Abstract
Megakaryocyte–erythrocyte progenitor (MEP) cells are potential in vivo targets of dengue virus (DENV); the virus has been found associated with megakaryocytes ex vivo and platelets during DENV-induced thrombocytopenia. We report here that DENV serotype 2 (DENV2) propagates well in human non-differentiated MEP cell lines (Meg01 and K562). In comparison to virus propagated in Vero cells, viruses from MEP cell lines had similar structure and buoyant density. However, differences in MEP-DENV2 stability and composition were suggested by distinct protein patterns in western blot analysis. Also, antibody neutralization of envelope domain I/II on MEP-DENV2 was reduced relative to that on Vero-DENV2. Infectious DENV2 was produced at comparable kinetics and magnitude in MEP and Vero cells. However, fewer virion structures appeared in electron micrographs of MEP cells. We propose that DENV2 infects and produces virus efficiently in megakaryocytes and that megakaryocyte impairment might contribute to dengue disease pathogenesis.
Keywords: Dengue virus, Megakaryocyte, Erythrocyte, Progenitor, Meg01, K562, MEP, Electron microscopy
1. Introduction
Dengue virus (DENV) is an increasing public health threat, largely because of its ability to transmit not only by Aedes aegypti, a tropical and subtropical vector, but also via Aedes albopictus, the more prevalent mosquito vector endemic in temperate zones (World Health Organization, 2015). Approximately 390 million people are infected annually, although most of these infections do not progress to the point of major clinical disease (Bhatt et al., 2013). Persons of a wide range of ages can become infected and experience a variety of clinical manifestations (from mild dengue fever to more severe dengue hemorrhagic fever/dengue shock syndrome) with approximately 22,000 deaths occurring annually (Tsai et al., 2012). DENV is also a heavy public health burden because no specific therapeutics are available; one vaccine recently became available, but it was approved only for previously exposed populations and is at most 61% protective (Pasteur, 2014). Moreover, in spite of its widespread recurrence and emphasis in the literature, a number of its basic biologic and pathologic DENV mechanisms remain to be fully elucidated.
During blood meal, mosquitoes inoculate DENV directly into the skin. But more importantly, when mosquitoes probe the skin, they can find blood vessels and deposit virus directly into the capillaries, releasing virus into circulation (O’Rourke, 1956; Styer et al., 2007) and exposing many different cell types to pathogen. Permissiveness has been investigated in various cell types: dendritic cells (Ader et al., 2004; Ho et al., 2001; Sun et al., 2009; Wu et al., 2000), monocytes/macrophages (Arevalo et al., 2009; Daughaday et al., 1981; Diamond et al., 2000; Tan and Chu, 2013; Theofilopoulos et al., 1976), endothelial cells (AbuBakar et al., 2014; Arevalo et al., 2009; Diamond et al., 2000), and B cells (Takasaki et al., 2001; Theofilopoulos et al., 1976). A number of these cell lineages can get infected and reprogrammed, and many of these events might even contribute to disease pathology (Butthep et al., 1993; Green and Rothman, 2006; Lee et al., 2013; Libraty et al., 2001; Nielsen, 2009). But while a cell type might be permissive to DENV infection, a separate issue is whether that cell can efficiently produce high titers of infectious virus. The infectiousness of virus released from a number of cell types has been questioned (AbuBakar et al., 2014; Marianneau et al., 1999; Mosquera et al., 2005), and thus the cellular target responsible for viremia in humans remains controversial.
DENV infection of bone marrow cell populations has been implicated in a number of previous reports. It was noted even in early studies that bone marrow resident cells change in morphology and frequency (Bierman and Nelson, 1965; Kho et al., 1972; La Russa and Innis, 1995; Nelson et al., 1964; Noisakran et al., 2012). Bone marrow-derived megakaryocyte–erythrocyte progenitor cells were permissive and yielded high DENV2 titers (1 × 105 FFU/mL and 1 × 108 genome copy number [GCN]/mL) (Basu et al., 2008; Clark et al., 2012; Nakao et al., 1989). Also, a recent publication reports a positive correlation between DENV titers in dengue fever patient plasma and circulating CD61+ (megakaryocyte marker) cell count numbers (Hsu et al., 2015). While not conclusive, these observations suggest that CD61+ cells might contribute to DENV replication in vivo, since DENV can be propagated ex vivo from CD61+ cells isolated from bone marrow of infected animals (Noisakran et al., 2012). Studies have indicated that megakaryocytes stain positive for viral antigen and antigen positivity correlates with peak infectious titer and virus-like particle (VLP) production (Basu et al., 2008; Clark et al., 2012; Noisakran et al., 2012). However, despite an association of DENV2 with the megakaryocyte, the cell types that initially encountered and took up the virus in these experiments were uncertain because the effect could be due to infection of any of several cell types capable of differentiating into megakaryocytes. Thus, it is not known if megakaryocytes can be infected directly by DENV.
In this investigation, we sought to examine further cells of the megakaryocytic lineage as potential DENV2 hosts. Because bone marrow samples are difficult to acquire, and because of the low frequency of megakaryocytes in the bone marrow in general, our investigations were conducted with megakaryocyte–erythrocyte progenitor (MEP) cell lines: Meg01 (Ogura et al., 1985), a megakaryocytic cell line that has rarely been used in DENV research, and K562 (Lozzio et al., 1981) a MEP cell line that has the ability to differentiate into megakaryocytes and has been used in a number of DENV studies. We characterized DENV2 replication and production in Meg01, K562, or Vero cell lines, a gold-standard tool in DENV investigations, and also studied the structure and antigenicity of viruses produced in cultures of these cells. In all cell lines examined, DENV2 propagated to similar titers with comparable kinetics and produced infectious virions of similar density and structure. However, our study also revealed that particular composition and antigenicity differences did exist. This work supports previous findings indicating that cells of the megakaryocyte–erythrocyte lineage were permissive to DENV infection and might contribute to DENV pathogenesis (Clark et al., 2012; Diamond et al., 2000; Nakao et al., 1989; Noisakran et al., 2012).
2. Results
2.1. DENV2 propagates efficiently and produces virus particles in MEP cell lines
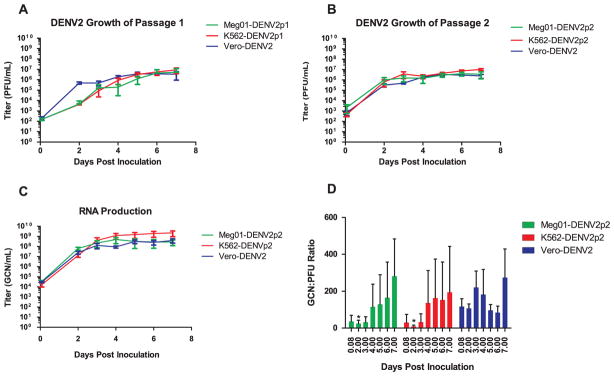
We examined virus growth kinetics with in vitro cell lines of the MEP lineage. Propagation of DENV2 in Meg01 or K562 cells was compared in parallel with Vero cells. All cells were inoculated with DENV2 that had been propagated previously in Vero cell monolayer cultures (Vero-DENV2) and cultured under similar conditions (Fig. 1A). Plaque assay analysis of passage 1 (p1) supernatants indicated that similar levels of infectious DENV2 were produced in all three cell lines, but virus growth in Meg01 and K562 cells appeared slightly delayed, reaching consistent titers of approximately 1 × 105 PFU/mL on day 4 after inoculation, at least 2 days after Vero-DENV2. To determine if slower growth was a consequence of the cell line or level of adaptation to the host, viruses Meg01-DENV2p1 and K562-DENV2p1 were passaged again in Meg01 or K562 cells, respectively, to yield suspensions designated Meg01-DENVp2 and K562-DENV2p2 (Fig. 1B). Meg01-DENV2p2 and K562-DENV2p2 grew with kinetics similar to those of Vero-DENV2, indicating that DENV2 can grow in these MEP cell lines equally well. Because of their similar replication kinetics, all further experiments were conducted with the p1 viral stocks.
