Abstract
Esophageal squamous cell carcinoma (ESCC) is a highly malignant tumor associated with a poor prognosis, and the molecular mechanisms underlying its formation and progression remain poorly understood. KIAA1522 is upregulated in various tumor tissues, but its function is unknown. Alterations in KIAA1522 expression and its implication in ESCC are currently unclear. In this study, an immunohistochemical analysis of ESCC tissues showed that KIAA1522 was highly expressed in 46% (157/342) of ESCC specimens and that its expression was inversely correlated with the degree of differentiation (P=0.03). Furthermore, small interfering RNA-mediated silencing of KIAA1522 revealed that overexpression of this protein reinforced malignant cell proliferation and anoikis resistance of ESCC cells in vitro. More importantly, KIAA1522 depletion significantly suppressed the growth of ESCC xenograft tumors and lung metastasis of ESCC cells in nude mice. At the molecular level, inhibition of KIAA1522 expression markedly reduced the phosphorylated extracellular signal-regulated kinase (ERK) levels in both suspended and adherent ESCC cells, suggesting that KIAA1522 might promote cell proliferation and survival via the ERK cascade. Taken together, these data suggest that upregulation of KIAA1522 might enhance tumorigenicity and metastasis of ESCC cells through potentiating the ERK activity. Thus, aberrant expression of KIAA1522 plays oncogenic roles in ESCC and might serve as a novel molecular target in ESCC treatment.
Keywords: esophageal squamous cell carcinoma, KIAA1522, tumor growth, anoikis, metastasis, extracellular signal-regulated kinase
Introduction
Esophageal carcinoma is one of the 10 most common human malignancies, and approximately half of new cases and deaths associated with this cancer are recorded in China.1,2 Esophageal squamous cell carcinoma (ESCC) is the predominant pathological type of esophageal cancer in Chinese patients and ranks as the fourth leading cause of cancer-related deaths.3 The overall 5-year survival rate of patients with ESCC is only ~20%.4 Therefore, investigating molecules that play important roles in the initiation and progression of ESCC not only help to elucidate its pathogenic mechanism but also might provide novel molecular markers and therapeutic targets for the diagnosis and treatment of ESCC.
The KIAA1522 gene was discovered via sequencing and encodes a large protein with unknown functions.5 Information in the Gene Expression Atlas and European Bioinformatics Institute databases shows that KIAA1522 mRNA expression is upregulated in various tumor tissues, including lung and breast cancer tissues. However, the relationship between abnormal KIAA1522 expression and malignant tumors remains unclear. Our recent studies showed that KIAA1522 expression is overexpressed in non-small cell lung cancer (NSCLC) and colorectal cancer (CRC) and is positively correlated with poor prognosis of patients with NSCLC and CRC.6–8 In addition, we found that KIAA1522 overexpression promotes the proliferation of NSCLC cells in vitro, suggesting that KIAA1522 is an oncogene.6 Particularly, Chen et al found that CpG islands in the promoter region of the KIAA1522 gene exhibit high frequencies of methylation in ESCC patients from a Chinese Kazakh population of Xinjiang,9 but the changes in the expression of this protein and its functional roles in ESCC remain to be determined.
In the present study, we analyzed the alterations in KIAA1522 protein expression and its clinical significance in ESCC through a tissue microarray (TMA)-immunohistochemistry (IHC) assay. Furthermore, we assessed the effects of KIAA1522 overexpression on the malignant phenotypes of esophageal cancer cells in vivo and in vitro.
Materials and methods
Tissue microarray and IHC analysis
Fresh tissues containing ESCCs and surgical margins histologic normal epithelia were collected at the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences (CAMS), Peking Union Medical College (PUMC), Beijing, China. All patients signed independent informed consent forms for the sampling and molecular analyses. TMAs containing primary ESCCs and surgical margins normal epithelia were constructed and incubated with anti-KIAA1522 antibody (Sigma, St Louis, MO, USA) as previously described.10 The results of the immunohistochemical staining were scored in a blinded manner. Positive staining was detected in the cytoplasm, and the protein expression level of KIAA1522 was rated 0 (negative), 1 (weakly positive), 2 (moderately positive), or 3 (strongly positive) according to the intensity of the staining. The highest score among all effective points represented the final score of the specimen. For the statistical analysis, all cases were grouped as either KIAA1522-positive (score range of 2–3) or KIAA1522-low-positive/negative (score range of 0–1). This study was approved by the Ethics Committee/Institutional Review Board of National Cancer Center/Cancer Hospital, CAMS and PUMC (No NCC2015G-06).
Cell culture
The human ESCC cell lines KYSE150 and KYSE510 were generously provided by Dr Shimada (Kyoto University, Kyoto, Japan). The cell lines were cultured in RPMI 1640 medium with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), penicillin (100 U/mL), and streptomycin (100 mg/mL). This study was approved by the Ethics Committee/Institutional Review Board of National Cancer Center/Cancer Hospital, CAMS and PUMC (No NCC2015G-06).
