Abstract
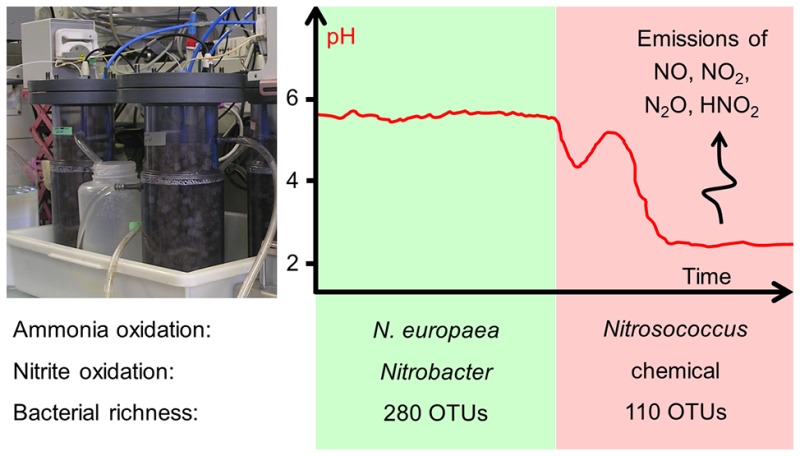
Ammonia oxidation decreases the pH in wastewaters where alkalinity is limited relative to total ammonia. The activity of ammonia oxidizing bacteria (AOB), however, typically decreases with pH and often ceases completely in slightly acidic wastewaters. Nevertheless, nitrification at low pH has been reported in reactors treating human urine, but it has been unclear which organisms are involved. In this study, we followed the population dynamics of ammonia oxidizing organisms and reactor performance in synthetic fully hydrolyzed urine as the pH decreased over time in response to a decrease in the loading rate. Populations of the β-proteobacterial Nitrosomonas europaea lineage were abundant at the initial pH close to 6, but the growth of a possibly novel Nitrosococcus-related AOB genus decreased the pH to the new level of 2.2, challenging the perception that nitrification is inhibited entirely at low pH values, or governed exclusively by β-proteobacterial AOB or archaea. With the pH shift, nitrite oxidizing bacteria were not further detected, but nitrous acid (HNO2) was still removed through chemical decomposition to nitric oxide (NO) and nitrate. The growth of acid-tolerant γ-proteobacterial AOB should be prevented, by keeping the pH above 5.4, which is a typical pH limit for the N. europaea lineage. Otherwise, the microbial community responsible for high-rate nitrification can be lost, and strong emissions of hazardous volatile nitrogen compounds such as NO are likely.
1. Introduction
Ammonia oxidation to nitrite, the first step of nitrification, is a biological process that releases protons. Ammonia oxidation can substantially decrease the pH in terrestrial and aquatic systems that do not contain sufficient alkalinity to buffer the proton release. This can for instance happen in acidic soils or wastewaters with a low alkalinity to total ammonia ratio.
Ammonia oxidizing bacteria (AOB) in wastewater treatment, however, are typically found to be acid-sensitive: the activity of AOB was found to decrease with pH and to completely cease at pH values slightly below pH 6.1 Occasional reports indicate that ammonia oxidation can still occur at lower pH. It was observed that ammonia oxidation proceeds at pH values of around 4 in engineered reactors containing synthetic wastewaters.2−5 In nitrified urine, the pH dropped to values as low as 2.5.6 The minimal pH value of 2.5 is stunning, as a lower pH limit of 2.9 was demonstrated for ammonia oxidation in acidic tea soils7 and as nitrification is not expected at pH values below 3 in acidic lakes.8 Ammonia oxidation in urine was shown to be due to biological activity.6 However, it is not clear which organisms were involved.
Low pH values can be reached during nitrification of urine. Stored human urine contains an alkalinity to total ammonia ratio of 1 mol·mol–1.9 A minimal molar ratio of 2 mol·mol–1 would be required for complete ammonia oxidation. Consequently, only 50% of the total ammonia in urine is oxidized until most of the alkalinity has been consumed and the pH has dropped substantially. The drop of pH to very low values is a concern for engineered reactors: at pH values below 4.5, nitrous acid (HNO2) decomposes chemically.6 It was observed that during chemical HNO2 decomposition around 16% of the transformed nitrogen was lost by volatilization, partially in the form of harmful gases (HNO2, nitric oxide, and nitrous oxide).6 Nitrogen losses in urine nitrification reactors at neutral pH values, where no chemical HNO2 conversion takes place, are expected to be negligible,10 except if nitrite accumulates.11 Hence, a fundamental knowledge on involved organisms and processes is important in order to prevent these strong off-gas emissions.
The main population of AOB in urine nitrification reactors at neutral pH values was found to be affiliated with the Nitrosomonas europaea lineage.12 Their activity was shown to cease at pH values close to 5.4.12 Hence, it has to be expected that a population shift from this acid-sensitive to other, acid-tolerant, AOB is responsible for low-pH nitrification in urine. A complete population shift from Nitrosomonas europaea to Nitrosomonas oligotropha has been observed in a reactor operated with synthetic low-strength nitrogen wastewater as the pH dropped from above 6 to 4.5.5 However, the wastewater used in these experiments contained far lower salt and total ammonia concentrations than the concentrations expected in urine.9Nitrosomonas oligotropha have a high ammonia affinity, but also a high salt sensitivity.13 Hence, it remains unclear whether these AOB are also selected in wastewater with high ammonia concentrations, such as urine.
Several AOB are better adapted to high salt concentrations, for example, the γ-proteobacterial AOB (e.g., genus Nitrosococcus).14 Based on morphological observations, AOB were hypothesized to be active at a pH value as low as 2.9 in acidic tea soils belong to the genus of Nitrosococcus.7 However, γ-proteobacterial AOB are predominantly found in marine environments15 and have not been detected in wastewater treatment reactors.16 Recent studies showed that ammonia oxidizing archaea (AOA) outnumber AOB at low pH values in the soil,17 and play a more important role than AOB in strongly acidic soils.18 While the relative abundance of AOA is low compared to the relative abundance of AOB in municipal wastewater treatment,19 the occurrence of AOA in wastewater reactors at low pH values has, to our knowledge, so far not been investigated.
The growth of bacteria in acidic environments requires specific adaptation mechanisms: bacteria need to keep their cell internal pH values close to neutrality against the extracellular pH, a phenomenon known as pH homeostasis.20 One known mechanism of pH homeostasis is the uptake of potassium ions, which allows for the inversion of the membrane potential and decreases the proton pressure on the cytoplasmic membrane.21
The aim of this study was to select for the ammonia oxidizing organisms that drive the pH in wastewater with high ammonia concentrations to very low values and to investigate how the selection of these organisms affect the reactor performance and the overall bacterial community structure. The bacterial population dynamics and reactor performance in wastewater with high ammonia concentrations were compared with parallel reactors operated using wastewater with low ammonia concentrations. The availability of potassium ions was altered to test its importance for bacterial survival at low pH.