Fig. 1.
Replication kinetics of DENV2 in Meg01, K562, and Vero cells. Cells were inoculated at an MOI=0.1 FFU/mL. Virus from Meg01, K562, and Vero cell supernatants acquired days 2–7 were quantified by either plaque assay or RT-qPCR. Time courses were done at least in triplicate and error bars represent SD. (A) Infectious virus titer time course of Vero-DENV2 passaged in the indicated cell lines. (B) Infectious virus titer time course of virus passaged a second time in the same cell line. Vero-DENV2 data is the same as (A). (C) Quantification of passage 2 virus in (B) by RT-qPCR. (D) GCN:PFU ratios (n=5). *p < 0.05 when compared with corresponding value from Vero-DENV2 using student’s t-test. Abbreviations: FFU=focus forming unit; GCN=genome copy number; MOI=multiplicity of infection; PFU=plaque forming units; RT-qPCR=reverse transcription-quantitative polymerase chain reaction; SD=standard deviation.
In addition to infectious titers, RNA genome copy number (GCN) quantification suggested that virus was released into the supernatant with comparable kinetics for all three cell lines tested, with K562-DENV2p2 yielding slightly higher values on days 6 and 7 (Fig. 1C). Although these cell lines appeared to release infectious virus and viral RNA with similar kinetics, GCN:PFU ratios differed slightly. Meg01 and K562 cells yielded lower GCN:PFU ratios at early time points, though only day 2 differences were significant (p=0.013 and p=0.012, respectively) (Fig. 1D). The mean ratios at this time point were 24.4 (Meg01-DENV2), 9.2 (K562-DENV2), and 107.4 (Vero-DENV2) (Table 1). Thus, Meg01 and K562 cell lines appeared to release fewer noninfectious virions than Vero cells at early time points of infection. Meg01-DENV2 and K562-DENV2 GCN:PFU ratios appeared to increase over time, suggesting an increase in the release of noninfectious virus at later time points or an increase in virus particle degradation over time (perhaps as a consequence of cell culture proteases). Vero-DENV2 also showed an increase in GCN:PFU ratio with time, except on days 5 and 6, when they dropped and then rose again on day 7 (Fig. 1D). The reason for this dip in GCN:PFU ratio is unknown but might be due to a second round of virus amplification.
Table 1.
Time course of GCN:PFU ratios of Meg01-DENV2p2, K562-DENV2p2, and Vero-DENV2 from cell supernatant.
| DPI | K562-DENV2p2 | Meg01-DENVp2 | Vero-DENV2 | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Mean | SD | Mean | SD | Mean | SD | |
| 0.083 | 22.7 | 44.5 | 36.1 | 33.3 | 117 | 94.9 |
| 2.000 | 9.21* | 6.88 | 24.4* | 18.3 | 107 | 53.1 |
| 3.000 | 32.0 | 45.1 | 32.0 | 29.2 | 221 | 199 |
| 4.000 | 136 | 176 | 115 | 124 | 182 | 305 |
| 5.000 | 162 | 212 | 129 | 161 | 96.9 | 69.2 |
| 6.000 | 153 | 206 | 165 | 193 | 84.3 | 78.3 |
| 7.000 | 194 | 249 | 282 | 202 | 274 | 348 |
Abbreviations: DPI=days post-inoculation; GCN=genome copy number; PFU=plaque-forming unit; and SD=standard deviation.
p < 0.05 compared with corresponding value from Vero-DENV2 using student’s t-test.
Meg01-DENV2, K562-DENV2, and Vero-DENV2 were compared for their ability to replicate in cells from human bone marrow tissue specimens. These viruses were isolated through sucrose gradients, quantified by RT-qPCR, and then propagated in human bone marrow tissue specimens. Virus production then was evaluated by an enzyme-linked immunosorbent assay specific for detection of DENV nonstructural protein-1 (NS-1). In these experiments, NS-1 peaked at similarly high levels (>4,000 ng/mL) in human bone marrow supernatants, irrespective of the cell type in which the inoculated virus had been produced (data not shown).
Electron microscopy analysis of concentrated supernatants from day 3 suggested that Meg01 and K562 cells released virus that appeared similar to Vero-DENV2 (Supplemental Fig. 1). Virions, identified by staining with 3H5 (envelope-specific) monoclonal antibody, were in the 50 nm range of size and had a “hairy” appearance.
2.2. DENV2-infected MEP cell lines synthesize lower numbers of virus-induced structures
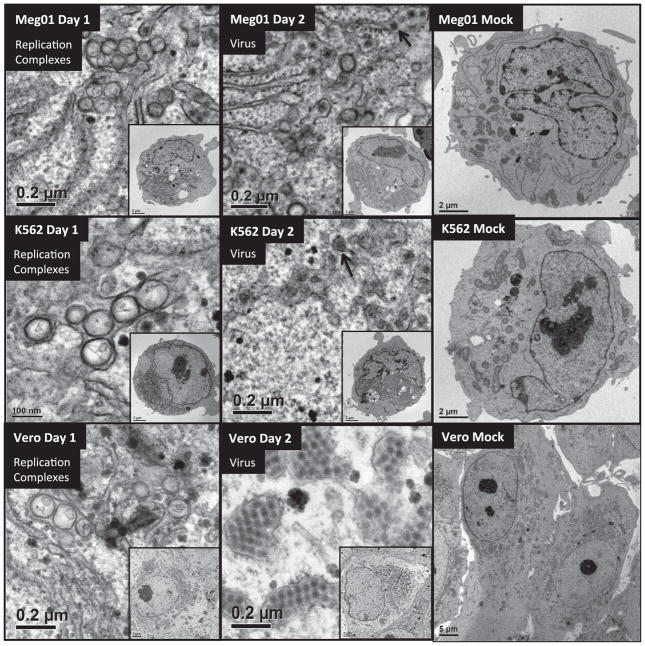
We examined the morphology of DENV2-infected MEP cells. Meg01, K562, or Vero cells were inoculated at a low multiplicity of infection (MOI), harvested on days 1 or 2, thin-sectioned, and imaged by electron microscopy (EM) (Fig. 2). DENV2-inoculated Meg01 and K562 cells produced virus particles and replication complex structures similar to DENV2-infected Vero cells, although there was some variability in the shape of replication complex shape. Meg01 and K562 replication complexes often appeared elongated/elliptical (data not shown).
Fig. 2.
EM imaging of DENV2-infected Meg01, K562, or Vero cells. Meg01, K562, or Vero cells were inoculated with DENV2 at low MOI or mock-infected and cell pellets or monolayers were fixed with glutaraldehyde and processed for thin-sectioning EM. (Top) Meg01, (Mid) K562, and (Bot) Vero depict the following structures (left-to-right): replication complexes from day 1 DENV2-infected cell; virus from day 2 DENV2-infected cell (arrows indicate virus); day 2 mock-infected cell. Insets show cell of origin. Abbreviations: EM=electron micrograph; MOI=multiplicity of infection.
