Abstract
Pseudomonads are cosmopolitan microbes able to produce a wide array of specialized metabolites. These molecules allow Pseudomonas to scavenge nutrients, sense population density, and enhance or inhibit growth of competing microbes. However, these valuable metabolites are typically characterized one-molecule-one-microbe at a time instead of inventoried in large numbers. To index and map the diversity of molecules detected from these organisms, 260 strains of ecologically diverse origins were subjected to mass spectrometry-based molecular networking. Molecular networking not only enables dereplication of molecules, but also sheds light on their structural relationships. Moreover, it accelerates discovery of new molecules. Herein, through indexing the Pseudomonas specialized metabolome, we report the molecular networking-based discovery of four molecules and their evolutionary relationships: a poaeamide analog, and a molecular sub-family of cyclic lipopeptides, the bananamides 1, 2, and 3. Analysis of their biosynthetic gene cluster shows that it constitutes a distinct evolutionary branch of the Pseudomonas cyclic lipopeptides. Through analysis of an additional 370 extracts of wheat-associated Pseudomonas, we demonstrate how the detailed knowledge from our reference index can be efficiently propagated to annotate complex metabolomic data from other studies akin to the way newly generated genomic information can be compared to data from public databases.
The production of specialized metabolites by Pseudomonas species leads to strain-specific biological activities.1, 2–5 For instance, siderophores and cyclic lipopeptides produced by strains of P. putida and P. fluorescens act as bioactive agents against susceptible plant and animal pathogens, thereby conveying protection and promoting plant growth, while toxins and glycolipids produced by P. syringae and P. aeruginosa contribute to virulence and pathogenicity.2, 6–10 A key challenge when analyzing isolates is the effort required to identify known molecules. The effort involved in determining activity, biochemical characterization, and structure elucidation is a huge expense in time and money and is a wealth of information that is not easily accessible. Other fields such as sequencing, have seen a large increase in the value of expensive data and has made the data searchable for the rest of the scientific public. Unlike what is done with sequencing data, we cannot take natural product data and compare and contrast this information to other previously collected data sets. However, due to increasing computational power, annotation of known molecules can be facilitated by creating a reference index.
Mass spectrometry (MS) has become an invaluable tool for natural product discovery due to its sensitivity and throughput. One challenge in specialized metabolite research is identifying known versus unknown metabolites detected by MS.11 Dereplication (the identification of known molecules) of natural products can be performed with the Dictionary of Natural Products, AntiBase, and MarinLit.11–14 However, these databases are behind paywalls, not searchable with raw data, and certainly not searchable with millions of data points at once. Global Natural Products Social Molecular Networking (GNPS), which utilizes tandem mass spectrometry (MS/MS) as a proxy for molecular structure enables dereplication, visualization of molecular space as a network, and enables propagation of chemical features to unidentified molecules.11, 15–17