RNA interference and transfection
The target sequences of KIAA1522-specific siRNA and non-silencing control siRNA (siCtrl) were as follows: siKIAA-A, 5′-GGCTGAGAATGACAAACAT-3′, siKIAA-B, 5′-CAUGACUCAUUUCCCAAAUTT-3′, siKIAA-C, 5′-CUCCUGUCAUCUCCAAAGATT-3′, siCtrl, 5′-UUCUCCGAACGUGUCACGU-3′. All siRNAs were chemically synthesized by Genepharma (Shanghai, China). Cells were transfected with siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, and cells were harvested 48 h after transfection.
Lentiviral pLKO-KIAA1522 shRNA (shKIAA) and pLKO-non-silencing shRNA control (shCtrl) constructs were generated by inserting the corresponding double-stranded oligonucleotides of siKIAA-A into the pLKO.1-puro lentiviral vector (Addgene, Cambridge, MA, USA). KYSE150 cells were infected with equal amounts of control shRNA virus or KIAA shRNA virus, and stable clones were selected with 2 μg/mL puromycin (Calbiochem, La Jolla, CA, USA) for 1 week.
Western blot analysis
Protein lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The blots were blocked and incubated with an antibody against KIAA1522 (Sigma), phosphorylated extracellular signal-regulated kinase (pERK) (Thr202/Tyr204), or extracellular signal-regulated kinase (ERK) (CST, Beverly, MI, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Kangcheng, Shanghai, China) was used as a loading control. After washing, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies (Applygen, Beijing, China). The signals were visualized using a super enhanced chemiluminescence (ECL) detection reagent (Applygen).
Colony formation assay
The proliferation potential of the cells was assessed by plating 500 cells in 6-well plates. After 2 weeks, the cells were fixed with methanol and stained with crystal violet.
Cell viability assay
ESCC cells (2×103/well) were seeded in 96-well plates with three replicates. The viable cells were quantified every 24 h using a Cell Counting Kit-8 (CCK-8; DojinDo, Kumamoto, Japan) according to the manufacturer’s recommended protocol. The absorbance at 450 nm was measured using an ELX808 microplate reader (BioTek Instruments, Winooski, VT, USA).
Cell cycle analysis
The cells were harvested and fixed in ice-cold 70% ethanol overnight at −20°C. After two washes, the cells were treated with ribonuclease A, stained with propidium iodide (PI), and then analyzed using a BD™ LSRII flow cytometer (BD Biosciences, San Jose, CA, USA).
Assessment of apoptosis and anoikis
Apoptotic cells were double-labeled with Annexin V-fluorescein isothiocyanate (FITC) and PI using an Annexin V/FITC kit (Beyotime Biotechnology, Shanghai, China) and analyzed by flow cytometry. The percentage of Annexin V-positive cells was calculated. For the measurement of anoikis, the cells were plated in dishes coated with polyhydroxylethylmethacrylate (PolyHEMA; Sigma) as described previously.11 After 20 h of growth in suspension, the cells were harvested for apoptosis detection.
Animal experiments
Animal protocols were approved by the Animal Center of the Institute of National Cancer Center/Cancer Hospital, CAMS and PUMC (NCC2015A013), and the National Institutes of Health Guide for the Care and Use of Laboratory Animals was followed. KYSE150 cells stably expressing shCtrl or shKIAA were used in animal experiments. For the tumor growth assay, the cells were inoculated subcutaneously into the flanks of 4-week-old female athymic nude mice (Nu/Nu; Vital River, Beijing, China). Each of the 10 mice in each group were injected with 2×106 cells. The subcutaneous (sc) tumors were measured every 3 days for 3 weeks, and the tumor volume was calculated using the following formula: V = 1/6π × length × width2. The mice were euthanized, and the average weight of tumor tissues was obtained.
For the tumor metastasis assay, six female 6- to 8-week old, nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Beijing HFK Bioscience, Beijing, China) were injected with 2×106 cells per animal via the tail vein. The mice were sacrificed 4 weeks after inoculation and the lung metastases were examined. The lung tissues were fixed in Bouin’s solution and the number of metastatic nodules was counted.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). Either the Kruskal–Wallis test or the Mann–Whitney U test was used to evaluate the association between the KIAA1522 protein levels and the clinicopathological characteristics. The differences in the results between groups were compared using Student’s t-test, one-way analysis of variance (ANOVA), or the nonparametric Kruskal–Wallis test. Kaplan–Meier survival curves were generated to determine the relationship between KIAA1522 levels and the overall survival of ESCC patients, and the differences between the curves were calculated using the log-rank test. P-values <0.05 were considered significant.
