Abstract
Notch signaling is adjusted to different physiological contexts by expression patterns of Notch ligands and receptors, as well as by posttranslational modifications that modulate the ligand/receptor affinity. In this issue of The EMBO Journal, Suckling et al (2017) show that an interaction of Notch ligands with membrane lipids promotes Notch binding and activation, thus proposing a new mode of Notch activity regulation.
Subject Categories: Signal Transduction, Structural Biology
Signaling by the Notch pathway affects practically all tissues during development. As Spyros Artavanis‐Tsakonas once said, half the world works on Notch, and the other half works on Notch but does not know it yet. The processes mediated by the Notch pathway range from determination of cell fates between sister cells during neurogenesis, through specification of tissue boundaries in fly wings, to coordination of cellular oscillators during somitogenesis (Artavanis‐Tsakonas et al, 1999). How is the Notch pathway tweaked to accommodate this diverse array of biological functions? One obvious way is through a combinatorial action of multiple receptors and ligands and allocation of different features and expression patterns to these receptors and ligands. Another tactic is by posttranslational modifications, such as O‐glycan modification of the Notch receptors by Fringe proteins, which alters their ligand‐binding affinity. The work by Suckling et al (2017) has identified yet a third strategy for achieving diversity in Notch signaling activity, through the binding of the N‐terminal tip of the ligand to the cell membrane (Fig 1).
Figure 1. Interaction between the C2 domain of Notch ligands and the phospholipid membrane modulates Notch signaling.
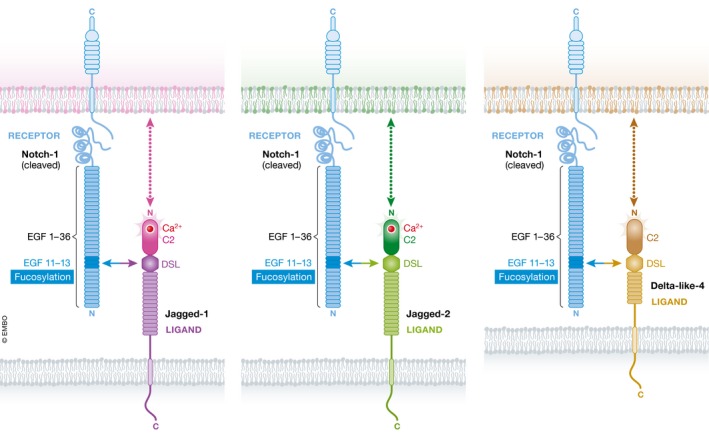
The interaction between Notch receptors in one cell (blue) and Notch ligands (Jagged‐1, purple; Jagged‐2, green; Delta‐like‐4, brown) in the neighboring cell is mediated by the EGF11‐13 domain in the Notch receptor and EGF1/DSL/C2 domains in the Notch ligands. Previous work showed that the receptor–ligand affinity varies between Notch ligands and is regulated by fucosylation of the Notch‐interacting domain (horizontal arrows). The work by Suckling et al (2017) shows evidence that Notch–ligand interaction can also be modulated by the contact between the ligand C2 domain and the phospholipids in the cell membrane that displays Notch (vertical dashed arrows). The variability in the structure between the C2 domains of the different ligands may affect its interaction with membranes containing different lipid compositions.
In recent years, there have been several important works providing detailed structural information on the interface between Notch receptors and their ligands (Chillakuri et al, 2013; Luca et al, 2015, 2017; Kovall et al, 2017). The structure of the receptor‐binding domain of Notch ligands contains three epidermal growth factor (EGF) repeats, a Delta/Serrate/Lag‐2 (DSL) domain, and at the N‐terminal end a C2 domain (also called MNNL domain—module at the N terminus of Notch ligand). Previous work from the Lea and Handford groups suggested that the C2 domain, a known phospholipid‐binding domain, promotes binding of the Notch ligand Jagged‐1 to the cell membrane (Chillakuri et al, 2013). The new work by Suckling et al (2017) provides structural and biochemical evidence that the C2 domain in different Notch ligands may differentially regulate the interaction with Notch‐expressing cells that have distinct plasma membrane phospholipid compositions. The researchers use analysis of newly solved structures of human ligands Jagged‐2 and Delta‐like‐4 and in vitro liposome binding assays to show that Jagged‐1, Jagged‐2, and Delta‐like‐4, but not Delta‐like‐1, interact with phospholipids. Furthermore, they show that binding of Notch1, and in some cases, Notch2, to the ligand‐binding domain, cooperatively enhances its interaction with liposomes, that is, binding to one interaction partner facilitated binding of the other. Thus, coupling between the binding to the receptor and the nature of the membrane environment in which the receptor is embedded may provide an effective means to modulate Notch signaling. The activity of ligands could be increased in cells or tissues that display membranes that are most suitable for binding the ligand C2 domain.