2. Materials and Methods
2.1. Reactor Operation under Continuous-Flow Regime
Reactor Configurations
Four moving bed biofilm reactors (MBBR) with a volume of 2 L each were operated under continuous-flow conditions. Each reactor was filled with 40% (volumetric ratio) K1 Kaldnes biofilm carriers with a specific surface area of 500 m2·m–3.22 The reactor temperature was adjusted to 25.4 ± 0.1 °C with a thermostat (F32, Julabo Labortechnik GmbH, Seelbach, Germany). To maintain constant nitrogen loading rates, as detailed below, reactors were supplied with influent at specific volumetric flow rates (REGLO Digital, ISMATEC, Wertheim, Germany). A sufficient mixing of biofilm carriers was ensured by aeration with pressurized, premoistened, ambient air at 35 NL·h–1 (22R1411/01807, Wisag, Fällanden, Switzerland). In combination with low nitrification rates, the high air flow maintained the dissolved oxygen close to saturation. Online pH monitoring, the setup for batch experiments, and the characteristics of the inoculum are described in the Supporting Information.
Influent Compositions
The experimental design consisted of four reactors fed with different synthetic influent solutions to investigate the effects of urine and wastewater matrices, and of potassium and sodium cations (Table 1). Two so-called urine reactors (UR) were supplied with influent that contained total ammonia and total salt concentrations similar to women’s urine,23 but varied in their potassium and sodium concentrations. Ammonia rather than urea was added to the synthetic solutions, because urea decomposes very quickly in urine collection and storage systems.24 Urea is therefore completely decomposed in most cases as it enters a urine treatment facility, for example, a nitrification reactor. Two wastewater reactors (WWR) were fed with a synthetic substrate containing lower total ammonia and total salt concentrations, and high potassium (WWR-K) or sodium (WWR-Na) concentrations. Influents with high potassium (UR-K, WWR-K) or sodium concentrations (UR-Na, WWR-Na) should provide information on the necessity of potassium for AOB growth at low pH values. The recipes of all synthetic influent solutions are given in Table S1. Micro- and macronutrients were added as specified in Table S2. The influent solutions did not contain organic substances. The liquid phase sampling and chemical analyses are described in the Supporting Information. The relative standard deviation for liquid phase analysis was below 4% for all compounds.
Table 1. Average Measured Concentrations of Ammonium and Accompanying Salts in the Reactor Influent Solutionsa.
| UR-K | UR-Na | WWR-K | WWR-Na | ||
|---|---|---|---|---|---|
| pH | 9.18 ± 0.06 | 9.32 ± 0.07 | 8.09 ± 0.34 | 8.16 ± 0.33 | |
| NH4–N | mg·L–1 | 1710 ± 140 | 1630 ± 90 | 149 ± 8 | 145 ± 16 |
| TIC | mgC·L–1 | 753 ± 60 | 695 ± 123 | 219 ± 10 | 211 ± 24 |
| PO4–P | mg·L–1 | 146 ± 6 | 138 ± 13 | 11.0 ± 1.5 | 11.4 ± 1.6 |
| Cl | mg·L–1 | 1740 ± 100 | 1550 ± 130 | 387 ± 22 | 381 ± 33 |
| Na | mg·L–1 | 5.59 ± 0.40 | 1160 ± 130 | 6.20 ± 0.62 | 424 ± 130 |
| K | mg·L–1 | 2100 ± 260 | <1 | 799 ± 35 | <1 |
| Alkalinityb | meq·L–1 | 123 | 130 | 19 | 19 |
The urine reactors (UR-K and UR-Na) contained high salts and high total ammonia concentrations; the wastewater reactors (WWR-K and WWR-Na) contained low salts and low total ammonia concentrations. Influent solutions to the urine reactors as well as the influent solutions to the wastewater reactors varied also in their sodium and potassium content. All influent solutions had alkalinity to ammonia ratios of less than 2 mol·mol–1.
Calculated.
Operational Conditions
All operational conditions were kept the same throughout the whole experimental duration, except for the nitrogen loading rates. During a start-up phase of 9 days, the urine and wastewater reactors were fed with a nitrogen loading rate of 355 ± 15 and 95 ± 5 mg NH4–N·L–1·d–1, respectively. The experiment was initiated (time point zero) by a decrease in the influent rates to 22.8 mL·d–1 (UR) and 101 mL·d–1 (WWR), resulting in nitrogen loading rates of 19 ± 2 (UR) and 8 ± 2 mg NH4–N·L–1·d–1 (WWR) and hydraulic retention times were 88 (UR) and 20 d (WWR). The influent rates were reduced, because results from previous studies on urine nitrification6,23 suggest that acid-tolerant AOB grow when the inflow stops or is very low. After decrease at time zero, the inflow rates were kept constant for the rest of the experiment to provide a constant input of substrate for the AOB. The reactors were operated over 300 days, but microbial analyses were limited to biomass samples up to day 160.
2.2. Analysis of Nitric Oxide (NO), Nitrous Oxide (N2O), and Nitrogen Dioxide (NO2) Concentrations in the Off-Gas
On day 246, the NO, N2O, and NO2 concentrations in the off-gas of all four reactors were analyzed by Fourier transform infrared (FTIR) spectroscopy (GASMET CX-4000, Temet Instruments, Helsinki). The instrument was equipped with a heated (40 °C) flow-through gas cell with a 9.8 m path length. The quantification limits for NO, N2O, and NO2 were 2, 0.2, and 1 ppm, respectively, and the expanded standard uncertainty is around 15% for NO and N2O and 25% for NO2 (95% confidence level).25
2.3. Molecular Biology and Numerical Methods
16S rRNA Gene-Based Amplicon Sequencing and Polymerase Chain Reaction (PCR)
Biomass sampling and extraction of genomic DNA is described in the Supporting Information. DNA extracts were sent to Research and Testing Laboratory (Lubbock, TX, USA) for 16S rRNA gene-based amplicon sequencing according to facility’s protocol26 adapted to the MiSeq Illumina desktop technology. The primer pair 341F (5′-CCTACGGGNGGCWGCAG-3′)/785R (5′-GACTACHVGGGTATCTAATCC-3′) was used to target the v3–v4 hypervariable region of the bacterial 16S rRNA gene pool.27In silico testing, analysis of samples with primers targeting archaea, analysis with quantitative polymerase chain reaction (qPCR) for the relative abundance of archaea and Nitrosococcus, as well as with qualitative PCR for AOA are described in the Supporting Information.
Bioinformatic Processing of Amplicon Sequencing Data Sets
The amplicon sequencing data sets were processed using the bioinformatics workflow implemented in the MIDAS field guide including taxonomic assignment using the RDP classifier28,29 against the MIDAS database54 of reference sequences curated from SILVA for wastewater environments. The relative abundance of operational taxonomic units (OTU) or phylotypes were estimated from the number of assigned sequence reads to total reads per sample.