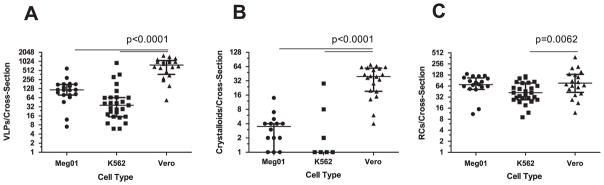
Virus particles appeared more numerous in DENV2-infected Vero cells, so virus particles from day 2 were enumerated. The analyses were performed using 20–27 cell cross-sections from each of the infected cell lines. Significant differences were observed between the infected MEP and Vero cell lines. Meg01 and K562 cells both produced fewer numbers of virus particles per cell cross-section (averaging 140.9 and 94.9, respectively) than did Vero cells per cross-section (764.2; p < 0.0001) (Fig. 3; Table 2). In addition, fewer crystalloids formed in infected Meg01 and K562 cell lines (p < 0.0001). The majority of K562 cells did not have a single virus cluster. Less variation in numbers of replication complexes was observed between MEP cells and Vero cells, although infected K562 cells had fewer complexes (average, 49.7) than Vero cells (average, 92.2; p=0.0062). In addition to the lower frequency of virions per cell, fewer numbers of MEP cells appeared to be infected. When evaluating cells on an entire EM grid square, the endoplasmic reticulum (ER) was distended in 17.5% of Meg01 and 19.2% of K562 cells, while most of the Vero cells (85.5%) appeared to contain virus-induced structures (Table 2).
Fig. 3.
Quantitative comparison of virus-induced structures in Meg01, K562, or Vero cells. Meg01, K562, or Vero cells were inoculated at a low MOI and cell pellets or monolayers from day 2 were fixed, thin-sectioned, stained, and analyzed. Cross-sections of 20 Meg01, 27 K562, or 20 Vero cells were evaluated for the formation of VPs, crystalloid structures, and RCs. (A) Concentration of VPs per cell cross-section. (B) Concentration of crystalloid structures per cross-section. (C) Concentration of RCs per cross-section. Bar indicates median, and whiskers show standard deviations. p Values were obtained using unpaired student’s t-test. Abbreviations: MOI=multiplicity of infection; RCs=replication complexes; VPs=virus particles.
Table 2.
Quantification of DENV2-induced structures from day 2 infected Meg01, K562, or Vero cells.
| Meg01 | K562 | Vero | ||||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Mean ± SD | Median (Q0,1,3,4) | Mean ± SD | Median (Q0,1,3,4) | Mean ± SD | Median (Q0,1,3,4) | |
| VPs per cross-section | 140.9 ± 125.1** | 116 (7, 78.8, 174.3, 593) | 94.9 ± 185** | 36 (6, 16.5, 64.5, 908) | 764.2 ± 443.2 | 769 (49, 393.8, 976.3, 1689) |
| Crystalloids per cross-Section | 3.3 ± 3.1** | 3.5 (0, 1, 4, 14) | 1.6 ± 5.5** | 0 (0, 0, 0.5, 28) | 35.5 ± 22.5 | 37 (0, 16.8, 54.8, 71) |
| VPs per crystalloid | 9.2 ± 5.7* | 8.8 (0, 6.4, 12.3, 23) | 2.6 ± 5.7** | 0 (0, 0, 0, 22) | 14.5 ± 6.6 | 12.9 (0, 10.8, 19.4, 27.4) |
| Replication complexes per cross-section | 70.4 ± 40.9 | 69.5 (0, 52.5, 103.5, 136) | 49.7 ± 29.9* | 42 (9, 28.5, 70.5, 117) | 92.2 ± 78.9 | 71 (12, 38.3, 120.8, 342) |
| Percent infected (number of EM grids analyzed) | 17.5% ± 5.8% (10)** | – | 19.2% 6% (11)** | – | 85.8% ± 3% (5) | – |
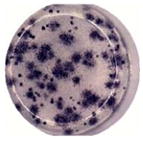
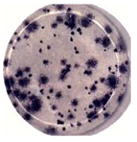
| Percent small focus Size | 100% | 98.8% | 65.8% | |||

|
|
|
|
|
|
|
Abbreviations: EM=electron micrograph; SD=standard deviation; Q=quartile; and VPs=virus particles.
p < 0.01 compared with the corresponding value from Vero cells using student’s t-test.
p < 0.0001 compared with the corresponding value from Vero cells using student’s t-test.
A link has been suggested between numbers of viruses produced by a cell and virus plaque diameter (Junjhon et al., 2008; Lee et al., 2010). DENV2 derived from Meg01 and K562 cells had more uniform sizes of small foci (Table 2) and plaques (data not shown), while DENV2 derived from Vero cells formed foci with various widths. It is important to note that other DENV strains were not examined in such a detailed manner, and thus, it is not known if reduced intracellular virion numbers correlate with reduced focus/plaque diameters for other strains grown in MEP cell lines.
Growth of a limited number of strains was examined in Meg01 and K562 cells. Production of prototypic strains from the three other DENV serotypes, DENV1 (Hawaii, gift from CDC), DENV3 (H87, gift from CDC), and DENV4 (Hawaii, gift from Dr. Duane Gubler), were tested in a limited number of experiments with Meg01 and K562 cells. Using a focus-forming unit assay (FFA) for quantification, all strains could be propagated in MEP cell lines but not reproducibly. Titers of 1 × 105 FFU/mL were obtained with all viruses in both MEP cell lines, except for DENV4 in Meg01, which only reached approximate 1 × 104 FFU/mL titers (data not shown). Reduced replication might be attributable to the strain type and the presence of defective interfering particles. The DENV4 strain gave rise to large foci when grown in Vero cells, but focus sizes varied when grown in MEP-DENV4 cells.
2.3. Minor differences in quantity and density observed with purified Vero-DENV2 and MEP-DENV2
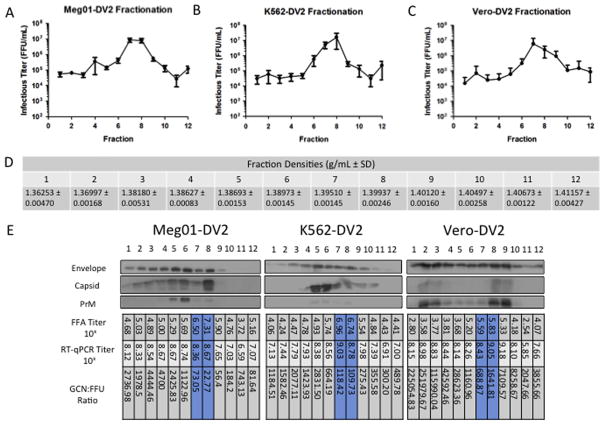
Virus was propagated on a larger scale in Meg01, K562, or Vero cells for 3 days, and supernatants were collected for virus purification. After fractionation through 0–35% potassium tartrate gradients and removal from gradient solutions, virus was assayed for infectivity by FFA. The infectious titers of fractions from all virus purifications performed are displayed (Fig. 4A–C). The data represent seven Meg01-DENV2, five K562-DENV2, and four Vero-DENV2 large-scale purifications. The highest infectious titers were found consistently in either fraction 7 or 8 at the approximate density of 1.39 g/mL (Fig. 5D), which differs from the density specified using cesium chloride gradients (1.22–1.24 g/mL) (Smith et al., 1970; Stevens and Schlesinger, 1965). Virus peaked in fraction 7 with 60% of the K562-DENV2 purifications (3 of 5), 50% of the Vero-DENV2 purifications (2 of 4), and 71% of the Meg01-DENV2 purifications (5 of 7). The variation in localization in fraction 7 vs 8 might be attributable to minor differences in gradient preparation rather than differences in virus density.
Fig. 4.
Characterization of gradient-fractionated Meg01-DENV2, K562-DENV2, or Vero-DENV2. Large-scale batches of DENV2 were propagated in (A) Meg01, (B) K562, or (C) Vero cells and then purified through potassium tartrate-glycerol gradients and fractionated. From each fraction, virus was analyzed by FFU and western blot assays. (A–C) Graphs indicate the mean concentration and SD of infectious virus from all replicates performed (Meg01-DENV2, n=7; K562-DENV2, n=5; and Vero-DENV2, n=4) per fraction. (D) Density readings for fractions 1–12 (n=3). (E) Western blot, FFA titer (FFU/mL), RT-qPCR titer (GCN/mL), and GCN:FFU ratio comparison from a representative DENV2 puri3cation from each cell line. Envelope, capsid, and prM proteins were detected with 4G2, 6F3-1, and GeneTex polyclonal antibody, respectively. Abbreviations: FFA=focus-forming unit assay; FFU=focus-forming unit; GCN=genome copy number; prM=premembrane; RT-qPCR=reverse transcriptase-quantitative polymerase chain reaction; SD=standard deviation.