These interactions may have relevance to human disorders, as some of the human disease‐associated mutations affecting Notch ligands were identified within the C2 domain. In fact, the structural analysis revealed that specific mutations associated with extrahepatic biliary atresia (EHBA), which is a rare disease of the liver and bile duct, occur at the phospholipid‐binding loops of the C2 domain of Jagged‐1. When recombinant proteins carrying these mutations were tested, their binding to Notch was unaltered but their lipid‐binding properties were impaired. This suggests that the capacity of the C2 domain to bind lipids is an essential element in regulation of Notch signaling, and its loss can lead to human disorders.
This research opens up several basic questions on the nature of Notch pathway receptor–ligand interactions. First, given the relatively large size of the ectodomain of Notch receptors, it is unclear how the phospholipid‐interacting domain of the ligands “reaches” the membrane of the Notch‐expressing cells (see Fig 1 for illustration). Moreover, the distance between the membranes of the interacting cells may actually increase during receptor–ligand interaction by the pulling forces exerted on Notch receptors by endocytosing ligands (Gordon et al, 2015; Seo et al, 2016). Second, the structural analysis shows that while the C2 domain of Jagged‐1 and Jagged‐2 contains Ca2+ ions, that of Delta‐like‐4 does not. It would be interesting to understand the role Ca2+ ions may play in the interaction between Notch ligands and the cell membrane. Finally, it would be instrumental to test whether distinct Notch ligands are indeed associated with different types of phospholipids. Future studies that will modify the C2 domain in model organisms should be able to test the spectrum of defects that are observed. The possible link between the lipid composition of target tissues and the added specificity provided by the ligand C2 domain opens exciting directions in Notch signaling research.
See also: RJ Suckling et al (August 2017)
Contributor Information
Ben‐Zion Shilo, Email: benny.shilo@weizmann.ac.il.
David Sprinzak, Email: davidsp@post.tau.ac.il.
References
- Artavanis‐Tsakonas S, Rand M, Lake R (1999) Notch signaling: cell fate control and signal integration in development. Science 284: 770 [DOI] [PubMed] [Google Scholar]
- Chillakuri CR, Sheppard D, Ilagan MX, Holt LR, Abbott F, Liang S, Kopan R, Handford PA, Lea SM (2013) Structural analysis uncovers lipid‐binding properties of Notch ligands. Cell Rep 5: 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, Blacklow SC (2015) Mechanical allostery: evidence for a force requirement in the proteolytic activation of notch. Dev Cell 33: 729–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Gebelein B, Sprinzak D, Kopan R (2017) The canonical notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev Cell 41: 228–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC (2015) Structural biology. Structural basis for Notch1 engagement of Delta‐like 4. Science 347: 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein‐Peikar M, Haltiwanger RS, Zhu C, Ha T, Garcia KC (2017) Notch‐Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science 355: 1320–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D, Southard KM, Kim JW, Lee HJ, Farlow J, Lee JU, Litt DB, Haas T, Alivisatos AP, Cheon J, Gartner ZJ, Jun YW (2016) A mechanogenetic toolkit for interrogating cell signaling in space and time. Cell 165: 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckling RJ, Korona B, Whiteman P, Chillakuri C, Holt L, Handford PA, Lea SM (2017) Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands. EMBO J 36: 2204–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]