Curating a Pseudomonas specialized metabolite index
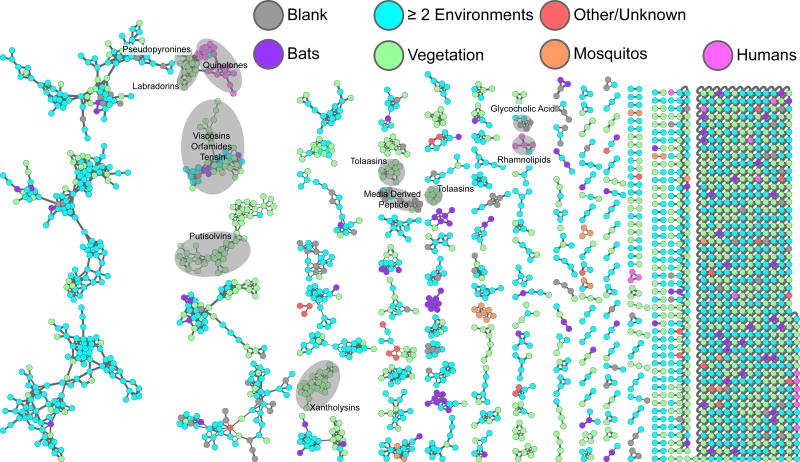
To create a reference metabolite index for Pseudomonas, multiple laboratories contributed to a collection of 260 Pseudomonads, isolated from locations around the globe and from a range of environmental niches. Bacteria were cultured and extracts subjected to LC-MS/MS (Supplementary Figure 1, Supplementary Tables 1 and 2). To generate a molecular map of the detectable metabolites, the LC-MS/MS data was subjected to molecular networking on GNPS and visualized in Cytoscape (Figure 1, Methods, and Supplementary Figure 2).17, 18 Characteristics of the samples such as environmental niche, molecular weight, geographic isolation, and species-specific molecule production were visualized in the network (Figure 1 and Supplementary Figure 2). We observe distinct molecules from environments even where fewer individual strains were analyzed. For example, of the less well represented Pseudomonas, the strain library contains 37 bat-, 3 mosquito-, and 3 human-associated Pseudomonas isolates and these environments show molecules not observed in other Pseudomonas.19 This observation shows a correlation between a strain’s environment and the molecules that are produced and begs the question of whether the environment dictates molecule production and whether certain molecules are required to thrive in specific niches.20, 21 The strain library consists of 21 different species of Pseudomonas, but is primarily composed of P. putida and P. fluorescens (68%, Supplementary Figure 2 and Supplementary Table 1). 5% of molecules are uniquely produced by P. putida, 10% are uniquely produced by P. fluorescens, and 65% of molecules are produced by two or more species, indicating that most are produced by multiple species (Supplementary Figure 3 and Supplementary Tables 1 and 2).4, 8, 22
Figure 1.
Pseudomonas molecular network—environmental isolation visualization. Known Pseudomonas compounds that were observed in our data were dereplicated and highlighted. Node colors represent environments from which the strains were isolated. Grey nodes: blank injections, media controls, and internal standard (glycocholic acid). Teal nodes: two or more environments. Red nodes: other or unknown environments. Purple nodes: bat environments. Green nodes: vegetation environments. Orange nodes: mosquito environments. Pink nodes: human environments.
One of the challenges associated with specialized metabolite discovery is the re-discovery of previously characterized metabolites. Previous data is often spread between multiple databases, primary literature, and lost amongst laboratory notebooks. While we aimed to use the dereplication feature of GNPS, where experimentally derived MS/MS are matched to spectra of annotated and curated MS/MS spectra within the GNPS database, at the start of this project GNPS and other public MS/MS libraries did not contain many of the Pseudomonas specialized metabolites present in the literature, with the exception of lipid annotations.1, 17 Therefore, we manually dereplicated MS and MS/MS spectra against the literature. Using the 2009 review by Gross and Loper as a reference, there are 119 natural products from pseudomonads that belong to 30 molecular families after including the xantholysin, rhamnolipid, labradorin, and pseudopyronine molecular families currently in the literature.1 