Fig. 5.
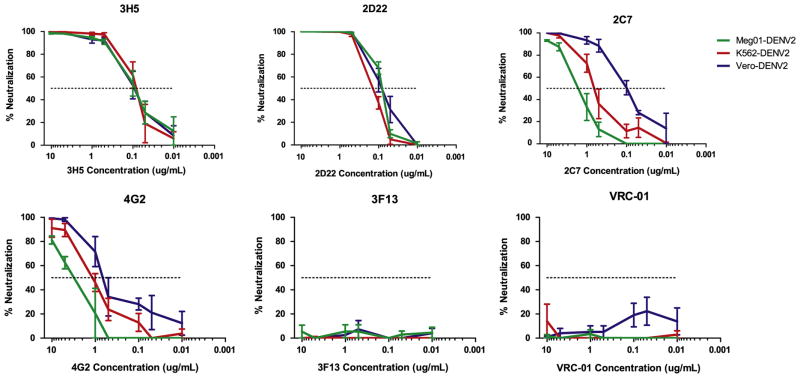
Neutralization assays of Meg01-DENV2, K562-DENV2, or Vero-DENV2 with monoclonal antibodies. Mouse (3H5 and 4G2) or human (2D22, 2C7, and 3F13) anti-DENV2 envelope antibodies or control anti-HIV envelope antibody (VRC-01) were tested for their neutralization capacity via plaque reduction neutralization assays. Graphs indicate the average percent neutralization with decreasing concentrations of antibody (n=3).
Average peak infectious titers for Vero-DENV2 (6.5 × 106 FFU/mL) were generally two times lower than those from the MEP cells lines (Meg01-DENV2, 1.2 ×107 FFU/mL; K562-DENV2, 1.8 × 107 FFU/mL), even though on average about twice as many cells were used to propagate Vero-DENV2. In representative virus purifications, the Vero-DENV2 peak titer was at least 10 times lower than MEP cell line titers (Meg01-DENV2, 2.1 × 107 FFU/mL; K562-DENV2, 9.2 × 106 FFU/mL; Vero-DENV2, 6.8 × 105 FFU/mL) (Fig. 5E), a difference that did not correspond with the starting cell populations. Vero-DENV2 might be considered less-stable through purification processes than MEP-DENV2. However, our observations from immuno-EM imaging experiments did not agree with that notion, based on the observation that Vero-DENV2 was the easiest virion to detect.
GCN titers and GCN:FFU ratios of purified fractions also were evaluated (Fig. 4E). In general, the lowest GCN:FFU ratios from purified and fractionated virus were found in fractions 7 and 8, corresponding with the infectious virus peaks. Similar to the results from viral supernatants, Meg01-DENV2 peaks had the lowest ratios (73.1 GCN/FFU [fraction 7] and 22.8 GCN/FFU [fraction 8]), and Vero-DENV2 had the highest ratios (689 GCN/FFU [fraction 7] and 1640 GCN/FFU [fraction 8]), suggesting again that markedly higher numbers of noninfectious virus are produced in Vero cells relative to MEP cells.
2.4. MEP-DENV2 structural protein fractionation patterns vary from that of Vero-DENV2
Western blots were performed with equal volumes of each fraction to compare protein content from fraction-to-fraction and to compare MEP-DENV2 with Vero-DENV2 (Fig. 4E). Envelope and premembrane (prM) proteins appeared more abundant in Vero-DENV2 than MEP-DENV2 samples in several fractions. On the other hand, capsid protein was more abundant in DENV2 produced in MEP cells.
In general, the amount of envelope protein correlated poorly with the titers of infectious virus, although infectious virus did correspond somewhat with the presence of capsid and prM proteins. These structural proteins from Vero-DENV2 purifications peaked in fractions 8 and 9, close to the infectious virus peaks in 7 and 8. (Note that in this purification similarly high titers [1 × 105 FFU/mL] were present in fraction 9.) A second focus of concentrated protein occurred in Vero-DENV2 fractions 2 and 3, which probably corresponds with the smaller virion structures reported in the literature (Allison et al., 2003; Ferlenghi et al., 2001; Ishikawa and Konishi, 2006; Junjhon et al., 2008). Capsid and prM proteins were present in K562-DENV2 and Meg01-DENV2 infectious virus peaks, although higher concentrations were found in adjacent fractions. Potentially, these bands (which are not present in Vero-DENV2 purifications) resulted from damaged virus particles migrating to slightly lower densities or alternatively resulted from differences in PrM cleavage in MEPs vs Vero. Additional investigations will be needed to delineate the exact origin for this difference.
2.5. Antigenicity of MEP-DENV2 differs at domain I/II of envelope protein
We examined antigenicity because post-translational modifications of viral proteins such as glycosylation are known to vary among host cell types (Bryant et al., 2007; Dejnirattisai et al., 2011; Lee et al., 2010). Neutralizing antibody concentrations were determined by plaque reduction neutralization assay (PRNA) for Meg01-DENV2, K562-DENV2, and Vero-DENV2 with a series of monoclonal antibodies: 3H5, 4G2, 2D22, 2C7, 3F13, and VRC-01 (Fig. 5).
Anti-DIII envelope monoclonal antibodies 3H5 (mouse-derived) and 2D22 (human-derived) neutralized all three viruses similarly. However, neutralization of MEP-DENV2 with anti-envelope DI/II antibodies 4G2 (mouse-derived) or 2C7 (human-derived) required higher levels of antibody. The half maximum inhibitory concentrations (IC50) of 2C7 for Meg01-DENV2 (4.1 μg/mL) and K562-DENV2 (0.45 μg/mL) were elevated markedly in comparison with the IC50 value for Vero-DENV2 (0.11 μg/mL, p < 0.05) (Table 3). The IC50 of 4G2 for Meg01-DENV2 (5.42 μg/mL) also was much higher than that for Vero-DENV2 (0.33 μg/mL, p=0.0011). Most antibodies were capable of neutralizing K562-DENV2 and Vero-DENV2 completely, but 2C7, 4G2, and 3H5 did not fully neutralize Meg01-DENV2. Neutralization with human polyclonal antibodies also was performed. Serum sample DF 3457 neutralized Meg01-DENV2 to a lesser extent, but no difference was detected with endemic plasma (Supplemental Fig. 2), or with plasma of a person from a DENV endemic country. In summary, modest differences in neutralization were observed with Meg01-DENV2, suggesting Meg01-produced DENV2 might be more resistant to neutralization, at least in vitro. As expected, the control non-neutralizing anti-dengue antibody 3F13 and HIV-specific monoclonal antibody VRC-01 did not neutralize dengue virus.
Table 3.