Phylogenetic and Numerical Analyses
The sequencing data were submitted to NCBI with the BioProject ID 293261. Data files were imported into the R package Phyloseq30 for further processing. Samples with a sequencing depth of less than 10 000 reads were removed from the sequencing data set. Sequencing depths were between 15 722 and 48 338 reads and a median sequencing depth of 42 488 reads was obtained per sample. Nonbacterial and chloroplast sequences were removed from the data set prior to analysis. Phyloseq was used for analysis and plotting of alpha diversity measures. For further analysis OTUs that did not have more than two reads in three or more samples were removed from the data set. Package vegan31 was used to perform Nonmetric Multidimensional Scaling with function metaMDS(). Function bioenv() was used to determine most relevant parameters to explain community variation.32 Function envfit() was used to fit the determined environmental variables to the ordination.
A Neighbor Joining phylogenetic tree was constructed in MEGA (version 6.06)33 using the Maximum Composite Likelihood model on a ClustalW alignment of OTU reference sequences best BLAST matches from NCBI and reference organism sequences obtained from RDP; 500 bootstrap resamplings were carried out to test the tree topology.
Analyses of variance (ANOVA) were conducted to assess the extent and significance of the effects of the two main factors of feed composition (synthetic urine versus synthetic wastewater) and monovalent cationic specie (K+ vs Na+) on microbial population dynamics, by analogy to Weissbrodt et al.34 Heatmaps of Spearman’s rank-order correlation coefficients were computed according to Weissbrodt et al.35 in order to delineate clusters of predominant OTUs (>5%) sharing similar dynamics in relationship with operational conditions and process responses.
3. Results
3.1. Nitrification Performance of MBBRs with Synthetic Urine and Synthetic Wastewater
Urine Reactors
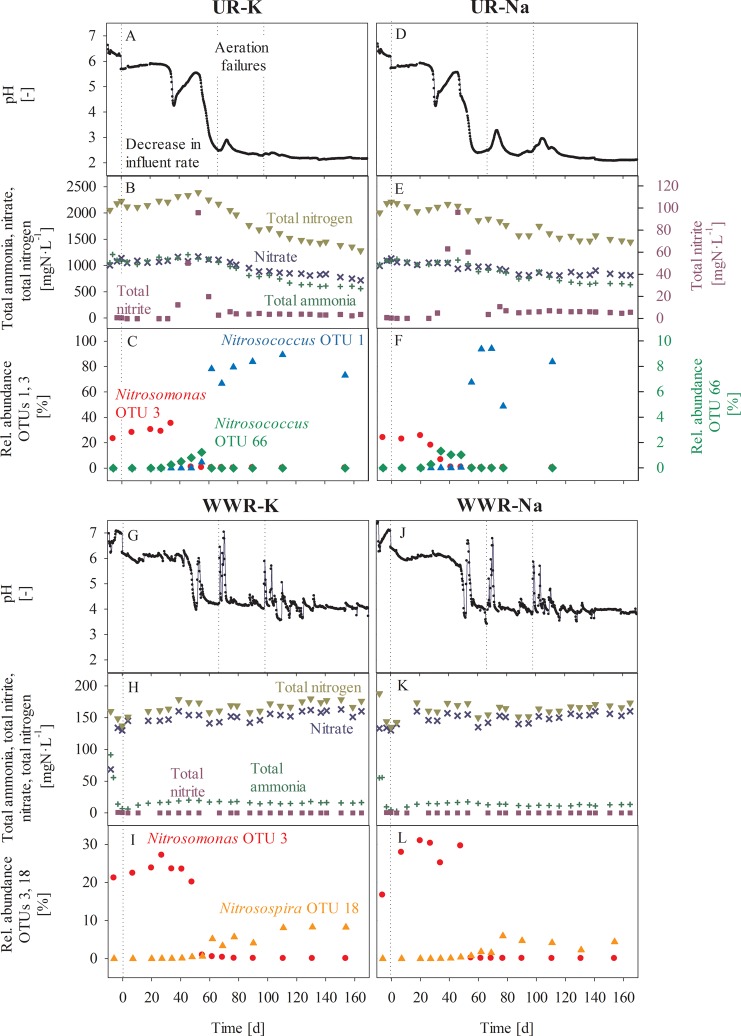
After the decrease in the influent loading (time point zero) the pH started to drop to a level of 4.3 after 30 (UR-K) and 25 days (UR-Na, Figure 1), respectively. As the reactor was continuously supplied with synthetic urine, such a pH drop can only be explained by an increased rate of NH3 oxidation and proton production by AOB.
Figure 1.
For each experimental condition pH and nitrogen species in the reactor (total ammonia, total nitrite, total nitrate, and total nitrogen) are shown together with the relative abundance of AOB. Results for the synthetic urine reactors (UR-K and UR-Na), and the synthetic wastewater reactors (WWR-K and WWR,Na) are presented in panels A–C, D–F, G–I, and J–L, respectively. Experimental conditions are described in more detail in the text and in Table 1. Sequencing samples from day 131 (both urine reactors), as well as 90 and 154 (UR-Na) were excluded due to the low sequencing depth.
In parallel to the pH drop, the total nitrite (NO2– and HNO2) concentrations increased. Subsequently, the pH increased again, which is a sign that AOB growth was slower due to an inhibition effect. HNO2 is a known inhibitor for AOB.36 Despite the high HNO2 concentrations (Figure S1) a second decrease of pH was observed after 52 (UR-K) and 46 days (UR-Na) to average pH values of 2.2 ± 0.1 (UR-K) and 2.3 ± 0.3 (UR-Na). During this phase, the total nitrite concentrations decreased from around 100 mgN·L–1 to 3.7 ± 0.8 (UR-K) and 5.9 ± 1.4 mgN·L–1 (UR-Na) and remained stable for the rest of this study. The pH increased only slightly after an aeration failure on days 68 and 98. Despite the low pH values, average ammonia oxidation rates of 13.8 ± 0.3 (UR-K) and 14.5 ± 0.8 mgN·L–1·d–1 (UR-Na) were maintained until day 160. These rates were slightly higher than the nitrification rates of 12.0 ± 0.8 (UR-K) and 11.8 ± 1.0 mgN·L–1·d–1 (UR-Na) observed before the second pH drop. After the well-controlled reactor operation of 160 days, the reactors were run for another 120 days. In this phase the reactor pH remained constant at the very low levels (results not shown), proving that AOB could also grow over long time-periods at such low pH values.
After the second pH drop, the total nitrogen concentration (sum of total ammonia, total nitrite, and nitrate) in the reactor decreased. Nitrogen losses accounted to 9.2 (UR-K) and 9.4 mgN·L–1·d–1 (UR-Na) corresponding to 53 and 50%, respectively (Figure 1). Off-gas measurement for NO, NO2, and N2O revealed that the losses from the reactor solution were mainly due to the volatilization of NO: 8.7 (UR-K) or 7.1 mgN·L–1·d–1 (UR-Na) were detected. NO2 and N2O were also detectable: NO2 was 1.3 or 1.6 mgN·L–1·d–1 in UR-K and UR-Na, whereas N2O accounted for 0.4 or 0.2 mgN·L–1·d–1, respectively. Total emissions of analyzed nitrogen compounds in the off-gas were 10.4 mgN·L–1·d–1 and 8.9 mgN·L–1·d–1, which corresponds well to the nitrogen losses in the liquid phase (Table S3). HNO2 emissions were not analyzed, but are expected to be small estimated from Henry’s Law. NO was thus the major compound produced at low pH in the urine reactors, followed by NO3– (Table S3).