Results
KIAA1522 expression is upregulated in ESCC cells
We determined the expression of KIAA1522 protein in 342 ESCC samples and 252 surgical margins normal epithelial tissues through TMA-IHC analysis. KIAA1522 was mainly localized in the cytoplasm of esophageal epithelia cells (Figure 1). Positive expression of KIAA1522 was observed in 46% (157/342) and 4% (11/252) of tumor and surgical margins normal tissues, respectively, showing a significant difference between tumor and normal tissues (P<0.0001). In addition, the KIAA1522 protein expression level was inversely correlated with the degree of differentiation of ESCC (P=0.03, Table 1). There was no significant correlation between KIAA1522 overexpression and overall survival of Chinese patients with ESCC (Figure S1).
Figure 1.
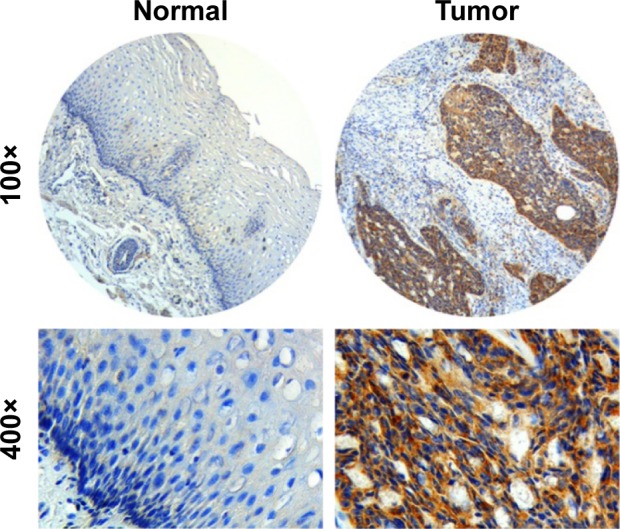
Representative IHC staining for the KIAA1522 protein in ESCC tumors and adjacent normal esophageal squamous epithelium.
Note: Original magnification: 100× and 400×.
Abbreviations: ESCC, esophageal squamous cell carcinoma; IHC, immunohisto-chemistry.
Table 1.
Association of KIAA1522 levels with clinicopathological features of 342 patients with ESCC
| Clinicopathological features | KIAA1522 expression
|
Total | P-value | |
|---|---|---|---|---|
| Negative (%) | Positive (%) | |||
| All patients | 185 (54) | 157 (46) | 342 | |
| Age (years) | 0.632 | |||
| <60 | 92 (55.4) | 74 (44.6) | 166 | |
| ≥60 | 93 (52.8) | 83 (47.2) | 176 | |
| Gender | 0.837 | |||
| Male | 135 (55.4) | 113 (44.6) | 248 | |
| Female | 50 (53.2) | 44 (46.8) | 94 | |
| TNM classification | ||||
| pT | 0.625* | |||
| T1 | 8 (66.7) | 4 (33.3) | 12 | |
| T2 | 105 (51.5) | 99 (48.5) | 204 | |
| T3 | 40 (56.3) | 31 (43.7) | 71 | |
| T4 | 32 (58.2) | 23 (41.8) | 55 | |
| pN | 0.361 | |||
| N0 | 37 (49.3) | 38 (50.7) | 75 | |
| N1 | 147 (55.5) | 118 (44.5) | 265 | |
| Histology grade | 0.030 | |||
| Good (G1) | 42 (66.7) | 45 (33.3) | 87 | |
| Moderate (G2) | 106 (51.5) | 96 (48.5) | 202 | |
| Poor (G3) | 36 (56.3) | 15 (43.7) | 51 | |
| AJCC6 stage | 0.921 | |||
| I | 14 (56.0) | 11 (44.0) | 25 | |
| II | 101 (53.2) | 89 (46.8) | 190 | |
| III | 69 (55.2) | 56 (44.8) | 125 | |
Note:
Fisher’s exact test.
Abbreviations: ESCC, esophageal squamous cell carcinoma; TNM, tumor, regional lymph node, distant metastasis; pT, pathologic T stage; pN, lymph node metastases; AJCC6, American Joint Committee on Cancer (Sixth Edition).
