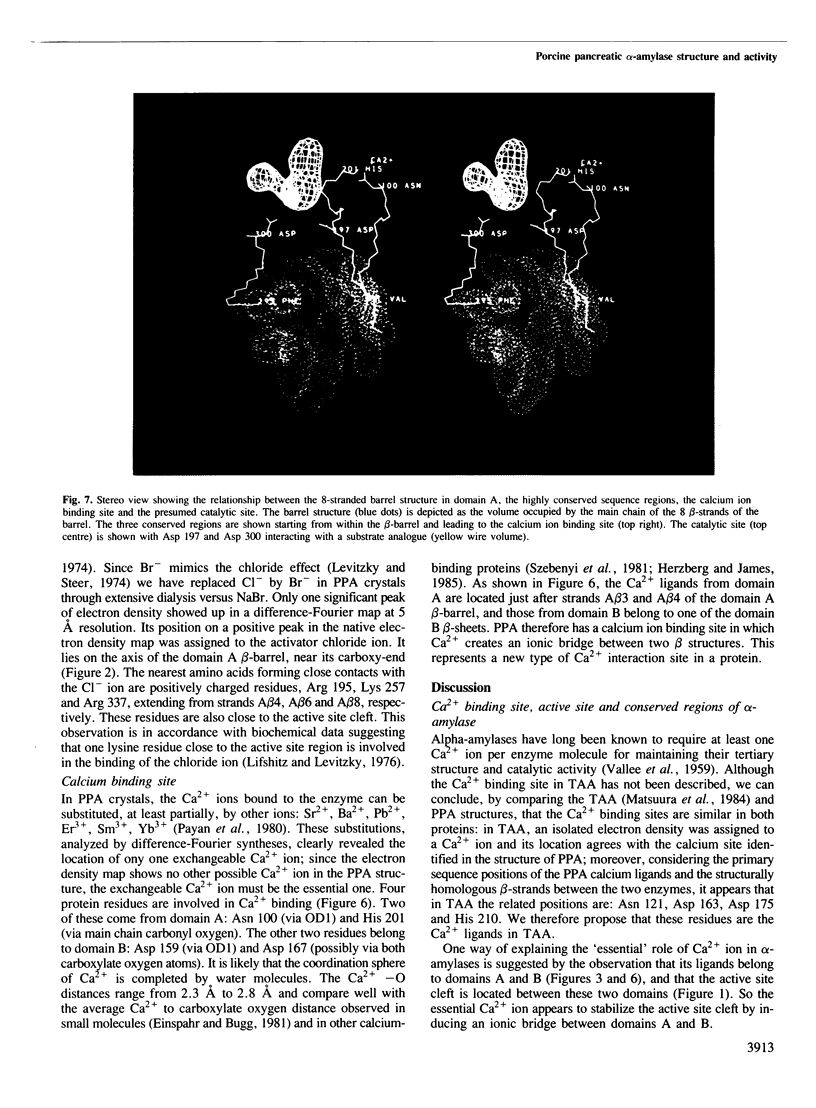
The crystal structure of porcine pancreatic alpha-amylase (PPA) has been solved at 2.9 A resolution by X-ray crystallographic methods. The enzyme contains three domains. The larger, in the N-terminal part, consists of 330 amino acid residues. This central domain has the typical parallel-stranded alpha-beta barrel structure (alpha beta)8, already found in a number of other enzymes like triose phosphate isomerase and pyruvate kinase. The C-terminal domain forms a distinct globular unit where the chain folds into an eight-stranded antiparallel beta-barrel. The third domain lies between a beta-strand and a alpha-helix of the central domain, in a position similar to those found for domain B in triose phosphate isomerase and pyruvate kinase. It is essentially composed of antiparallel beta-sheets. The active site is located in a cleft within the N-terminal central domain, at the carboxy-end of the beta-strands of the (alpha beta)8 barrel. Binding of various substrate analogues to the enzyme suggests that the amino acid residues involved in the catalytic reaction are a pair of aspartic acids. A number of other residues surround the substrate and seem to participate in its binding via hydrogen bonds and hydrophobic interactions. The 'essential' calcium ion has been located near the active site region and between two domains, each of them providing two calcium ligands. On the basis of sequence comparisons this calcium binding site is suggested to be a common structural feature of all alpha-amylases. It represents a new type of calcium-protein interaction pattern.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banks W., Greenwood C. T. Mathematical models for the action of alpha-amylase on amylose. Carbohydr Res. 1977 Aug;57:301–315. doi: 10.1016/s0008-6215(00)81939-4. [DOI] [PubMed] [Google Scholar]
- Bause E., Hettkamp H., Legler G. Conformational aspects of N-glycosylation of proteins. Studies with linear and cyclic peptides as probes. Biochem J. 1982 Jun 1;203(3):761–768. doi: 10.1042/bj2030761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaupoil-Abadie B., Raffalli M., Cozzone P., Marchis-Mouren G. Determination of the carbohydrate content of porcine pancreatic amylase. Biochim Biophys Acta. 1973 Feb 28;297(2):436–440. doi: 10.1016/0304-4165(73)90090-1. [DOI] [PubMed] [Google Scholar]
- Blake C. C. Exons and the evolution of proteins. Int Rev Cytol. 1985;93:149–185. doi: 10.1016/s0074-7696(08)61374-1. [DOI] [PubMed] [Google Scholar]
- Blundell T., Lindley P., Miller L., Moss D., Slingsby C., Tickle I., Turnell B., Wistow G. The molecular structure and stability of the eye lens: x-ray analysis of gamma-crystallin II. Nature. 1981 Feb 26;289(5800):771–777. doi: 10.1038/289771a0. [DOI] [PubMed] [Google Scholar]
- Carrell H. L., Rubin B. H., Hurley T. J., Glusker J. P. X-ray crystal structure of D-xylose isomerase at 4-A resolution. J Biol Chem. 1984 Mar 10;259(5):3230–3236. [PubMed] [Google Scholar]
- Chan Y., Braun P. J., French D., Robyt J. F. Porcine pancreatic alpha-amylase hydrolysis of hydroxyethylated amylose and specificity of subsite binding. Biochemistry. 1984 Nov 20;23(24):5795–5800. doi: 10.1021/bi00319a019. [DOI] [PubMed] [Google Scholar]