Wastewater Reactors
In the wastewater reactors the pH decreased after around 40 days (Figure 1). Total nitrite concentrations in the reactor remained below the detection limit of 0.015 mgN·L–1 in almost all samples and were thus clearly lower than in the urine reactors. In contrast to urine reactors, nitrogen losses from the liquid phase were negligible (Table S3). A new pH level of 4.2 ± 0.4 (WWR-K) and 4.0 ± 0.4 (WWR-Na) was reached. Nitrification rates of 8.2 ± 0.6 (WWR-K) and 8.0 ± 0.5 mgN·L–1·d–1 (WWR-Na) were retained, which is similar to the nitrification rate before the pH drop (7.9 ± 1.1 and 8.7 ± 0.8 mgN·L–1·d–1).
The chemical speciation model PhreeqC was used to calculate the minimal pH values, which would be reached, if all ammonia was converted to nitrate. In the synthetic wastewater solutions, the minimal pH value would be 2.6, while the synthetic urine solutions would allow the pH to decrease to a minimal value of 0.9 (see the Supporting Information for further details). The buffer capacity of the influent is therefore sufficiently low in both solutions to allow for reaching very low pH values during nitrification.
Low Impact of Monovalent Cations
The two urine reactors showed very similar reactor behavior, as did the two wastewater reactors: the difference in K+ and Na+ content had little effect (Figure 1). Potassium concentrations in the reactors fed with sodium-rich influent were higher than expected from the influent composition (Table 1): 25.8 ± 21.3 mg·L–1 and 13.2 ± 4.4 mg·L–1 in UR-Na and WWR-Na, respectively (Table S4), likely due to the leakage of potassium ions from the pH electrodes. The potassium levels were, however, still more than 80 and 60 times lower compared to the potassium reactors UR-K and WWR-K, respectively.
3.2. Shifts in Nitrifying Populations
Urine Reactors
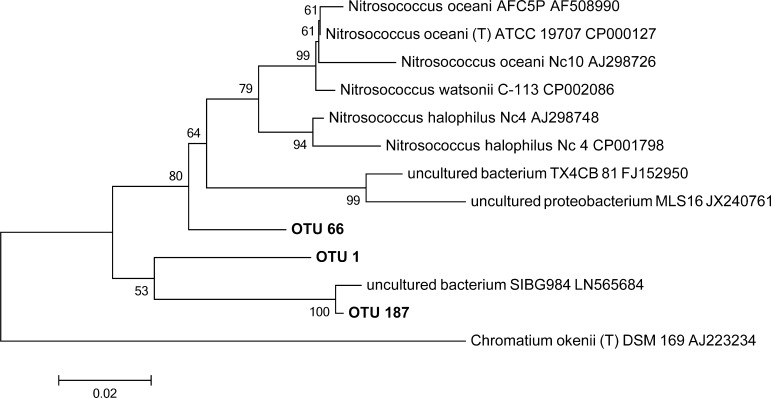
Nitrosomonas OTU 3 was the most abundant AOB in the beginning of the experiment (>15% relative abundance according to 16S rRNA gene sequencing results) and the relative abundance of all other AOB was below 0.2%. According to BLAST, Nitrosomonas OTU 3 affiliates with the Nitrosomonas europaea lineage. As soon as the pH in the urine reactors decreased, the relative abundance of Nitrosomonas OTU 3 declined to values below 0.5% (Figure 1). Concomitantly with this first pH decrease, the relative abundance of OTU 66 sequence, which according to BLAST showed the greatest similarity to Nitrosococcus oceani (95% identity) increased to above 1%. However, the relative abundance of OTU 66 decreased again with the second pH drop, whereas the closely related OTUs 1 and 187, with 93% BLAST similarity to Nitrosococcus halophilus strain Nc4, increased strongly. OTU 1 reached maximal relative abundances of 94% and remained the only AOB with relative abundance of more than 0.5% until the end of the experiment. OTU 187 is not shown in Figure 1, because its abundance was considerably lower than the abundance of OTU 1. The dynamics of the Nitrosococcus-related OTU 1 was also confirmed by a TaqMan qPCR assay designed to specifically quantify this OTU (Figure S2). A de novo phylogenetic tree indicated that Nitrosococcus OTU 1 clustered separately from known Nitrosococcus sequences, while the rare OTU 187 was 99% similar to an environmental sequence retrieved from leaf cutter ant nests (Figure 2). Although these results would have to be confirmed, for example, by full-length 16S rRNA gene sequences and other indicators, this suggests that the sequences of OTU 1 belong to an undescribed species, possibly even a new genus.
Figure 2.
Neighbor Joining tree of Nitrosococcus-like sequences based on 16S rRNA gene-based amplicon sequencing and reference sequences, based on 421 nucleotide positions. Numbers indicate % of 500 bootstrapped tree topologies supporting the displayed phylogeny. Scale indicates substitutions per position. Chromatium okenii was included as an outgroup within the class of γ-Proteobacteria.
Bradyrhizobiaceae OTU 2, an abundant sequence that was assigned by our pipeline to the family of Bradyrhizobiaceae, showed 100% identity to Nitrobacter (Nitrobacter sp. 219, AM286375.1). OTU 2 was abundant at the beginning of the experiment, but disappeared in the urine reactors after the second pH drop (Figure 3, and S3). The absence of nitrite oxidizing bacteria (NOB) in the urine reactors was confirmed with batch experiments demonstrating no nitrite oxidation (Figure S4).
Figure 3.
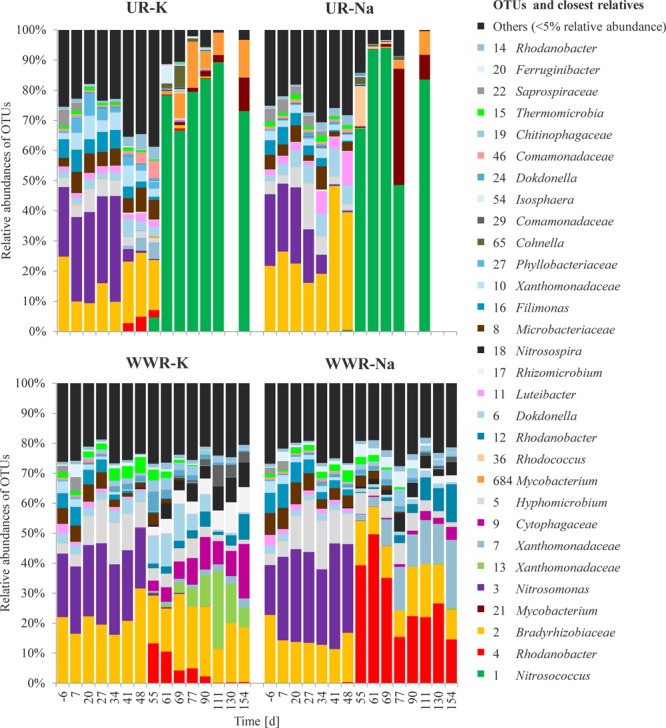
Dynamics of OTUs that displayed relative abundances above 5% of the bacterial community over the experimental period in the four reactors operated with synthetic urine (UR-K and UR-Na) and synthetic wastewater (WWR-K and WWR-Na). Their relative abundances and closest neighbors were retrieved from the high-resolution MiSeq data sets of 16S rRNA gene-based amplicon sequencing and after mapping against MIDAS. These phylotypes were identified at family (-aceae suffix) and genus levels.