We observed 9 of these families, or 30% of the Pseudomonas molecular families described (Figure 1, Supplementary Figures 4 and 5, and Supplementary Tables 1–4). However, lack of molecular observation may be due to several reasons. The strain(s) responsible for the production of a compound is not in our Pseudomonas collection. The compounds are not produced in high enough titer to be observed. The extraction conditions used here, while broad, may not be suitable for some compounds, and the current chromatography conditions select for more hydrophobic molecules.
For the observed non-peptidic molecules, examining MS/MS spectra for characteristic mass shifts can shed light on structural information; mass shifts of 162 or 176 Daltons (Da) suggest sugar moieties while mass shifts of 14, 28, or 42 Da suggest lipid or alkyl side chains.23 Characteristic mass shifts were combined with accurate mass measurements, information about the samples (e.g. bacterial genus and species) and were compared to literature values. Based on the Metabolomics Standards Initiative’s reporting standards, manual dereplication of non-peptidic molecules resulted in level 2 — putatively annotated compounds — of known human-associated Pseudomonas metabolites for the rhamnolipid and quinolone molecular families, as well as the labradorin and pseudopyronine molecular families from vegetation-associated Pseudomonas (Figure 1, Supplementary Figure 3, and Supplementary Tables 1–4).24–28 The rhamnolipids were structurally distinct molecules produced by human-associated strains. Rhamnolipids behave as biosurfactants, promote uptake and biodegradation of substrates, and act as immune modulators and virulence factors.29 Similarly to the rhamnolipids, the quinolones were produced by human-associated strains and behave as quorum signals that coordinate biofilm formation, virulence, and antibiotic resistance.30 Conversely, the labradorins and pseudopyronines are produced by vegetation-associated Pseudomonas, where the original characterization are from a phytopathogen and plant-derived pseudomonads but can also be retrieved from a marine sponge-derived Pseudomonas. Both have antimicrobial properties.25–27, 31, 32
For peptidic molecules, MS/MS spectra yield fragment ions with mass differences corresponding to amino acid monomers, where consecutive mass differences represent a de novo peptide sequence tag.33 As with non-peptidic molecules, accurate masses and amino acid sequence tags can be compared to literature. We were able to identify a number of peptide molecular families, including viscosin/WLIP/massetolides, orfamides, putisolvins, xantholysins, and tolaassins (Figure 1, Supplementary Figure 3).3–5, 24, 34–37 All of these compounds are involved in motility, behave as biosurfactants, and have anti-microbial, anti-parasitic and anti-biofilm activities.3–5, 24, 34–39 All the MS/MS spectra associated with these annotations are publicly available at http://gnps.ucsd.edu.17
Since molecular networking clusters molecules based on structural similarity, a single match to the GNPS Pseudomonas library allows for propagation of that structure through an entire molecular family. Of the metabolites we dereplicated, three seemingly separate molecular families (quinolones, labradorins, and pseudopyronines) cluster together due to similarities in alkyl side chain fragmentation. More specifically, the alkyl side chains are adjacent to an olefin and attached to a heterocyclic moiety that does not readily fragment, thereby resulting in clustering primarily due to alkyl fragmentation. Even though these molecules share a molecular family, due to the inherent gas phase behavior defined by their chemical structure, the families are separated into sub-families based on the subtleties of their structural differences.