- Elödi P., Móra S., Krysteva M. Investigation of the active center of porcine-pancreatic amylase. Eur J Biochem. 1972 Jan 21;24(3):577–582. doi: 10.1111/j.1432-1033.1972.tb19720.x. [DOI] [PubMed] [Google Scholar]
- Elödi P. Role of histidyl residues in the activity of porcine pancreatic amylase. Acta Biochim Biophys Acad Sci Hung. 1972;7(3):241–245. [PubMed] [Google Scholar]
- Glick Z., Bray G. A. Effects of acarbose on food intake, body weight and fat depots in lean and obese rats. Pharmacol Biochem Behav. 1983 Jul;19(1):71–78. doi: 10.1016/0091-3057(83)90314-3. [DOI] [PubMed] [Google Scholar]
- Goldman A., Ollis D. L., Steitz T. A. Crystal structure of muconate lactonizing enzyme at 3 A resolution. J Mol Biol. 1987 Mar 5;194(1):143–153. doi: 10.1016/0022-2836(87)90723-6. [DOI] [PubMed] [Google Scholar]
- Granger M., Abadie B., Mazzei Y., Marchis-Mouren G. Enzymatic activity of TNB blocked porcine pancreatic amylase. FEBS Lett. 1975 Feb 1;50(2):276–278. doi: 10.1016/0014-5793(75)80507-2. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Bovey R., Young R. A. Tissue-specific expression of mouse-alpha-amylase genes: nucleotide sequence of isoenzyme mRNAs from pancreas and salivary gland. Cell. 1980 Aug;21(1):179–187. doi: 10.1016/0092-8674(80)90125-7. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Herzberg O., James M. N. Common structural framework of the two Ca2+/Mg2+ binding loops of troponin C and other Ca2+ binding proteins. Biochemistry. 1985 Sep 24;24(20):5298–5302. doi: 10.1021/bi00341a004. [DOI] [PubMed] [Google Scholar]
- Ihara H., Sasaki T., Tsuboi A., Yamagata H., Tsukagoshi N., Udaka S. Complete nucleotide sequence of a thermophilic alpha-amylase gene: homology between prokaryotic and eukaryotic alpha-amylases at the active sites. J Biochem. 1985 Jul;98(1):95–103. doi: 10.1093/oxfordjournals.jbchem.a135279. [DOI] [PubMed] [Google Scholar]
- Kluh I. Amino acid sequence of hog pancreatic alpha-amylase isoenzyme I. FEBS Lett. 1981 Dec 28;136(2):231–234. doi: 10.1016/0014-5793(81)80624-2. [DOI] [PubMed] [Google Scholar]
- Lebioda L., Hatada M. H., Tulinsky A., Mavridis I. M. Comparison of the folding of 2-keto-3-deoxy-6-phosphogluconate aldolase, triosephosphate isomerase and pyruvate kinase. Implications in molecular evolution. J Mol Biol. 1982 Dec 5;162(2):445–458. doi: 10.1016/0022-2836(82)90537-x. [DOI] [PubMed] [Google Scholar]
- Levitzki A., Steer M. L. The allosteric activation of mammalian alpha-amylase by chloride. Eur J Biochem. 1974 Jan 3;41(1):171–180. doi: 10.1111/j.1432-1033.1974.tb03257.x. [DOI] [PubMed] [Google Scholar]
- Lifshitz R., Levitzki A. Identity and properties of the chloride effector binding site in hog pancreatic alpha-amylase. Biochemistry. 1976 May 4;15(9):1987–1993. doi: 10.1021/bi00654a028. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y., Brändén C. I. Structure of glycolate oxidase from spinach. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6855–6859. doi: 10.1073/pnas.82.20.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- Mavridis I. M., Hatada M. H., Tulinsky A., Lebioda L. Structure of 2-keto-3-deoxy-6-phosphogluconate aldolase at 2 . 8 A resolution. J Mol Biol. 1982 Dec 5;162(2):419–444. doi: 10.1016/0022-2836(82)90536-8. [DOI] [PubMed] [Google Scholar]