KIAA1522 overexpression enhances the proliferation ability and tumorigenicity of ESCC cells
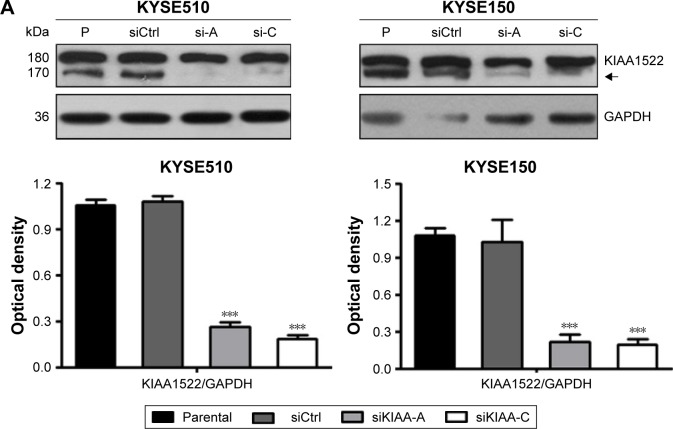
To assess the effect of KIAA1522 overexpression on the malignant phenotypes of esophageal cancer cells, we silenced KIAA1522 expression in the ESCC cell lines KYSE510 and KYSE150 using specific siRNA. Transient transfection of siRNA effectively inhibited KIAA1522 expression in both cell lines, resulting in a prominent reduction in the proliferation and colony-forming ability of these cells in vitro but no obvious effect on the cell cycle distribution and apoptosis (Figures 2 and S2).
Figure 2.
Inhibition of KIAA1522 expression suppresses the proliferation and colony formation ability of ESCC cells.
Notes: The ESCC cell lines KYSE510 and KYSE150 were transiently transfected with two independent KIAA1522-specific siRNA (si-A and si-C) or negative control siRNA (siCtrl) for 48 h. (A) The expression levels of KIAA1522 protein were detected by immunoblotting. The arrow indicates the specific band of KIAA1522 protein located around 170 kDa. Relative ratios of absorbance for KIAA1522 to GAPDH were plotted. The data are presented as the mean ± SEM. ***P<0.001. (B) Cell proliferation was determined by CCK-8 assay. Data are mean ± SEM. ***P<0.001. (C) Colony formation assay. Representative results are shown, and the colony number was plotted. The data are presented as the mean ± SEM from three separate experiments. ***P<0.001.
Abbreviations: CCK-8, cell counting kit-8; Ctrl, control; ESCC, esophageal squamous cell carcinoma; GAPDH, glyceral dehyde 3-phosphate dehydrogenase; P, parental; SEM, standard error of the mean; si, siRNA; si-A, siKIAA-A; si-c, siKIAA-C.
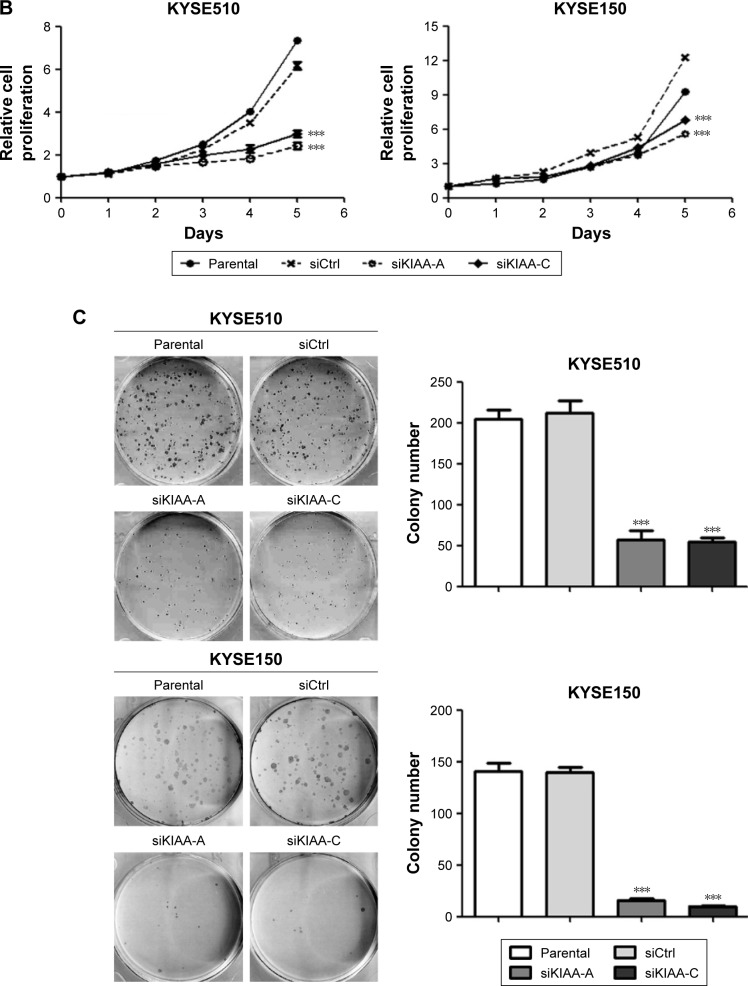
We subsequently investigated the effect of KIAA1522 on tumor growth in vivo. To this end, stable strains of KYSE510 and KYSE150 cells expressing KIAA1522-specific shRNA (denoted as shKIAA in this paper) or the negative control shRNA (denoted as shCtrl) were established. Similarly, the knockdown of KIAA1522 expression by shRNA in both cell lines markedly reduced the KIAA1522 expression level and colony formation capacity (Figure 3A and B). More importantly, KIAA1522 downregulation significantly repressed the growth of xenograft tumors derived from KYSE150 cells in nude mice (Figure 3C–E).