Discovery of specialized metabolites
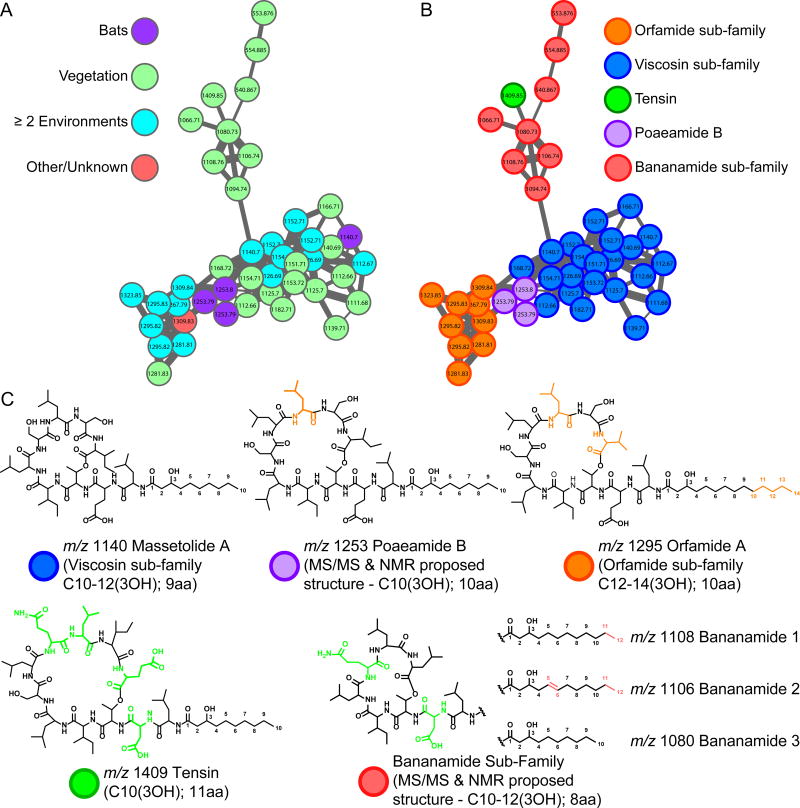
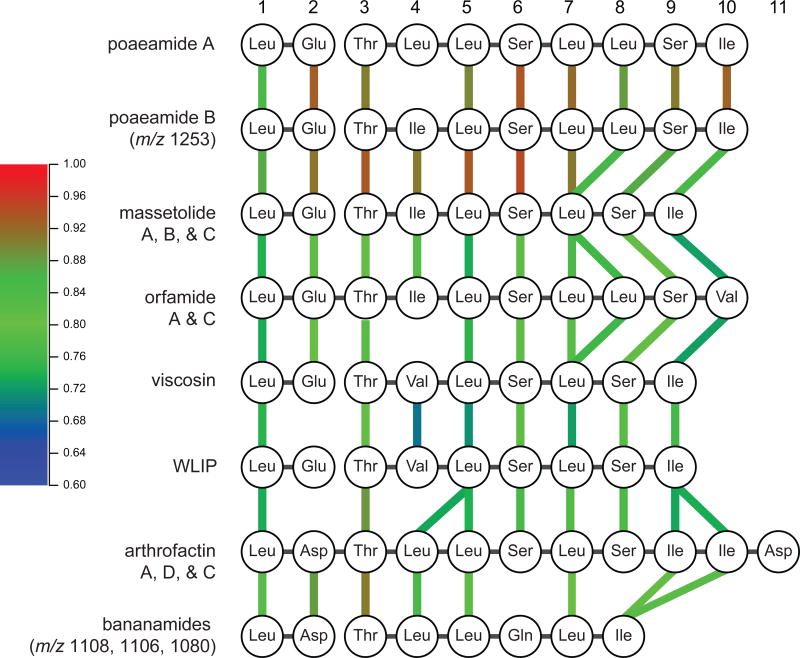
Sub-families are also observed in other molecular families. Figure 2 demonstrates a molecular family comprising related peptides including viscosin, white line inducing principle (WLIP), viscosinamide, massetolides A-F, orfamides A-C, and tensin.4, 34, 40 Differences due to amino acid substitution and varying fatty acid chains leads to the formation of sub-families. Further analysis of the viscosin molecular sub-families led to the identification of two uncharacterized members: a molecule at m/z 1253 and the sub-family at m/z 1108, 1106, 1094, 1080 and 1066, a sub-family most similar to tensin and massetolide A.
Figure 2.
A) The nonribosomal cyclic lipopeptides molecular family with environmental isolation mapping. Node colors delineate the environments from which the specific molecules of the cyclic lipopeptide family originate. Purple nodes: bat environments. Green nodes: vegetation environments. Teal nodes: two or more environments. Red nodes: other or unknown environments. B) The cyclic lipopeptide molecular family with node colors representing sub-families. Orange nodes: molecules from the orfamide sub-family. Blue nodes: molecules from the viscosin sub-family. Green node: tensin. Purple nodes: poaeamide B. Red nodes: molecules from the bananamide sub-family. C) Representatives from each sub-family were selected to show structural similarities. Massetolide A (from the viscosin sub-family) was chosen as a starting point and drawn in black. All other molecules were drawn in black where structural similarities to massetolide A existed. Portions of molecules highlighted in colors corresponding to the sub-family and illustrate the structural differences in comparison to massetolide A.
m/z 1253, produced solely by Pseudomonas synxantha CR32, was isolated from the bat species Myotis mystacinus in the Hranice Abyss of the Czech Republic. The MS/MS analysis yielded an amino acid sequence tag of Glu-Dhb-Ile/Leu-Ile/Leu-Ser-Ile/Leu-Ile/Leu-Ser-Ile/Leu a tag is similar to orfamide A (m/z 1295) and massetolide A (m/z 1140) (Figure 3). Compared to orfamide A, m/z 1253 substitutes an Ile/Leu for Val in the 10th position. Compared to massetolide A, m/z 1253 contains an additional Ile/Leu (Figures 2–4). m/z 1253 was isolated and NMR confirmed the identity of the amino acid residues predicted from MS/MS and revealed a C10-3-hydroxy fatty acid tail (Supplementary Figure 6, Supplementary Table 5). m/z 1253 is similar to poaeamide A from Pseudomonas poae, and therefore call m/z 1253, poaeamide B (Figure 4).22
Figure 3.
MS/MS comparison of select molecules from the cyclic lipopeptide molecular family. Molecules are arranged by increasing m/z. Sequence tags for each molecule are shown and color coded according to amino acid monomer. All molecules form a lactone linkage with the C-terminus of isoleucine (massetolide A and poaeamide B), leucine (bananamides 1–3), valine (orfamide A), or glutamic acid (tensin) to threonine (observed in the MS/MS spectra as DHB).
Figure 4.