The DNA yield per carrier was determined as an estimator for total biomass. The overall DNA yield from urine reactor carriers decreased very strongly after the second pH drop (Figure S3). The high relative abundance of Nitrosococcus OTU 1 was thus at least partly due to a strong biomass decay. However, when using the DNA yield and the relative abundance of OTU 1 to estimate the total abundance of this group, then this value increased from below 0.01 to average values of 0.8 μg DNA·carrier–1 after the second pH drop, indicating that OTU 1 was actually growing. This was further confirmed by qPCR analysis of OTU 1 abundance (Figure S2).
Wastewater Reactors
Similar to the urine reactors, the relative abundance of Nitrosomonas OTU 3 decreased below 0.5% as the pH in the wastewater reactors started to drop (Figure 1). Instead, the relative abundance of Nitrosospira sp. (OTU 18) increased to maximal values of 8%. Nitrobacter-like sequences from the family of Bradyrhizobiaceae remained constant over the whole experimental duration (Figure 3, and S3), indicating that NOB remained viable under the low pH conditions in the wastewater reactors, which was also confirmed in batch experiments (Figure S4). DNA yield per carrier remained relatively constant in the wastewater reactors (Figure S3).
Low Abundance of Archaea
AOA were not detected in any of the low pH reactors by any of the primer pairs used for the 16S rRNA gene-based amplicon sequencing. AOA were also not detected with the AOA-specific PCR assay37 (Figure S5). qPCR for overall abundance of archaea compared to bacteria also failed to detect archaea in the low pH urine reactors, and showed that archaea never exceeded a relative abundance of more than 0.7% at any time in any of the reactors (Figure S6).
3.3. Shifts in Overall Bacterial Community Compositions
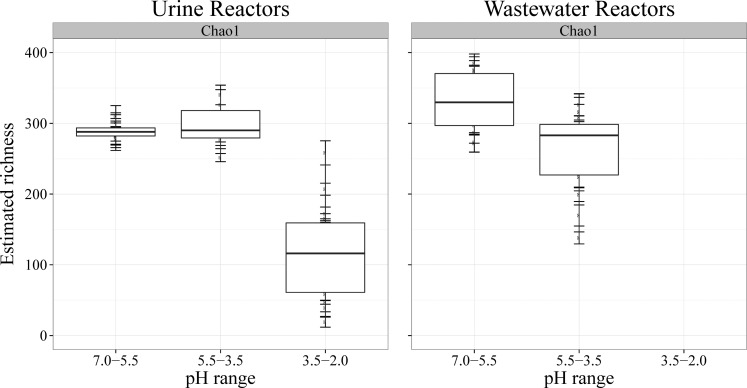
The estimated Chao1 richness of the sequencing data sets was correlated to the pH ranges in the reactors (Figure 4). Whereas the richness remained at around 280 OTUs during the first pH drop to 4.3 in the urine reactors, it decreased dramatically to 110 OTUs as the pH dropped to average values of 2.2. The richness in the wastewater reactors decreased only slightly from around 340 to 280 OTUs as the pH regime shifted from above pH 5.5 to average values of 4.1, which corresponds well with the richness in the urine reactors in the same pH range (pH 5.5–3.5).
Figure 4.
Chao 1 estimated richness for the urine and wastewater reactors as a function of the reactor pH. Samples were divided into three pH ranges: 7.0 to 5.5, 5.5 to 3.5, and 3.5 to 2.0. The wastewater reactors did not reach pH levels below 3.5. Number of samples per pH range for urine reactors: 9 (pH 7.0 to 5.5), 6 (5.5 to 3.5), 11 (3.5 to 2.0). Wastewater reactors: 14 (7.0 to 5.5), 16 (5.5 to 3.5), 0 (3.5 to 2.0).
Urine and wastewater reactors originally contained very similar microbial communities that differentiated increasingly over the course of the experiment, as represented in the nonmetric multidimensional scaling analysis (Figure 5). pH and HNO2 showed the best correlation of the tested environmental variables (pH, HNO2, NO2–, NH3, NH4+, and total salts) with community structure (spearman correlation coefficients: 0.74 for pH, 0.59 for HNO2).
Figure 5.
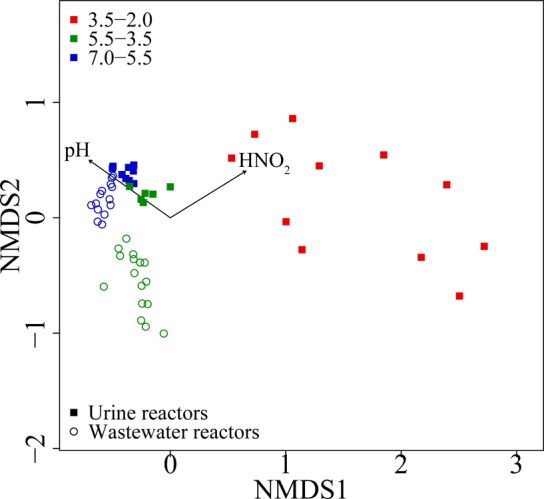
Nonmetric multidimensional scaling analysis (NMDS) of the community structure for all biomass samples and the fitted environmental variables pH and HNO2. Proximity in the NMDS plot indicates similarity in the composition of microbial communities of the samples. Microbial communities in the two reactor types were very similar after inoculation, but differentiated over time as the pH dropped. The drop to highly acidic conditions in urine reactors went along with a shift to a very distinct community that was correlated also with the increase in HNO2 (Spearman correlation coefficients: 0.74 for pH, 0.59 for HNO2).
The heatmap of Spearman’s rank-order correlations delineated three major clusters of coevolving predominant OTUs (>5%). Nitrosococcus OTU 1, Nitrosomonas OTU 3, Nitrosospira OTU 18 belonged to one cluster each. Hardly any OTUs clustered together with Nitrosococcus OTU 1, except of the two OTUs 21 and 684 affiliated with the genusMycobacterium (Figure S7). These two OTUs reached maximal abundances of 38.3% (OTU 21) and 15.1% (OTU 684) in the urine reactors after the second pH drop (Figure 3).