Figure 3.
Knockdown of KIAA1522 expression reduces the tumorigenicity of esophageal carcinoma cells in nude mice.(p)(p)Notes: KYSE510 and KYSE150 cells were infected with lentiviruses expressing KIAA1522 shRNA (shKIAA) or negative control shRNA (shCtrl), and stable cell clones were selected with puromycin. (A) Western blot analysis of KIAA1522 expression levels in stable cell clones. The arrow indicates the position of KIAA1522 specific band. Relative ratios of absorbance for KIAA1522 to GAPDH were plotted. The data are presented as the mean ± SEM. ***P<0.001. (B) Colony formation assay. (C–E) KYSE150-shKIAA or shCtrl stable cell clones were implanted subcutaneously into nude mice (n=10 per group), the tumor volumes were measured every 3 days for 3 weeks, and sc tumors were excised 21 days after inoculation. (C) Tumor growth curves. The data are presented as the mean ± SEM. **P<0.01, ***P<0.001. (D) Tumor weight. ***P<0.001; (E) Photographs of sc tumors. Scale bar =1 cm.(p)(p)Abbreviations: Ctrl, control; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; sc, subcutaneous; sh, shRNA.
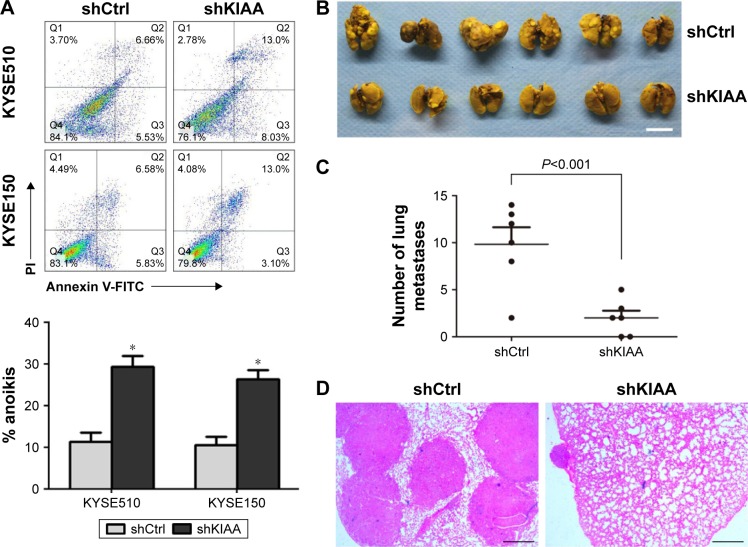
KIAA1522 upregulation inhibits anoikis of ESCC cells
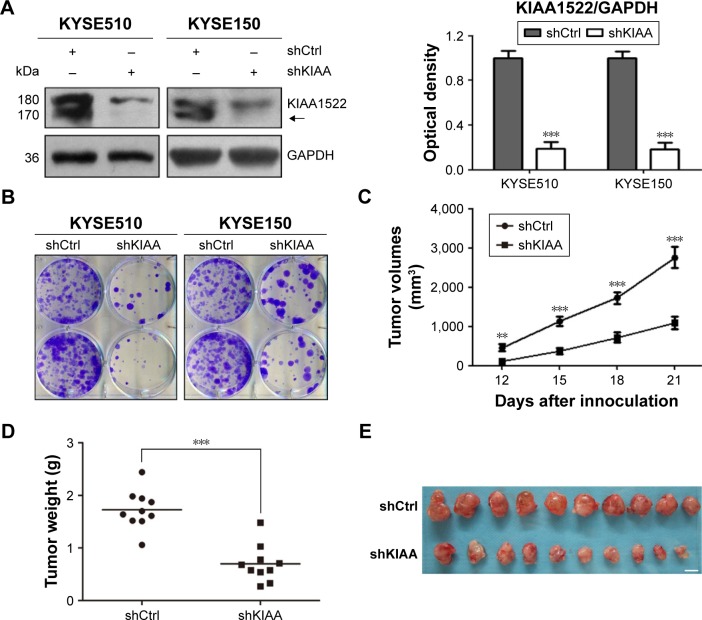
Intriguingly, we found that the KIAA1522 protein levels in ESCC cells were apparently increased upon detachment from the extracellular matrix (ECM; Figure 4A). On the basis of our observations, we speculated that detachment-induced upregulation of KIAA1522 might be responsible for the susceptibility of ESCC cells to anoikis, that is, detachment-induced apoptotic death. We then assessed the effect of KIAA1522 on the ability of ESCC cells to resist anoikis. The results showed that depletion of KIAA1522 by siRNA significantly induced apoptosis in KYSE510 and KYSE150 cells cultured in suspension compared with that measured in parental or non-silencing siRNA-transfected cells (Figure 4B and C). Therefore, our data suggest that upregulation of KIAA1522 is capable of protecting ESCC cells from anoikis during cell detachment.