Neutralization capacity (measured by IC50, IC90, and maximum neutralization) of antibodies against Meg01-DENV2, K562-DENV2, or Vero-DENV2.
| Antibody | Meg01-DENV2 | K562-DENV2 | Vero-DENV2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| IC50 (μg/mL) | IC90 (μg/mL) | Max (%) | IC90 (μg/mL) | IC90 (μg/mL) | Max (%) | IC50 (μg/mL) | IC90 (μg/mL) | Max (%) | |
| 2D22 | 0.12 | 1.54 | 100 | 0.18 | 1.88 | 100 | 0.1 | 1.48 | 100 |
| 2C7 | 4.14* | 8.12 | 94.1 | 0.45* | 5.27 | 100 | 0.11 | 1.93 | 100 |
| 4G2 | 5.42** | 10.0 | 87.3 | 0.87 | 13.6 | 98.8 | 0.33 | 5.73 | 100 |
| 3H5 | 0.1 | 1.91 | 99.2 | 0.11 | 1.62 | 100 | 0.11 | 1.86 | 100 |
| 3F13 | NN | NN | 16.7 | NN | NN | 2.13 | NN | NN | 12.1 |
| VRC101 | NN | NN | 10.4 | NN | NN | 42.3 | NN | NN | 38.6 |
Abbreviations: IC50=half maximal inhibitory concentration; IC90=90% maximal inhibitory concentration; and NN=non-neutralizing.
p<0.05 compared with the corresponding value from Vero-DENV2 using student’s t-test.
p=0.0011 compared with the corresponding value from Vero-DENV2 using student’s t-test.
3. Discussion
Megakaryocytes and platelets are dysfunctional in dengue patients, and direct infection of megakaryocytes is one potential attributing factor that might explain this phenomenon. To examine whether cells of the megakaryocyte–erythrocyte lineage can be directly infected by DENV, we took advantage of the readily available Meg01 megakaryoblast and the related K562 erythroid cell lines to assess DENV2 viral growth and virus particle characteristics and compared them with those from the Vero epithelial cell line that is typically used to propagate DENV. Our data suggest DENV2 production in Meg01 and K562 is more efficient than that in Vero, based on their lower viral GCN:PFU ratios and reduced virus particle levels despite easily quantifiable infectious virus. Also, despite similar levels of infectious virus in day 3 cell supernatants, EM analyses of unconcentrated supernatants failed to reveal MEP-DENV2 particles, suggesting that EM particle:infectious virus ratios also might be reduced in these cell lines. This observation is not surprising. DENV particles have rarely been documented directly from patient and rhesus macaque tissues; the virions that have been imaged were found inside of platelets (Noisakran et al., 2009; Noisakran et al., 2012). Because little work has been done to characterize DENV particles directly produced in human patients, the potential that in vivo virus structure differs from cell culture virus remains a viable possibility. Alternative virus structures with different protein content have been suggested for DENV produced in vivo (Hsu et al., 2015).
Ultrastructural studies have indicated that different DENV-infected cell lines display unique features (e.g., convoluted structures are absent in the insect cell line C6/36 and crystalloid structures rarely form in cell lines) (Junjhon et al., 2014); thus analyses of the megakaryocyte–erythrocyte lineage were conducted. Our previous report evaluating mature megakaryocytes indicated that they produced DENV2, with an abundance of virus-like particles in the cytoplasm (Clark et al., 2012). However, EM analyses with Meg01 and K562 suggested a far more controlled production of classical virus particles – 50 nm electron dense structures within ER-derived vesicles. Although the differentiated megakaryocytes contained virus and crystalloid structures, it is possible that a number of the virus-induced structures observed in that report might have been polysomes (strings of ribosomes linked together by mRNA). Polysomes (also known as dense particles) are electron dense and approximately the same size as the virus core (Hase et al., 1987; Ko et al., 1979; Sriurairatna et al., 1973); they are indicative of high levels of protein production and were numerous in the cytoplasm of DENV2-infected Vero cells in this study. In contrast, we did not observe abundant polysome-like structures in Meg01 or K562 cells, which could reflect reduced viral protein production and account for the lower levels of virus particle assembly relative to Vero cells.
This study supports the concept that abundant virion production is not required for high infectious titers. Although high levels of viral protein production can be observed in kidney epithelial (Vero) cells, these observations should be evaluated cautiously because kidney cells are not likely natural targets of DENV infection in vivo. Kidney cell lines have a tendency to produce noninfectious subviral virions, while western blot results of MEP-DENV2 did not indicate the presence of these types of particles. DENV2 protein production in MEP cell lines appeared to be coordinated, leading to lower amounts of excessive viral protein production, thereby reducing the likelihood of immune recognition. Many mutations associated with reduced virus production have already been identified (Junjhon et al., 2008; Lee et al., 2010; Pryor et al., 2004; Yoshii et al., 2004) and could potentially play a role in shaping virus particle production in MEP cells.
In addition to differences in virus particle production, we also found minor variations in virus composition and structure. While the three cell lines examined propagated virus of comparable morphology and density, MEP-DENV2 appeared to have less prM protein. DENV is known to be unique among the flaviviruses for its inherently inefficient prM cleavage process, which is facilitated by a mutation in the prM trypsin cleavage recognition site that inhibits cleavage (Junjhon et al., 2008). Less prM was noted in purified fractions of MEP-DENV2, which could indicate more proficient cleavage and virus maturation, potentially explaining the efficient infectious virus production observed in this report. Also, antigenic composition presented subtle differences. In particular, we observed differences in neutralization with envelope domain I/II antibodies, which could be significant since many potent anti-DENV neutralizing antibodies produced in humans are directed against this epitope, and poorly neutralizing domain I/II antibodies do not correlate with protection (Beltramello et al., 2010; de Alwis et al., 2012; Shrestha et al., 2010; Smith et al., 2012; Wahala et al., 2009). In one example, in vitro neutralization of Vero-DENV2 was demonstrated clearly with serum from vaccinated volunteers; however, no protection was observed against this serotype in vaccine recipients (Sabchareon et al., 2012). Potentially, this discrepancy might be attributable to differential antigenicity of the envelope domain I/II protein epitopes displayed on Vero-DENV2 and on in vivo-DENV2. Data in this report suggests that vaccine recipient serum might neutralize Meg01-DENV2 differently from Vero-DENV2. The importance of these differences in antigenicity remains to be fully elucidated, but in vivo protection in the aforementioned vaccine study could have been predicted better potentially with neutralization assays involving virus propagated in a target cell line, such as Meg01-DENV2. Additional work examining Meg01-DENV antigenicity and the role of the megakaryocyte lineage in DENV pathogenesis is warranted. This new system for propagating infectious DENV provides a new tool for the design of dengue vaccines and for the evaluation of antiviral compounds.
4. Materials and methods
4.1. Virus and cells
The DENV strain used in these experiments was 16681 (DENV serotype 2) originally grown in Vero-E6 cells. This virus is referred to here as Vero-DENV2. Stocks of Vero-DENV2 were propagated once in Meg01 cells (Meg01-DENV2) or K562 cells (K562-DENV2). Meg01 cells were a gift from Dr. Ofori-Acquah at Emory University. Vero and K562 cells were grown in RPMI medium (Cellgro, Manassas, VA) with 10% fetal bovine serum (FBS) (Atlanta Biologicals) and penicillin-streptomycin (PS) (Cellgro), while Meg01 was cultured in RPMI with PS and 20% FBS. (Meg01 had poor growth kinetics at low cell densities; high FBS concentrations were used to ensure continuous doubling and permissiveness). All infected cells were maintained in RPMI medium supplemented with PS 10% FBS medium, unless otherwise specified. For imaging studies, we used exosome-free FBS (prepared by centrifuging FBS at 100,000g for 18 h and passing through a 0.2 μm cellulose acetate filter unit [Corning]).
4.2. Comparison of virus growth kinetics in different cell lines
Vero-DENV2 was propagated in 2 ×106 cells of Meg01, K562, or Vero cells by inoculation with an MOI of 0.1 FFU/cell. Cells were incubated with virus for 2 h in a 15 mL polypropylene conical tube in a CO2 incubator at 37 °C, washed three times with D-PBS (Lonza, Walkersville, MI), and resuspended to a final concentration of 5 × 105 cells/mL with RPMI 10% FBS (K562 and Vero cells) or RPMI 20% FBS (for Meg01 cells) in T25 flasks (Corning). Medium was added and aliquots were taken daily from day 2–7. Samples were analyzed via plaque assay and RT-quantitative PCR.