Analyses of variance (ANOVA) conducted on the population profiles further confirmed that the liquid matrix (i.e., synthetic urine or synthetic wastewater) was the main factor for the selection of Nitrosococcus, Nitrosospira, and Bradyrhizobiaceae affiliates (maximal F-values of 940, 1930, and 136, respectively; P-values of 0.02, 0.01, and 0.05, respectively), rather than the type of monovalent cation (i.e., K+ or Na+; maximal F-values of 1, 1, and 11, respectively; P-values of 0.5, 0.5, 0.2, respectively).
4. Discussion
4.1. Selection of AOB Populations
Low affinity for NH3 has been hypothesized to be the reason for growth cessation of Nitrosomonas europaea at low pH values, as the availability of NH3 decreases with decreasing pH.5 However, rather than NH3 limitation, a direct effect of the high proton concentration on the energy conservation is the likely reason for the low pH limit of 5.4 of AOB from the Nitrosomonas europaea lineage.12 Correspondingly, Nitrosomonas OTU 3 disappeared in this study as soon as the pH dropped to values below 5.4 in all experiments (Figure 1).
The low pH selected for γ-proteobacterial AOB or Nitrosospira sp. in the urine and wastewater reactors, respectively (Figure 1). Cultured Nitrosococcus species grow optimally at salt concentrations of 300–700 mmol·L–1 NaCl depending on the species,14 while at least the Nitrosospira sp. of Nitrosospira briensis are characterized by a maximum salt tolerance of 250 mmol·L–1 only.13 The Nitrosococcus-related organisms in the urine reactors appear to share this trait of a high salt tolerance as they were apparently better adapted to the salinity of 300 mmol·L–1 in the urine reactors, whereas Nitrosospira sp. were better adapted to the 45 mmol·L–1 in the wastewater reactors and could not thrive in the urine reactors. The different NH3 concentrations may have been an additional selection criterion. However, the similar NH3 affinity constants of 6–11 μmol·L–1 for Nitrosospira(38) and 8.1 μmol·L–1 for Nitrosococcus oceani(39) stress salt tolerance as a major selection criterion.
The shift from Nitrosococcus OTU 66 to OTU 1 corresponds to an increase in the HNO2 concentrations (Figure S1) and is thus likely due to a higher HNO2 tolerance of OTU 1. These traits, in particular acid and HNO2 tolerance, ultimately allowed Nitrosococcus OTU 1 to drive the system to a new stable state in which it dominated the bacterial community. Nitrosospira OTU 18 may be less resistant to extreme environments and did therefore not cause such strong acidification.
4.2. Nitrosococcus OTU 1 Causes, and Grows in, Environments with Low pH Values and High HNO2 Concentrations
The decrease in pH and increase in HNO2 levels caused by the growth of Nitrosococcus OTU 1 corresponded with the decrease in microbial richness and overall DNA yields per carrier (Figures 4, and S3). A strong influence of pH on microbial diversity has been reported for soils: soil pH was the major factor determining the richness of soil bacterial communities.40 Low environmental pH values decrease the intracellular pH value in bacteria, which in turn compromises enzyme activity, as well as protein and DNA stability.41 Low intracellular pH values also hamper the energy generation in certain bacteria, for example, AOB affiliating with the Nitrosomonas europaea lineage (Section 4.1).12 pH homeostasis is therefore an essential requirement for the survival of bacteria at low pH values.20 HNO2 impedes pH homeostasis under acidic conditions as it diffuses passively across the cytoplasmic membrane and decreases the intracellular pH value.42 HNO2 also inhibits enzymes43 and it decomposes to NO (section 4.3), which is another toxic compound for bacteria.44 It is therefore not surprising that most of the bacteria did not survive these toxic conditions.
Nitrosococcus OTU 1 and Mycobacterium OTUs 21 and 684, however, still managed to grow (Figure 3, and Figure S3). The uptake of potassium ions to inverse the membrane potential is a known pH homeostasis mechanism.21 The potassium concentration, however, did not have a significant impact on the reactor performance or the microbial community in our experiments (Figures 1, and 3), indicating that either still sufficient potassium was available in the reactors fed with sodium-rich influent or that sodium ions were used instead. Sodium ions have been found to increase the activity of Thiobacillus thiooxidans at low pH values, but the positive influence of sodium was less pronounced than the one for potassium.45 The Gram-positive bacteria of the genus Mycobacterium are also known to have lipid-rich cell walls, which play an important role in their resistance to acids.46 Highly impermeable cell membranes are another prerequisite for bacterial growth at low pH values as they reduce the leakage of protons.20 Thus, acid tolerance can be due to a large variety of factors and the presence of potassium or possibly sodium alone does not determine, whether the acid tolerant bacteria grow in.
4.3. Biological versus Chemical Nitrite Oxidation
The NOB of the genus Nitrospira have been reported to be active in engineered reactors at minimum pH values between 3.2 and 4.5.2−5 The NOB of the genus Nitrobacter have been widely detected in acidic soils (pH values as low as 3)47 and were also observed at average pH values of 4.1 in the synthetic wastewater in this study (Figure 1). It is possible that in some studies, chemical nitrite oxidation was wrongly interpreted as NOB activity. Nevertheless, it is likely that acumulated HNO2 rather than pH alone inhibited Nitrobacter sp. in the urine reactors with the first pH drop to 4.3 and caused the accumulation of total nitrite.
Despite the apparent absence or inactivity of NOB, total nitrite remained low once the pH dropped to pH levels below 2.5 (Figure 1), indicating nitrite conversion. At low pH values, HNO2 is chemically converted to NO3–, involving several volatile intermediates, such as NO, NO2, and N2O3.6 Van Cleemput and Baert48 observed experimentally that NO is the major gaseous decomposition product, while NO3– production was favored under conditions in which NO was not stripped, which corresponds very well with the results in this study: strong emissions of NO were observed due to the strong aeration in the MBBR, while NO3– concentrations decreased during the course of the experiment. NO can also be produced by AOB via the nitrifier denitrification pathway;49 however, McKenney et al.50 found that emissions due to the chemical process are dominant at pH values below 4.5. NO is an unwanted nitrification byproduct as it impacts human health and is considered to be the main precursor of ground-level tropospheric ozone in rural areas.51
4.4. Implications for Wastewater Treatment
With our results we show that γ-proteobacterial AOB and Nitrosospira sp. are important players in wastewaters with high and low ammonia content, respectively, and can cause strong pH decreases. This finding challenges the perception that low pH nitrification is either not possible or dominated by AOA. The growth of γ-proteobacterial AOB is more critical than the growth of Nitrosospira sp., as γ-proteobacterial AOB acidify the wastewater more strongly allowing for the chemical decomposition of HNO2 (Figure 1). The selection of γ-proteobacterial AOB may not only be a risk in urine nitrification reactors, but also during the treatment of other wastewaters with high ammonia concentrations with limited alkalinity, e.g., digester supernatant, animal wastewaters, or landfill leachate. Besides reports on low pH nitrification with human urine in biofilm systems6 and with suspended biomass systems,23,52 nitrification at pH values below 5 has also been observed in poultry manure.53 This study shows that nitrification of urine, manure, digester supernatant, or another wastewater with high ammonia content is prone to low pH values, if the ratio of alkalinity to total ammonia is less than 2. When nitrifiying such solutions, any decrease of the pH far below the typical limit of 5.412 should be prevented. Otherwise, acid-tolerant γ-proteobacterial AOB will grow in, which has two detrimental consequences: first, the loss of the microbial community, which is responsible for high-rate nitrification at neutral pH, and, second, the emission of hazardous volatile nitrogen compounds such as NO, N2O, NO2, and HNO2.