- McKnight G. L., O'Hara P. J., Parker M. L. Nucleotide sequence of the triosephosphate isomerase gene from Aspergillus nidulans: implications for a differential loss of introns. Cell. 1986 Jul 4;46(1):143–147. doi: 10.1016/0092-8674(86)90868-8. [DOI] [PubMed] [Google Scholar]
- Nakajima R., Imanaka T., Aiba S. Nucleotide sequence of the Bacillus stearothermophilus alpha-amylase gene. J Bacteriol. 1985 Jul;163(1):401–406. doi: 10.1128/jb.163.1.401-406.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishide T., Nakamura Y., Emi M., Yamamoto T., Ogawa M., Mori T., Matsubara K. Primary structure of human salivary alpha-amylase gene. Gene. 1986;41(2-3):299–304. doi: 10.1016/0378-1119(86)90110-1. [DOI] [PubMed] [Google Scholar]
- O'Donnell M. D., McGeeney K. F., FitzGerald O. Effect of free and conjugated bile salts on alpha-amylase activity. Enzyme. 1975;19(3):129–139. doi: 10.1159/000458984. [DOI] [PubMed] [Google Scholar]
- Pasero L., Mazzéi-Pierron Y., Abadie B., Chicheportiche Y., Marchis-Mouren G. Complete amino acid sequence and location of the five disulfide bridges in porcine pancreatic alpha-amylase. Biochim Biophys Acta. 1986 Jan 30;869(2):147–157. doi: 10.1016/0167-4838(86)90289-x. [DOI] [PubMed] [Google Scholar]
- Phillips D. C., Sternberg M. J., Thornton J. M., Wilson I. A. An analysis of the structure of triose phosphate isomerase and its comparison with lactate dehydrogenase. J Mol Biol. 1978 Feb 25;119(2):329–351. doi: 10.1016/0022-2836(78)90440-0. [DOI] [PubMed] [Google Scholar]
- Pierrot M., Astier J. P., Abadie B., Marchis-Mouren G. Preliminary x-ray investigation on a new crystalline variety of porcine pancreatic alpha-amylase. FEBS Lett. 1977 Jul 1;79(1):105–108. doi: 10.1016/0014-5793(77)80360-8. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Robyt J. F., French D. The action pattern of porcine pancreatic alpha-amylase in relationship to the substrate binding site of the enzyme. J Biol Chem. 1970 Aug 10;245(15):3917–3927. [PubMed] [Google Scholar]
- Rogers J. C. Conserved amino acid sequence domains in alpha-amylases from plants, mammals, and bacteria. Biochem Biophys Res Commun. 1985 Apr 16;128(1):470–476. doi: 10.1016/0006-291x(85)91702-4. [DOI] [PubMed] [Google Scholar]
- Schibler U., Pittet A. C., Young R. A., Hagenbüchle O., Tosi M., Gellman S., Wellauer P. K. The mouse alpha-amylase multigene family. Sequence organization of members expressed in the pancreas, salivary gland and liver. J Mol Biol. 1982 Mar 5;155(3):247–266. doi: 10.1016/0022-2836(82)90004-3. [DOI] [PubMed] [Google Scholar]
- Schneider G., Lindqvist Y., Brändén C. I., Lorimer G. Three-dimensional structure of ribulose-1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum at 2.9 A resolution. EMBO J. 1986 Dec 20;5(13):3409–3415. doi: 10.1002/j.1460-2075.1986.tb04662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigner C., Prodanov E., Marchis-Mouren G. On porcine pancreatic alpha-amylase action: kinetic evidence for the binding of two maltooligosaccharide molecules (maltose, maltotriose and o-nitrophenylmaltoside) by inhibition studies. Correlation with the five-subsite energy profile. Eur J Biochem. 1985 Apr 1;148(1):161–168. doi: 10.1111/j.1432-1033.1985.tb08820.x. [DOI] [PubMed] [Google Scholar]