Evolutionary relationships between the assembly lines of the newly identified cyclic lipopeptides and structurally related molecules, based on their constituent phylogenetic groups of adenylation domains, as indicated in Supplementary Figure 12. Each NRPS module is represented by its A-domain, along with its substrate specificity. Colored lines connect A-domains in adjacently depicted assembly lines that belong to the same phylogenetic group; note that some clades contain multiple substrates due to recent evolution of the substrate specificity of an individual member (e.g., Val-10 in the orfamide BGC), and that some substrate specificities are split across two clades (e.g., Glu-2 of the WLIP BGC originates from a different Glu-specific clade),. Color gradient represents a cosine similarity score between two given A-domains.
m/z’s 1108, 1106, 1094, 1080 and 1066 which we now call the bananamides, could not be dereplicated. The bananamides are named as such because they are only found to be produced by P. fluorescens collected from the banana rhizoplane in the wetlands of Galagedara, Sri Lanka.41 Analysis of the MS/MS data of m/z 1108, 1106, and 1080 yielded the amino acid tag Asp-Dhb-Ile/Leu-Ile/Leu-Gln-Ile/Leu-Ile/Leu. The molecule at m/z 1066 yielded a sequence tag Asp-Dhb-Ile/Leu-Ile/Leu-Gln-Ile/Leu-Val, where the 14 Dalton difference between m/z 1066 and 1080 is due to substitution of Ile/Leu for Val. Bananamides 1, 2, and 3 (m/z 1108, 1106, and 1080) were purified and NMR validated the sequence tag observed by MS/MS (Figure 3, Supplementary Figures 7–9, and Supplementary Table 6). MS and integrated proton values provide evidence for a C12 3-hydroxy fatty acid in m/z 1108, while m/z 1080 contains a C10 3-hydroxy fatty acid (Supplementary Figure 7 and 9 and Supplementary Table 6). m/z 1106 shows two olefinic protons with COSY correlations to two methylene protons that come from a C12 3-hydroxy unsaturated fatty acid at the fifth position (Figure 2, Supplementary Figure 8 and Supplementary Table 6). Such unsaturations haveonly been observed in a few Pseudomonas cyclic lipopeptides.42–47 Compared to massetolide A, the bananamides substitute the Glu with an Asp, the first Ser residue with a Gln, and an Ile with a Leu. An equivalent of the second Ser residue is absent. While compared to tensin, the bananamides lack the Ser, Glu, and one of the Leu residues (Figure 2 and 3).4
The automated peptidogenomics platform, Pep2Path, was attempted to match the MS/MS sequence tag data from poaeamide B and the bananamides to gene cluster families of public genome sequences. 16, 48 None were found. Therefore the genomes of P. synxantha CR32 and P. fluorescens BW11P2 were sequenced and subjected to antiSMASH analysis.49 antiSMASH revealed a nonribosomal peptide synthetase (NRPS) gene cluster predicted to make the poaeamide B (Figure 3, Supplementary Figure 10, and Supplementary Table 7),22 while the BW11P2 genome revealed a single NRPS gene cluster predicted to incorporate the amino acids consistent with the bananamide core peptide (Figure 3, Supplementary Figure 11, and Supplementary Table 8). The details of the biosynthetic gene cluster for poeaemide B and the bananamides is summarized in the following MiBIG50 links http://mibig.secondarymetabolites.org/repository/BGC0001346/index.html#cluster-1 and http://mibig.secondarymetabolites.org/repository/BGC0001347/index.html#cluster-2 (Figure 4, Methods, and Supplementary Figures 12–15). The Methods, Supplementary Figure 12–15 and Figure 4 outline the evolutionary relationships among the BGCs and reveal that the structural relations observed in the metabolite index are mirrored in the genetic relationships of the BGCs of these molecules.