Acknowledgments
This study was funded by the Bill and Melinda Gates Foundation and was conducted as part of the VUNA project (www.eawag.ch/vuna, Grant No. OPP1011603). The authors like to thank Karin Rottermann and Claudia Bänninger for the chemical analyses, Bettina Sterkele and Hanspeter Zöllig for the laboratory support, and Mads Albertsen for sequence processing guidance.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.7b00392.
Additional materials and methods; recipes for synthetic influent solutions; nitrogen balance for urine and wastewater reactors; chemical concentrations in the reactors; batch experiments for NOB; DNA mass and average copy numbers of AOB; microbiological measurements of AOA; heatmaps of Spaearman’s rank-order correlation of phylotypes and environmental conditions (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Painter H. A., Nitrification in the treatment of sewage and waste-waters. In Nitrification; Prosser J. I., Ed.; IRL Press: Oxford, 1986; Vol. 20, pp 185–211. [Google Scholar]
- Gieseke A.; Tarre S.; Green M.; de Beer D. Nitrification in a biofilm at low pH values: role of in situ microenvironments and acid tolerance. Appl. Environ. Microbiol. 2006, 72 (6), 4283–4292. 10.1128/AEM.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarre S.; Beliavski M.; Denekamp N.; Gieseke A.; de Beer D.; Green M. High nitrification rate at low pH in a fluidized bed reactor with chalk as the biofilm carrier. Water Sci. Technol. 2004, 49 (11–12), 99–105. [PubMed] [Google Scholar]
- Tarre S.; Green M. High-rate nitrification at low pH in suspended- and attached-biomass reactors. Appl. Environ. Microbiol. 2004, 70 (11), 6481–6487. 10.1128/AEM.70.11.6481-6487.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarre S.; Shlafman E.; Beliavski M.; Green M. Changes in ammonia oxidiser population during transition to low pH in a biofilm reactor starting with Nitrosomonas europaea. Water Sci. Technol. 2007, 55 (8–9), 363–368. 10.2166/wst.2007.278. [DOI] [PubMed] [Google Scholar]
- Udert K. M.; Larsen T. A.; Gujer W. Chemical nitrite oxidation in acid solutions as a consequence of microbial ammonium oxidation. Environ. Sci. Technol. 2005, 39 (11), 4066–4075. 10.1021/es048422m. [DOI] [PubMed] [Google Scholar]
- Hayatsu M. The lowest limit of pH for nitrification in tea soil and isolation of an acidophilic ammonia oxidizing bacterium. Soil Sci. Plant Nutr. 1993, 39 (2), 219–226. 10.1080/00380768.1993.10416993. [DOI] [Google Scholar]
- Jeschke C.; Falagán C.; Knöller K.; Schultze M.; Koschorreck M. No nitrification in lakes below pH 3. Environ. Sci. Technol. 2013, 47 (24), 14018–14023. 10.1021/es402179v. [DOI] [PubMed] [Google Scholar]
- Udert K. M.; Larsen T. A.; Gujer W. Fate of major compounds in sourceseparated urine. Water Sci. Technol. 2006, 54 (11–12), 413–420. 10.2166/wst.2006.921. [DOI] [PubMed] [Google Scholar]
- Udert K. M.; Fux C.; Münster M.; Larsen T. A.; Siegrist H.; Gujer W. Nitrification and autotrophic denitrification of source-separated urine. Water Sci. Technol. 2003, 48 (1), 119–30. [PubMed] [Google Scholar]
- Schreiber F.; Wunderlin P.; Udert K. M.; Wells G. F. Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies. Front. Microbiol. 2012, 3, 372–372. 10.3389/fmicb.2012.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumasoli A.; Morgenroth E.; Udert K. M. Modeling the low pH limit of Nitrosomonas eutropha in high-strength nitrogen wastewaters. Water Res. 2015, 83, 161–170. 10.1016/j.watres.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Koops H.-P.; Purkhold U.; Pommerening-Röser A.; Timmermann G.; Wagner M., The Lithoautotrophic Ammonia-Oxidizing Bacteria. In The Prokaryotes; Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., Eds.; Springer: New York, 2006; pp 778–811. [Google Scholar]
- Koops H. P.; Böttcher B.; Möller U. C.; Pommerening-Röser A.; Stehr G. Description of a new species of Nitrosococcus. Arch. Microbiol. 1990, 154 (3), 244–248. 10.1007/BF00248962. [DOI] [Google Scholar]
- Ward B. B.; O’Mullan G. D. Worldwide distribution of Nitrosococcus oceani, a marine ammonia-oxidizing gamma-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl. Environ. Microbiol. 2002, 68 (8), 4153–7. 10.1128/AEM.68.8.4153-4157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. H.; Daims H.; Lemmer H.. FISH Handbook for Biological Wastewater Treatment: Identification and Quantification of Microorganisms in Activated Sludge and Biofilms; FISH: New York, 2009. [Google Scholar]
- Nicol G. W.; Leininger S.; Schleper C.; Prosser J. I. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 2008, 10 (11), 2966–78. 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Zhang L.-M.; Hu H.-W.; Shen J.-P.; He J.-Z. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 2012, 6 (5), 1032–1045. 10.1038/ismej.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G. F.; Park H.-D.; Yeung C.-H.; Eggleston B.; Francis C. A.; Criddle C. S. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ. Microbiol. 2009, 11 (9), 2310–2328. 10.1111/j.1462-2920.2009.01958.x. [DOI] [PubMed] [Google Scholar]
- Slonczewski J. L.; Fujisawa M.; Dopson M.; Krulwich T. A. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 2009, 55, 1–317. 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C.; Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007, 15 (4), 165–171. 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Rusten B.; Eikebrokk B.; Ulgenes Y.; Lygren E. Design and operations of the Kaldnes moving bed biofilm reactors. Aquacultural Engineering 2006, 34 (3), 322–331. 10.1016/j.aquaeng.2005.04.002. [DOI] [Google Scholar]
- Fumasoli A.; Etter B.; Sterkele B.; Morgenroth E.; Udert K. M. Operating a pilot-scale nitrification/distillation plant for complete nutrient recovery from urine. Water Sci. Technol. 2016, 73 (1), 215–222. 10.2166/wst.2015.485. [DOI] [PubMed] [Google Scholar]
- Udert K. M.; Larsen T. A.; Biebow M.; Gujer W. Urea hydrolysis and precipitation dynamics in a urine-collecting system. Water Res. 2003, 37 (11), 2571–82. 10.1016/S0043-1354(03)00065-4. [DOI] [PubMed] [Google Scholar]
- Mohn J.; Zeeman M. J.; Werner R. A.; Eugster W.; Emmenegger L. Continuous field measurements of δ13C–CO2 and trace gases by FTIR spectroscopy. Isot. Environ. Health Stud. 2008, 44 (3), 241–251. 10.1080/10256010802309731. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Wolcott R.; Dowd S., Tag-Encoded FLX Amplicon Pyrosequencing for the Elucidation of Microbial and Functional Gene Diversity in Any Environment. In High-Throughput Next Generation Sequencing; Kwon Y. M., Ricke S. C., Eds.; Humana Press, 2011; Vol. 733, pp 129–141. [DOI] [PubMed] [Google Scholar]
- Herlemann D. P. R.; Labrenz M.; Jürgens K.; Bertilsson S.; Waniek J. J.; Andersson A. F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5 (10), 1571–1579. 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y.; Wang Q.; Cole J. R.; Rosen G. L. Using the RDP Classifier to Predict Taxonomic Novelty and Reduce the Search Space for Finding Novel Organisms. PLoS One 2012, 7 (3), e32491. 10.1371/journal.pone.0032491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Garrity G. M.; Tiedje J. M.; Cole J. R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Applied and environmental microbiology 2007, 73 (16), 5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy S. J.; Saunders A. M.; Albertsen M.; Nierychlo M.; McIlroy B.; Hansen A. A.; Karst S. M.; Nielsen J. L.; Nielsen P. H.. MiDAS: the field guide to the microbes of activated sludge. Database (Oxford); 2015, bav062, doi: 10.1093/database/bav062. [DOI] [PMC free article] [PubMed]
- McMurdie P. J.; Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 2013, 8 (4), e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J.; Blanchet F. G.; Kindt R.; Legendre P.; Minchin P. R.; O’Hara R. B.; Simpson G. L.; Solymos P.; Henry M.; Stevens H.; Wagner H.. Vegan: Community Ecology Package, R package version 2.2-1; http://CRAN.R-project.org/package=vegan. 2015.