- Steer M. L., Levitzki A. The metal specificity of mammalian -amylase as revealed by enzyme activity and structural probes. FEBS Lett. 1973 Apr 1;31(1):89–92. doi: 10.1016/0014-5793(73)80079-1. [DOI] [PubMed] [Google Scholar]
- Straus D., Gilbert W. Genetic engineering in the Precambrian: structure of the chicken triosephosphate isomerase gene. Mol Cell Biol. 1985 Dec;5(12):3497–3506. doi: 10.1128/mcb.5.12.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D. I., Levine M., Muirhead H., Stammers D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979 Oct 15;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Szebenyi D. M., Obendorf S. K., Moffat K. Structure of vitamin D-dependent calcium-binding protein from bovine intestine. Nature. 1981 Nov 26;294(5839):327–332. doi: 10.1038/294327a0. [DOI] [PubMed] [Google Scholar]
- Takkinen K., Pettersson R. F., Kalkkinen N., Palva I., Söderlund H., Käriäinen L. Amino acid sequence of alpha-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J Biol Chem. 1983 Jan 25;258(2):1007–1013. [PubMed] [Google Scholar]
- Tappy L., Buckert A., Griessen M., Golay A., Jéquier E., Felber J. Effect of trestatin, a new inhibitor of pancreatic alpha-amylase, on starch metabolism in man. Int J Obes. 1986;10(3):185–192. [PubMed] [Google Scholar]
- Thoma J. A. Models for depolymerizing enzymes. Application to alpha-amylases. Biopolymers. 1976 Apr;15(4):729–746. doi: 10.1002/bip.1976.360150411. [DOI] [PubMed] [Google Scholar]
- Thoma J. A. Models for depolymerizing enzymes: criteria for discrimination of models. Carbohydr Res. 1976 May;48(1):85–103. doi: 10.1016/s0008-6215(00)83517-x. [DOI] [PubMed] [Google Scholar]
- VALLEE B. L., STEIN E. A., SUMERWELL W. N., FISCHER E. H. Metal content of alpha-amylases of various origins. J Biol Chem. 1959 Nov;234:2901–2905. [PubMed] [Google Scholar]
- Yamazaki H., Ohmura K., Nakayama A., Takeichi Y., Otozai K., Yamasaki M., Tamura G., Yamane K. Alpha-amylase genes (amyR2 and amyE+) from an alpha-amylase-hyperproducing Bacillus subtilis strain: molecular cloning and nucleotide sequences. J Bacteriol. 1983 Oct;156(1):327–337. doi: 10.1128/jb.156.1.327-337.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Galizzi A., Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. doi: 10.1093/nar/11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuuki T., Nomura T., Tezuka H., Tsuboi A., Yamagata H., Tsukagoshi N., Udaka S. Complete nucleotide sequence of a gene coding for heat- and pH-stable alpha-amylase of Bacillus licheniformis: comparison of the amino acid sequences of three bacterial liquefying alpha-amylases deduced from the DNA sequences. J Biochem. 1985 Nov;98(5):1147–1156. doi: 10.1093/oxfordjournals.jbchem.a135381. [DOI] [PubMed] [Google Scholar]
- Zelwer C., Risler J. L., Brunie S. Crystal structure of Escherichia coli methionyl-tRNA synthetase at 2.5 A resolution. J Mol Biol. 1982 Feb 15;155(1):63–81. doi: 10.1016/0022-2836(82)90492-2. [DOI] [PubMed] [Google Scholar]