4.3. Plaque assay and plaque reduction neutralization assay
Cells were seeded into either 6-or 12-well plates (Falcon, Durham, NC) the day before the experiment. For regular plaque assays, virus was 10-fold serially diluted in medium. Medium was removed from plates, virus dilutions applied in duplicate, and incubated at 37 °C for 1 h.
For Plaque Reduction Neutralization Assays (PRNAs), cells were seeded in a similar manner. Antibodies were serially diluted in RPMI 5% FBS medium. Viruses also were diluted in RPMI 5% FBS and mixed equal-volume with the antibody dilutions. A no-antibody control (~1000 PFU/reaction) was treated in a similar manner and used as the virus titration control. Samples were incubated in cell culture incubators at 37 °C for 1 h. After the incubation period, virus was diluted to a final volume of 1 mL and 5% of the reaction was applied to wells. Additional medium was added to cover the cells, and plates were incubated at 37 °C for 30 min.
For both plaque assays and PRNAs, cells and inocula were overlaid with 1.5% methylcellulose 1500 cps (MP Biomedicals, Solon, OH) medium (0.5X RPMI, 5% FBS, PS, pH 8.0) and incubated at 37 °C for 12 days. With PRNAs involving polyclonal human antibodies, the same medium except with a different methylcellulose (1.0% of 1500–5600 cps [Fisher Science Education]) was used and harvested on day 7. Plaques were visualized by staining monolayers with 0.1% crystal violet (Sigma-Aldrich) in 20% methanol before counting.
4.4. Reverse transcription-quantitative polymerase chain reaction
RNA was isolated from virus supernatants or concentrates with QIAamp viral RNA mini kit or EZ-1 virus mini kit v2.0 (Qiagen) using the manufacturer’s protocol. RNA was reverse transcribed into cDNA and amplified in a one-step reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay with LightCycler 480 RNA Master Hydrolysis Probe (Roche) using primers (DENV2U and DVL1) and probe (DVP1) for 40 cycles of 95 °C (15 sec) and 60 °C (1 min) on LightCycler 480 II (Roche), similar to a previous publication (Houng et al., 2001).
4.5. Antibodies
Mouse monoclonal antibodies 4G2 and 3H5 (CTK Biotech, San Diego, CA) specific to DENV2 envelope proteins were used in various assays. Anti-capsid 6F3-1 hybridoma supernatant and anti-polyclonal prM antibody (Genetex) were used in western blot assays. Human monoclonal antibodies (2C7, 2D22, and 3F13) were used in PRNAs. Convalescent patient serum samples used in PRNAs was provided by Dr. Chokephaibulkit, Dr. Pattanapanyasat, and Patcharee Songprakhone from Siriraj Hospital in Bangkok, Thailand. Endemic plasma (or plasma from a healthy donor native to a country endemic for DENV) was obtained through Emory University's blood donation program. VRC-01 (Mapp Biopharmaceutical, San Diego, CA), a human anti-HIV envelope monoclonal antibody, was used in PRNA as a negative control.
4.6. Virus purification
Vero-DENV2, K562-DENV2, and Meg01-DENV2 were propagated in a similar manner as described for the growth kinetics experiments. Vero cells: T162 flasks (Falcon, Durham, NC) were seeded with cells days before and about 4.8 × 108 cells were inoculated at an MOI of 0.1 FFU/cell with Vero-DENV2 stock virus. Inocula were removed and replaced with 45 mL of RPMI PS 10% FBS (exosome-free) medium. Meg01 or K562: 1–4 ×108 cells were inoculated at an MOI of 0.02 FFU/cell. Cells were incubated in T162 flasks for 1–2 h. Cells were washed three times with RPMI PS medium and resuspended to a final volume of 5 ×105 cells/mL in RPMI PS 10% exosome-free FBS. After 3 days of propagation, supernatant was clarified at 3,000 rpms for 30 min. Supernatant was treated with polyethylene glycol (PEG) 8,000 (Fisher BioReagents, Fair Lawn, NJ) solution (final concentration: 8% PEG 8,000, 1 M NaCl, 5 mM EDTA, pH 8.5) overnight. Virus was concentrated with a Beckman Optima L-70K ultracentrifuge at 12,000 rpm in SW28Ti rotors for 25 min and resuspended in TNE buffer (50 mM Tris–HCl, 75 mM NaCl, 5 mM EDTA, pH 8.0). Concentrated virus was fixed in 2% paraformaldehyde (Sigma-Aldrich) in TNE buffer (final pH 7.0) for EM. Continuous potassium tartrate dibasic hemihydrate (Sigma-Aldrich, St. Louis, MO) (0–35% w/w)-glycerol (30–12.5% w/w) gradients were formed with Gradient Master IP 107 (BioComp) using glycerol program 10–20% (v/v) in 14 × 89 mm ultraclear tubes. Concentrates were centrifuged in an SW41Ti rotor at 40 K rpm for 16–18 h. Twelve fractions were isolated by pipette, starting from the top of the gradient. An aliquot of each fraction was taken from some gradients (Meg01-mock, K562-mock, and Vero-mock) and averaged to determine the buoyant density with a Bausch & Lomb refractometer. Fractions were diluted with TNE buffer and centrifuged in the SW28Ti rotor at 28 K rpm for 1.5 h. Virus was resuspended in TNE buffer and aliquoted for further analyses.
4.7. Negative-staining immuno-EM and thin-sectioning EM
For immuno-EM, samples were fixed in 2% paraformaldehyde in TNE buffer and given to the Robert P. Apkarian integrated electron microscopy core service at Emory. Samples were applied to carbon-coated grids, incubated with DENV2 envelope-specific primary antibody (3H5), gold-conjugated anti-mouse secondary antibody, and tungsten stained.
For thin-sectioning EM, DENV2-infected K562, Meg01, and Vero cells at 1 and 2 days post-inoculation were washed twice with D-PBS, fixed in 4% glutaraldehyde in phosphate buffer overnight, and given to the EM core. The cells were processed for thin-sectioning EM as previously reported (Noisakran et al., 2009). Using IMOD imaging software (http://bio3d.colorado.edu/imod/), multiple images of different sections of the same cell were acquired and assembled together into one continuous cell image. A total of 20 Meg01, 27 K562, and 20 Vero single cell cross-sections were examined. In cell image analyses, a virus particle was defined as a circular electron dense object in the 30–60 nm range that appeared to be enclosed within the ER or an ER-derived membrane vesicle. Replication complexes were larger, circular, mostly-empty objects that also were enclosed within ER-derived membranes. Crystalloids were defined as a cluster of at least five virions that were not aligned linearly.
4.8. Focus-forming unit assay
Flat bottom 96-well plates (Celltreat) were seeded with 2 × 104 Vero cells per well the day before titration. Medium was removed from 96-well plates and 10-fold serial dilutions of virus samples were applied in duplicate. Plates were incubated for 1–2 h at 37 °C. Subsequently, cells and inocula were overlayed with 1.5% methylcellulose cps 1,500 medium (1X EMEM [Lonza, Walkersville, MI], 5% FBS, 2 mM L-glutamine, 10 mM HEPES, PS) and incubated for 3 days. Cells were washed three times in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.5), and fixed in 3.7% formaldehyde for 1 h at room temperature or overnight at 4 °C. Cells were permeabilized for 10 min with 1% triton X-100 (Acros) in PBS and washed five times with PBS. Monolayers were blocked with 2% normal goat serum (Jackson Immuno Research) in PBS for 1 h and then incubated with 10 μg/mL 4G2 in PBS for 1 h at 37 °C. After three washes, monolayers were incubated with goat anti-mouse IgG-HRP human absorbed antibody (Southern Biotech) in PBS-Tween 20 for 1 h at 37 °C. After three washes, foci were incubated in DAB (0.6 mg/mL 3,3′-diaminobenzidine tetrahydrochloride [Sigma-Aldrich], 0.08% NiCl2, 0.01% H2O2 in PBS) until development was complete.