- Clarke K. R.; Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar. Ecol.: Prog. Ser. 1993, 92, 205–219. 10.3354/meps092205. [DOI] [Google Scholar]
- Tamura K.; Stecher G.; Peterson D.; Filipski A.; Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbrodt D. G.; Schneiter G. S.; Fürbringer J.-M.; Holliger C. Identification of trigger factors selecting for polyphosphate- and glycogen-accumulating organisms in aerobic granular sludge sequencing batch reactors. Water Res. 2013, 47 (19), 7006–7018. 10.1016/j.watres.2013.08.043. [DOI] [PubMed] [Google Scholar]
- Weissbrodt D. G.; Shani N.; Holliger C. Linking bacterial population dynamics and nutrient removal in the granular sludge biofilm ecosystem engineered for wastewater treatment. FEMS Microbiol. Ecol. 2014, 88 (3), 579–595. 10.1111/1574-6941.12326. [DOI] [PubMed] [Google Scholar]
- Vadivelu V. M.; Keller J.; Yuan Z. Free ammonia and free nitrous acid inhibition on the anabolic and catabolic processes of Nitrosomonas and Nitrobacter. Water Sci. Technol. 2007, 56 (7), 89–97. 10.2166/wst.2007.612. [DOI] [PubMed] [Google Scholar]
- Francis C. A.; Roberts K. J.; Beman J. M.; Santoro A. E.; Oakley B. B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (41), 14683–8. 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q. Q.; Bakken L. R. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 1999, 30, 171–186. 10.1111/j.1574-6941.1999.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Ward B. B. Kinetic studies on ammonia and methane oxidation by Nitrosococcus oceanus. Arch. Microbiol. 1987, 147 (2), 126–133. 10.1007/BF00415273. [DOI] [Google Scholar]
- Fierer N.; Jackson R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (3), 626–631. 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P.; Tramonti A.; De Biase D. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiology Reviews 2014, 38 (6), 1091–1125. 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- Mortensen H. D.; Jacobsen T.; Koch A. G.; Arneborg N. Intracellular pH Homeostasis Plays a Role in the Tolerance of Debaryomyces hansenii and Candida zeylanoides to Acidified Nitrite. Applied and environmental microbiology 2008, 74 (15), 4835–4840. 10.1128/AEM.00571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Oehmen A.; Lim M.; Vadivelu V.; Ng W. J. The role of nitrite and free nitrous acid (FNA) in wastewater treatment plants. Water Res. 2011, 45 (15), 4672–82. 10.1016/j.watres.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Zumft W. G. The biological role of nitric oxide in bacteria. Arch. Microbiol. 1993, 160 (4), 253–264. 10.1007/BF00292074. [DOI] [PubMed] [Google Scholar]
- Suzuki I.; Lee D.; Mackay B.; Harahuc L.; Oh J. K. Effect of Various Ions, pH, and Osmotic Pressure on Oxidation of Elemental Sulfur by Thiobacillus thiooxidans. Appli. Environ. Microbiol. 1999, 65 (11), 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal O. H.; Nathan C. F.; Ehrt S. Acid Resistance in Mycobacterium tuberculosis. J. Bacteriol. 2009, 191 (15), 4714–4721. 10.1128/JB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W.; Kowalchuk G. A. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol. Biochem. 2001, 33 (7–8), 853–866. 10.1016/S0038-0717(00)00247-9. [DOI] [Google Scholar]
- Van Cleemput O.; Baert L. Nitrite: a key compound in N loss processes under acid conditions?. Plant Soil 1984, 76 (1–3), 233–241. 10.1007/BF02205583. [DOI] [Google Scholar]
- Wrage N.; Velthof G. L.; van Beusichem M. L.; Oenema O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 2001, 33 (12–13), 1723–1732. 10.1016/S0038-0717(01)00096-7. [DOI] [Google Scholar]
- McKenney D. J.; Lazar C.; Findlay W. J. Kinetics of the Nitrite to Nitric Oxide Reaction in Peat. Soil Science Society of America Journal 1990, 54 (1), 106–112. 10.2136/sssaj1990.03615995005400010016x. [DOI] [Google Scholar]
- Medinets S.; Skiba U.; Rennenberg H.; Butterbach-Bahl K. A review of soil NO transformation: Associated processes and possible physiological significance on organisms. Soil Biol. Biochem. 2015, 80 (0), 92–117. 10.1016/j.soilbio.2014.09.025. [DOI] [Google Scholar]
- Schielke S.Decentralised urine treatment with the nitritation/anammox process. Ph.D. Thesis, Swiss Federal Institute of Technology Zurich, Switzerland, 2015. [Google Scholar]
- Prakasam T. B. S.; Loehr R. C. Microbial nitrification and denitrification in concentrated wastes. Water Res. 1972, 6 (7), 859–869. 10.1016/0043-1354(72)90038-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- McIlroy S. J.; Saunders A. M.; Albertsen M.; Nierychlo M.; McIlroy B.; Hansen A. A.; Karst S. M.; Nielsen J. L.; Nielsen P. H.. MiDAS: the field guide to the microbes of activated sludge. Database (Oxford); 2015, bav062, doi: 10.1093/database/bav062. [DOI] [PMC free article] [PubMed]