Figure 4.
KIAA1522 depletion enhances anoikis in ESCC cells.
Notes: (A) KYSE510 and KYSE150 cells were cultured in suspension for the indicated times, and the KIAA1522 protein levels were detected by Western blotting. Relative ratios of absorbance for KIAA1522 to GAPDH were plotted. The data are presented as the mean ± SEM. **P<0.01. The arrows in images A and B indicate the position of KIAA1522 specific band. (B–D) KYSE150 and KYSE510 cells were transiently transfected with KIAA1522 siRNA for 48 h and then cultured on polyHEMA-coated dishes for 20 h. (B) KIAA1522 expression was determined by Western blot. (C) Anoikis was measured by Annexin V-FITC/PI staining and flow cytometry analysis. Representative results are shown (upper panel), and the percentages of apoptotic cells were plotted (lower panel). The data are presented as the mean ± SEM. **P<0.01, ***P<0.001.
Abbreviations: Ctrl, control; ESCC, esophageal squamous cell carcinoma; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NC, non-silencing; P, parental; PI, propidium iodide; polyHEMA, polyhydroxylethylmethacrylate; si, siRNA; si-A, siKIAA-A; si-B, siKIAA-B; si-c, siKIAA-C.
Anoikis is a specific apoptotic death due to loss or improper cell adhesion, including cell–ECM adhesion and cell–cell adhesion.12 Bioinformatics analysis predicted that KIAA1522 is an adhering protein. Intriguingly, we observed that suspended ESCC cells tend to adhere together and form a bigger cell mass, but inhibition of KIAA1522 expression led to suspended ESCC cells dispersed into multiple smaller cell masses, indicating that cell–cell adhesion was disrupted in KIAA1522 depleted cells (Figure S3).
KIAA1522 overexpression promotes the lung metastasis of esophageal carcinoma cells
Because anoikis resistance is a malignant phenotype closely associated with the metastatic potential of malignant tumors,13,14 we subsequently tested whether the inhibition of KIAA1522 affects the metastatic ability of ESCC cells in vivo. First, we confirmed that stable knockdown of KIAA1522 markedly impaired anoikis resistance of KYSE150 and KYSE510 cells (Figure 5A). Then, we performed a lung metastasis experiment involving the inoculation of nude mice with KYSE150-shKIAA or shCtrl cells via the tail vein. As a result, KIAA1522 knockdown significantly abrogated the ability of KYSE150 cells to form metastatic foci in the lungs of nude mice (Figure 5B–D), indicating that KIAA1522 expression promotes tumor metastases in vivo.
Figure 5.
KIAA1522 promotes lung metastasis of KYSE150 cells in NOD/SCID mice.
Notes: (A) KYSE510 and KYSE150 cells transfected with shKIAA or shCtrl were cultured on polyHEMA-coated dishes for 20 h and subjected to Annexin V-FITC/PI staining followed by flow cytometry analysis. Representative results are shown, and degrees of apoptosis were plotted. The data are presented as mean ± SEM. *P<0.05. (B–D) Nude mice were injected with 2×106 KYSE150-shCtrl or KYSE150-shKIAA stable cell clones via the tail vein. After 4 weeks, the mice were sacrificed. (B) The lungs were subjected to Bouin’s fixation and photographed. Scale bar =1 cm. (C) Visible lung metastases nudes were counted and plotted. (D) The sections of the lungs were stained with H&E. Scale bar =200 μm.
Abbreviations: Ctrl, control; NOD/SCID, nonobese diabetic/severe combined immunodeficiency; polyHEMA, polyhydroxylethylmethacrylate; sh, shRNA.
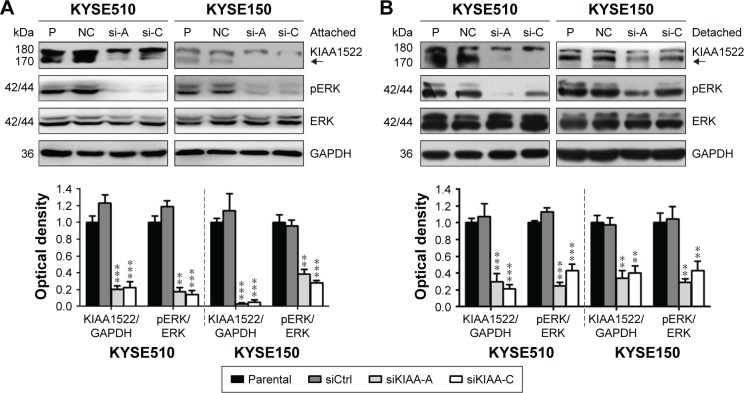
KIAA1522 activates extracellular signal-regulated kinase (ERK) signaling in ESCC cells
To further explore the relevant molecular mechanism, we detected the changes in signaling pathways related to proliferation and survival in ESCC cells after KIAA1522 inhibition. Specifically, depletion of KIAA1522 expression distinctly abrogated ERK activity in both adherent and suspension-cultured ESCC cells (Figure 6), indicating that KIAA1522 facilitates the proliferation and survival of ESCC cells at least partially through activating the ERK signaling pathway.