4.9. Western blot
Purified virus fractions (1–12) were diluted in 4x SDS-PAGE loading buffer (160 mM Tris, 6.4% SDS, 20% glycerol), loaded onto 10% or 12% separating SDS-polyacrylamide (Bio-Rad) gels and run in Tris-Glycine-SDS (TGS) running buffer at 90 v for 2–3 h with EPS 1001 power supply (General Electric). For western blots with 4G2, samples were not heated or reduced; for blots with 6F3-1, samples were heated; and for prM, samples were heated and reduced with β-mercaptoethanol. Gels were transferred to methanol-pretreated PVDF membranes (Bio-Rad) in transfer buffer (2.5 mM Tris, 19.2 mM glycine, 20% methanol) for 15–17 h at 30 v. Membranes were blocked for 1 h at room temperature with blocking buffer (5% milk in PBS-Tween 20). Membranes were incubated with 4G2 (10 μg/mL, 1 h), 6F3-1 (neat, 2 h), prM antibody (1:1,000, 2 h) in blocking buffer. After five washes with PBS-Tween 20, membranes were incubated 1 h with 1:1,000 dilution of appropriate secondary anti-mouse or anti-rabbit IgG-AP conjugated antibody. After washing, a 30-min incubation with Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega, Madison, WI) allowed for visualization of viral antigens.
4.10. Funding information
Funders played no role in study design, data collection, interpretation, preparation or the decision to submit the work for publication. This research received no specific grant from any funding agency in the public, commercial, or nonprofit sector. RFS is supported in part from the Department of Veterans Affairs.
Supplementary Material
Acknowledgments
Dr. Wright, Dr. Yi, Dr. Hampton, and staff from the Emory University Robert P. Apkarian Integrated Electron Microscopy Core provided EM expertise and processed and imaged virus and virus-infected cells.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2016.03.024.
References
- AbuBakar S, Shu MH, Johari J, Wong PF. Senescence affects endothelial cells susceptibility to dengue virus infection. Int J Med Sci. 2014;11:538–544. doi: 10.7150/ijms.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ader DB, Celluzzi C, Bisbing J, Gilmore L, Gunther V, Peachman KK, Rao M, Barvir D, Sun W, Palmer DR. Modulation of dengue virus infection of dendritic cells by Aedes aegypti saliva. Viral Immunol. 2004;17:252–265. doi: 10.1089/0882824041310496. [DOI] [PubMed] [Google Scholar]
- Allison SL, Tao YJ, O’Riordain G, Mandl CW, Harrison SC, Heinz FX. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J Virol. 2003;77:11357–11366. doi: 10.1128/JVI.77.21.11357-11366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White AM, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci USA. 2012;109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo MT, Simpson-Haidaris PJ, Kou Z, Schlesinger JJ, Jin X. Primary human endothelial cells support direct but not antibody-dependent enhancement of dengue viral infection. J Med Virol. 2009;81:519–528. doi: 10.1002/jmv.21408. [DOI] [PubMed] [Google Scholar]
- Basu A, Jain P, Gangodkar SV, Shetty S, Ghosh K. Dengue 2 virus inhibits in vitro megakaryocytic colony formation and induces apoptosis in thrombopoietin-inducible megakaryocytic differentiation from cord blood CD34+cells. FEMS Immunol Med Microbiol. 2008;53:46–51. doi: 10.1111/j.1574-695X.2008.00399.x. [DOI] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierman HR, Nelson ER. Hematodepressive virus diseases of Thailand. Ann Intern Med. 1965;62:867–884. doi: 10.7326/0003-4819-62-5-867. [DOI] [PubMed] [Google Scholar]
- Bryant JE, Calvert AE, Mesesan K, Crabtree MB, Volpe KE, Silengo S, Kinney RM, Huang CY, Miller BR, Roehrig JT. Glycosylation of the dengue 2 virus E protein at N67 is critical for virus growth in vitro but not for growth in intrathoracically inoculated Aedes aegypti mosquitoes. Virology. 2007;366:415–423. doi: 10.1016/j.virol.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Butthep P, Bunyaratvej A, Bhamarapravati N. Dengue virus and endothelial cell: a related phenomenon to thrombocytopenia and granulocytopenia in dengue hemorrhagic fever. Southeast Asian J Trop Med Public Health. 1993;24(1):246–249. [PubMed] [Google Scholar]
- Clark KB, Noisakran S, Onlamoon N, Hsiao HM, Roback J, Villinger F, Ansari AA, Perng GC. Multiploid CD61+cells are the pre-dominant cell lineage infected during acute dengue virus infection in bone marrow. Plos One. 2012;7:e52902. doi: 10.1371/journal.pone.0052902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughaday CC, Brandt WE, McCown JM, Russell PK. Evidence for two mechanisms of dengue virus infection of adherent human monocytes: trypsin-sensitive virus receptors and trypsin-resistant immune complex receptors. Infect Immun. 1981;32:469–473. doi: 10.1128/iai.32.2.469-473.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Webb AI, Chan V, Jumnainsong A, Davidson A, Mon-gkolsapaya J, Screaton G. Lectin switching during dengue virus infection. J Infect Dis. 2011;203:1775–1783. doi: 10.1093/infdis/jir173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J Virol. 2000;74:7814–7823. doi: 10.1128/jvi.74.17.7814-7823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, Harrison SC, Rey FA, Fuller SD. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell. 2001;7:593–602. doi: 10.1016/s1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis. 2006;19:429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- Hase T, Summers PL, Eckels KH, Baze WB. An electron and immunoelectron microscopic study of dengue-2 virus infection of cultured mosquito cells: maturation events. Arch Virol. 1987;92:273–291. doi: 10.1007/BF01317484. [DOI] [PubMed] [Google Scholar]
- Ho LJ, Wang JJ, Shaio MF, Kao CL, Chang DM, Han SW, Lai JH. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J Immunol. 2001;166:1499–1506. doi: 10.4049/jimmunol.166.3.1499. [DOI] [PubMed] [Google Scholar]
- Houng HS, Chung-Ming Chen R, Vaughn DW, Kanesa-thasan N. Development of a fluorogenic RT-PCR system for quantitative identification of dengue virus serotypes 1–4 using conserved and serotype-specific 3′ noncoding sequences. J Virol Methods. 2001;95:19–32. doi: 10.1016/s0166-0934(01)00280-4. [DOI] [PubMed] [Google Scholar]
- Hsu AY, Wu SR, Tsai JJ, Chen PL, Chen YP, Chen TY, Lo YC, Ho TC, Lee M, Chen MT, Chiu YC, Perng GC. Infectious dengue vesicles derived from CD61+cells in acute patient plasma exhibited a diaphanous appearance. Sci Rep. 2015;5:17990. doi: 10.1038/srep17990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Konishi E. Mosquito cells infected with Japanese encephalitis virus release slowly-sedimenting hemagglutinin particles in association with intracellular formation of smooth membrane structures. Microbiol Immunol. 2006;50:211–223. doi: 10.1111/j.1348-0421.2006.tb03788.x. [DOI] [PubMed] [Google Scholar]
- Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol. 2014;88:4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junjhon J, Lausumpao M, Supasa S, Noisakran S, Songjaeng A, Saraithong P, Chaichoun K, Utaipat U, Keelapang P, Kanjanahaluethai A, Puttikhunt C, Kasinrerk W, Malasit P, Sittisombut N. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J Virol. 2008;82:10776–10791. doi: 10.1128/JVI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho LK, Wulur H, Himawan T. Blood and bone marrow changes in dengue haemorrhagic fever. Paediatr Indones. 1972;12:31–39. [PubMed] [Google Scholar]
- Ko KK, Igarashi A, Fukai K. Electron microscopic observations on Aedes albopictus cells infected with dengue viruses. Arch Virol. 1979;62:41–52. doi: 10.1007/BF01314902. [DOI] [PubMed] [Google Scholar]
- La Russa VF, Innis BL. Mechanisms of dengue virus-induced bone marrow suppression. Baillieres Clin Haematol. 1995;8:249–270. doi: 10.1016/s0950-3536(05)80240-9. [DOI] [PubMed] [Google Scholar]
- Lee E, Leang SK, Davidson A, Lobigs M. Both E protein glycans adversely affect dengue virus infectivity but are beneficial for virion release. J Virol. 2010;84:5171–5180. doi: 10.1128/JVI.01900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IK, Hsieh CJ, Chen RF, Yang ZS, Wang L, Chen CM, Liu CF, Huang CH, Lin JW, Chen YH, Yang KD, Liu JW. Increased production of interleukin-4, interleukin-10, and granulocyte-macrophage colony-stimulating factor by type 2 diabetes’ mononuclear cells infected with dengue virus, but not increased intracellular viral multiplication. Biomed Res Int. 2013;2013:965853. doi: 10.1155/2013/965853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libraty DH, Pichyangkul S, Ajariyakhajorn C, Endy TP, Ennis FA. Human dendritic cells are activated by dengue virus infection: enhancement by gamma interferon and implications for disease pathogenesis. J Virol. 2001;75:3501–3508. doi: 10.1128/JVI.75.8.3501-3508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozzio BB, Lozzio CB, Bamberger EG, Feliu AS. A multipotential leukemia cell line (K-562) of human origin. Proc Soc Exp Biol Med. 1981;166:546–550. doi: 10.3181/00379727-166-41106. [DOI] [PubMed] [Google Scholar]
- Marianneau P, Steffan AM, Royer C, Drouet MT, Jaeck D, Kirn A, Deubel V. Infection of primary cultures of human Kupffer cells by Dengue virus: no viral progeny synthesis, but cytokine production is evident. J Virol. 1999;73:5201–5206. doi: 10.1128/jvi.73.6.5201-5206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera JA, Hernandez JP, Valero N, Espina LM, Anez GJ. Ultra-structural studies on dengue virus type 2 infection of cultured human monocytes. Virol J. 2005;2:26. doi: 10.1186/1743-422X-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao S, Lai CJ, Young NS. Dengue virus, a flavivirus, propagates in human bone marrow progenitors and hematopoietic cell lines. Blood. 1989;74:1235–1240. [PubMed] [Google Scholar]
- Nelson ER, Bierman HR, Chulajata R. Hematologic Findings in the 1960 Hemorrhagic Fever Epidemic (Dengue) in Thailand. Am J Trop Med Hyg. 1964;13:642–649. doi: 10.4269/ajtmh.1964.13.642. [DOI] [PubMed] [Google Scholar]
- Nielsen DG. The relationship of interacting immunological components in dengue pathogenesis. Virol J. 2009;6:211. doi: 10.1186/1743-422X-6-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noisakran S, Onlamoon N, Hsiao HM, Clark KB, Villinger F, Ansari AA, Perng GC. Infection of bone marrow cells by dengue virus in vivo. Exp Hematol. 2012;40:250–259. e254. doi: 10.1016/j.exphem.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noisakran S, Gibbons RV, Songprakhon P, Jairungsri A, Ajariyakhajorn C, Nisalak A, Jarman RG, Malasit P, Chokephaibulkit K, Perng GC. Detection of dengue virus in platelets isolated from dengue patients. Southeast Asian J Trop Med Public Health. 2009;40:253–262. [PubMed] [Google Scholar]
- O’Rourke F. Observations on pool and capillary feeding in Aedes aegypti. Nature. 1956;177:1087–1088. [Google Scholar]
- Ogura M, Morishima Y, Ohno R, Kato Y, Hirabayashi N, Nagura H, Saito H. Establishment of a novel human megakaryoblastic leukemia cell line, MEG-01, with positive Philadelphia chromosome. Blood. 1985;66:1384–1392. [PubMed] [Google Scholar]
- Pasteur S. Sanofi Pasteur’s dengue vaccine candidate successfully completes final landmark phase iii clinical efficacy study in Latin America. Sanofi Pasteur 2014 [Google Scholar]
- Pryor MJ, Azzola L, Wright PJ, Davidson AD. Histidine 39 in the dengue virus type 2 M protein has an important role in virus assembly. J Gen Virol. 2004;85:3627–3636. doi: 10.1099/vir.0.80283-0. [DOI] [PubMed] [Google Scholar]
- Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. 2012;380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O’Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. Plos Pathog. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE., Jr Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol. 2012;86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Brandt WE, Swanson JL, McCown JM, Buescher EL. Physical and biological properties of dengue-2 virus and associated antigens. J Virol. 1970;5:524–532. doi: 10.1128/jvi.5.4.524-532.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriurairatna S, Bhamarapravati N, Phalavadhtana O. Dengue virus infection of mice: morphology and morphogenesis of dengue type-2 virus in suckling mouse neurones. Infect Immun. 1973;8:1017–1028. doi: 10.1128/iai.8.6.1017-1028.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TM, Schlesinger RW. Studies on the nature of dengue viruses. I. Correlation of particle density, infectivity, and RNA content of type 2 virus. Virology. 1965;27:103–112. doi: 10.1016/0042-6822(65)90147-9. [DOI] [PubMed] [Google Scholar]
- Styer LM, Kent KA, Albright RG, Bennett CJ, Kramer LD, Bernard KA. Mosquitoes inoculate high doses of West Nile virus as they probe and feed on live hosts. Plos Pathog. 2007;3:1262–1270. doi: 10.1371/journal.ppat.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Fernandez S, Marovich MA, Palmer DR, Celluzzi CM, Boonnak K, Liang Z, Subramanian H, Porter KR, Sun W, Burgess TH. Functional characterization of ex vivo blood myeloid and plasmacytoid dendritic cells after infection with dengue virus. Virology. 2009;383:207–215. doi: 10.1016/j.virol.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Takasaki T, Takada K, Kurane I. Electron microscopic study of persistent dengue virus infection: analysis using a cell line persistently infected with Dengue-2 virus. Intervirology. 2001;44:48–54. doi: 10.1159/000050030. [DOI] [PubMed] [Google Scholar]
- Tan TY, Chu JJ. Dengue virus-infected human monocytes trigger late activation of caspase-1, which mediates pro-inflammatory IL-1beta secretion and pyroptosis. J Gen Virol. 2013;94:2215–2220. doi: 10.1099/vir.0.055277-0. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN, Brandt WE, Russell PK, Dixon FT. Replication of dengue-2 virus in cultured human lymphoblastoid cells and subpopulations of human peripheral leukocytes. J Immunol. 1976;117:953–961. [PubMed] [Google Scholar]
- Tsai JJ, Liu LT, Chang K, Wang SH, Hsiao HM, Clark KB, Perng GC. The importance of hematopoietic progenitor cells in dengue. Ther Adv Hematol. 2012;3:59–71. doi: 10.1177/2040620711417660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology. 2009;392:103–113. doi: 10.1016/j.virol.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Dengue and Severe Dengue Fact Sheet N117, Dengue and Severe Dengue. World Health Organization; Geneva, Switzerland: 2015. [Google Scholar]
- Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Frankel SS. Human skin Langerhans cells are targets of dengue virus infection. Nat Med. 2000;6:816–820. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- Yoshii K, Konno A, Goto A, Nio J, Obara M, Ueki T, Hayasaka D, Mizutani T, Kariwa H, Takashima I. Single point mutation in tick-borne encephalitis virus prM protein induces a reduction of virus particle secretion. J Gen Virol. 2004;85:3049–3058. doi: 10.1099/vir.0.80169-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.