Figure 6.
KIAA1522 activates ERK signaling pathway in ESCC cells.
Notes: (A) The ESCC cell lines KYSE510 and KYSE150 were transiently transfected with KIAA1522-specific siRNA or control non-silencing siRNA for 48 h. (B) Transiently transfected cells were cultured on polyHEMA-coated dishes for 20 h, and the cell lysates were immunoblotted for the indicated proteins. The arrows indicate the position of KIAA1522 specific band. Relative ratios of absorbance for KIAA1522 to GAPDH, pERK to total ERK were plotted. The data are presented as the mean ± SEM. **P<0.01, ***P<0.001.
Abbreviations: Ctrl, control; ERK, extracellular signal-regulated kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NC, non-silencing; P, parental; pERK, phosphorylated ERK; polyHEMA, polyhydroxylethylmethacrylate; si, siRNA; si-A, siKIAA-A; si-c, siKIAA-C.
Discussion
KIAA1522 is an unannotated gene, and very few studies performed to date have examined it. Through this study, we demonstrate that KIAA1522 expression is upregulated in 46% of ESCC specimens and that KIAA1522 overexpression not only substantially promotes the malignant proliferation of ESCC cells in vivo and in vitro but also increases their metastatic potential by enhancing anoikis resistance. Furthermore, our current data show that KIAA1522 potentiates ERK activity in ESCC cells under both adherent and suspension culture conditions.
KIAA1522 is a newly cloned gene, and its overexpression has been observed in several human cancers.5 However, the alterations in KIAA1522 expression in esophageal carcinoma are currently unclear. In the present study, a TMA-IHC analysis revealed that the KIAA1522 protein levels are elevated in 46% of ESCC specimens compared with surgical margins normal esophageal mucosa. In addition, KIAA1522 expression was found to be inversely correlated with ESCC differentiation. These data imply that KIAA1522 might be a pro-oncogene in ESCC.
Few studies have investigated KIAA1522 functions and relevant molecular mechanisms in malignant tumors. Our previous data reveal that KIAA1522 overexpression promotes the proliferation of human NSCLC cells in vitro, suggesting that it acts as an oncogene.6 Consistently, this study demonstrates that KIAA1522 overexpression contributes to the malignant proliferation of ESCC cells in vitro and in vivo.
Anoikis is a particular type of programmed cell death induced by the detachment of normal cells from the ECM and plays important roles in development and homeostasis.12,15 Anoikis resistance allows the survival of malignant tumor cells in blood vessels and the lymphatic system and their metastasis to distant sites, and thus contributes to tumor progression.12,14 In this study, we show that the knockdown of KIAA1522 expression significantly impairs the anoikis resistance of ECSS cells cultured in suspension, and our data imply that KIAA1522 might play a role in cell–cell adhesion. More importantly, KIAA1522 depletion prominently inhibits the lung metastasis of ESCC cells in nude mice. Collectively, our current data suggest that KIAA1522 overexpression promotes the metastasis of ESCC cells by enhancing their resistance to anoikis.
At the molecular level, the inhibition of KIAA1522 expression evidently reduces pERK levels in both adherent and suspension-cultured ESCC cells, suggesting that high KIAA1522 expression enhances the proliferation and metastasis potentials of ESCC cells by activating the ERK cascade. Consistently, we have found that KIAA1522 triggers ERK phosphorylation in NSCLC cells.6 Altogether, our data suggest that ERK is a vital and common downstream signal of KIAA1522 in multiple malignancies.
In summary, this study shows that KIAA1522 protein expression is upregulated in a subset of ESCC tissues and that KIAA1522 overexpression might promote tumor formation and progression of ESCC cells by enhancing their malignant proliferation ability and anoikis resistance through the activation of the ERK signaling pathway. Therefore, KIAA1522 might serve as a target in the treatment of ESCC.
Supplementary materials
Kaplan–Meier overall survival curve according to the KIAA1522 levels in ESCC patients.
Notes: Overall survival is shown for the 115 patients with ESCC based on KIAA1522 levels as measured by immunohistochemistry. P-values were calculated by log-rank test.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Effects of KIAA1522 knockdown on the apoptosis and cell cycle distribution of ESCC cells.
Notes: KYSE510 and KYSE150 cells were transiently transfected with KIAA1522-specific siRNA or control non-silencing siRNA. Cells were collected after 48 h and subjected to the analysis. The results of a flow cytometry analysis of apoptosis (A) and cell cycle distribution (B) are shown.
Abbreviations: Ctrl, control; ESCC, esophageal squamous cell carcinoma; si, siRNA.
KIAA1522 depletion inhibits cell–cell adhesion.
Notes: KYSE150 and KYSE510 cells were transiently transfected with KIAA1522 siRNA or control non-silencing siRNA for 48 h and then seeded on polyHEMA-coated dishes. The cell morphology was observed under the phase. Scale bars =50 μm.
Abbreviations: Ctrl, control; polyHEMA, polyhydroxylethylmethacrylate; si, siRNA.
Acknowledgments
This work was supported by the National Natural Science Foundation (81330052, 81520108023, and 81572841), the Beijing Natural Science Foundation (7151008), and the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-001).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Schweigert M, Dubecz A, Stein HJ. Oesophageal cancer–an overview. Nat Rev Gastroenterol Hepatol. 2013;10(4):230–244. doi: 10.1038/nrgastro.2012.236. [DOI] [PubMed] [Google Scholar]
- 5.Nagase T, Kikuno R, Ishikawa K, Hirosawa M, Ohara O. Prediction of the coding sequences of unidentified human genes. XVII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7(2):143–150. doi: 10.1093/dnares/7.2.143. [DOI] [PubMed] [Google Scholar]
- 6.Liu YZ, Yang H, Cao J, et al. KIAA1522 is a novel prognostic biomarker in patients with non-small cell lung cancer. Sci Rep. 2016;6:24786. doi: 10.1038/srep24786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YZ, Jiang YY, Wang BS, et al. A panel of protein markers for the early detection of lung cancer with bronchial brushing specimens. Cancer Cytopathol. 2014;122(11):833–841. doi: 10.1002/cncy.21461. [DOI] [PubMed] [Google Scholar]
- 8.Liu TT, Liang JW, Yu J, et al. Significance in expression of KIAA1522 in colorectal cancer. Carcino Terato Mutagen. 2017;29(2):87–90. 95. [Google Scholar]
- 9.Chen Y, Yin D, Li L, Deng YC, Tian W. Screening aberrant methylation profile in esophageal squamous cell carcinoma for Kazakhs in Xinjiang area of China. Mol Biol Rep. 2015;42(2):457–464. doi: 10.1007/s11033-014-3788-z. [DOI] [PubMed] [Google Scholar]
- 10.Shang L, Liu HJ, Hao JJ, et al. A panel of overexpressed proteins for prognosis in esophageal squamous cell carcinoma. PLoS One. 2014;9(10):e111045. doi: 10.1371/journal.pone.0111045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu SG, Wang BS, Jiang YY, et al. Atypical protein kinase Ciota (PKCiota) promotes metastasis of esophageal squamous cell carcinoma by enhancing resistance to anoikis via PKCiota-SKP2-AKT pathway. Mol Cancer Res. 2011;9(4):390–402. doi: 10.1158/1541-7786.MCR-10-0359. [DOI] [PubMed] [Google Scholar]
- 12.Taddei ML, Giannoni E, Fiaschi T, Chiarugi P. Anoikis: an emerging hallmark in health and diseases. J Pathol. 2012;226(2):380–393. doi: 10.1002/path.3000. [DOI] [PubMed] [Google Scholar]
- 13.Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19(11):1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- 14.Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett. 2008;272(2):177–185. doi: 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 15.Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833(12):3481–3498. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan–Meier overall survival curve according to the KIAA1522 levels in ESCC patients.
Notes: Overall survival is shown for the 115 patients with ESCC based on KIAA1522 levels as measured by immunohistochemistry. P-values were calculated by log-rank test.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Effects of KIAA1522 knockdown on the apoptosis and cell cycle distribution of ESCC cells.
Notes: KYSE510 and KYSE150 cells were transiently transfected with KIAA1522-specific siRNA or control non-silencing siRNA. Cells were collected after 48 h and subjected to the analysis. The results of a flow cytometry analysis of apoptosis (A) and cell cycle distribution (B) are shown.
Abbreviations: Ctrl, control; ESCC, esophageal squamous cell carcinoma; si, siRNA.
KIAA1522 depletion inhibits cell–cell adhesion.
Notes: KYSE150 and KYSE510 cells were transiently transfected with KIAA1522 siRNA or control non-silencing siRNA for 48 h and then seeded on polyHEMA-coated dishes. The cell morphology was observed under the phase. Scale bars =50 μm.
Abbreviations: Ctrl, control; polyHEMA, polyhydroxylethylmethacrylate; si, siRNA.