Applying the Pseudomonas specialized metabolite index
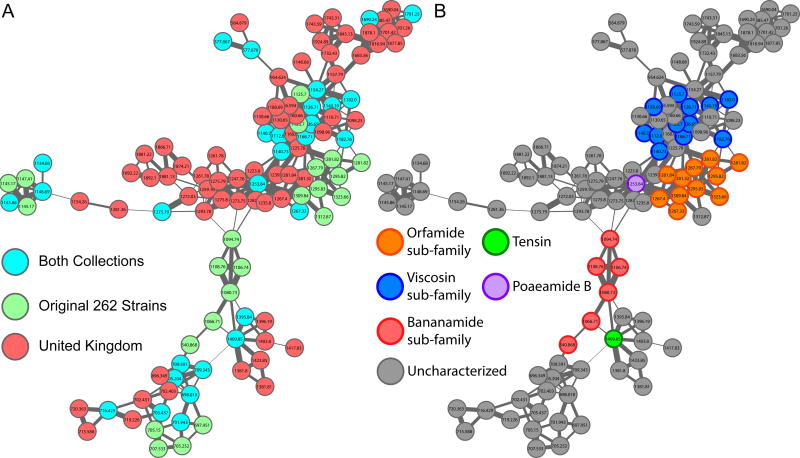
The Pseudomonas metabolite index was then used to examine an alternate Pseudomonas dataset. We compared the metabolite index curated from the original 260 strains with an additional 370 wheat-associated Pseudomonas extracts obtained from the United Kingdom to determine if our index aided molecular annotations (Supplementary Figures 4, 5, and 15 and Supplementary Tables 1 and 2). Twenty eight percent of detectable features are unique to our original collection, 39% are unique to the additional samples, with 33% of molecules overlapping between both collections (Supplementary Figure 16). Our current Pseudomonas index contains 9 annotated molecular families. By adding the additional 370 samples, 7 out of the 9 molecular families increase in the number of contributing samples and are automatically annotated. The lipopeptide molecular family focused on here (Figure 2) was produced by 34 strains out of the original 260 strains and increased to 97 contributing strains upon addition of the 370 additional UK samples (Figure 5). The same sub-families from Figure 2 are observed and highlighted in Figure 5, however, the addition of the UK samples increases the size of the overall molecular family and reveals many uncharacterized analogs (Figure 5). Poaeamide B, which was only produced by a single strain from the original 260, is now produced by a total of 45 strains. Conversely, the bananamides are still only identified in a single strain. Indexing specialized metabolites enabled us to determine the frequency of molecular detection in large bacterial collections. The molecular family, upon addition of the 370 extracts, reveals that many uncharacterized analogs are associated with the known sub-families and even provides insight that additional sub-families remain to be discovered. Ultimately, indexing known Pseudomonas compounds into GNPS allows for quick matching of these molecules when analyzing alternate datasets, thereby increasing the speed in which molecular characterization can take place from large culture collections. The effectiveness of the index will only increase as molecular knowledge continues to be added to MS/MS spectra in GNPS.
Figure 5.
The cyclic lipopeptide molecular family upon addition of the 370 wheat-associated samples from the United Kingdom. A) Node colors delineate which molecules are found from which Pseudomonas collection. Red nodes: additional 370 wheat-associated United Kingdom samples. Green nodes: original 260 Pseudomonas strains. Teal nodes: molecules that are produced by Pseudomonas from both collections. B) The nonribosomal cyclic lipopeptide molecular family with node colors representing sub-families. Orange nodes: molecules from the orfamide sub-family. Blue nodes: molecules from the viscosin sub-family. Green node: the molecule tensin. Purple nodes: poaeamide B. Red nodes: molecules from the bananamide sub-family.
Conclusion
(Re)-discovery and (re)-characterization of molecules and their evolutionary relationships is a time consuming and costly process, sometimes taking person-years to characterize a single molecule. The cost of dereplication, molecular annotation, and structure elucidation is not often disclosed in manuscripts, however when we do consider these costs, it is clear that the scientific community must organize this type of information for efficient reutilization and make the data searchable. The structural prediction of poaeamide B and bananamides 1–3 took 79 days and cost roughly $38,000 for all four molecules; this includes mass spectrometry costs and personnel salaries. The structural validation of poaeamide B after structural prediction based on MS/MS patterns and molecular family relationships to other known cyclic lipopeptides were already established, took 355 days and costs $86,000, while structural validation for bananamides 1–3 took 90 days and cost $25,000. These costs are small compared to other molecules that have been discovered. In the past, discovery of these molecules would be published, however the data and knowledge of the data would not be searchable in the same way gene sequences are searchable, thereby making annotation of metabolomics data from microbes always a time consuming process. In comparison, if we had four genes of interest we could search these genes in public databases and know: which organisms contain these sequences, which of these sequences is most similar, and know whether or not the sequence/sequence products have been characterized experimentally. All of this analysis could be accomplished in an afternoon. Indexing reference metabolomes and molecules provides similar capabilities that are currently the norm in sequence comparisons. For this reason, we believe in the importance of developing searchable indexes that are publicly accessible allows researchers to begin probing questions associated with evolutionary relatedness, uniqueness of molecules, and chemical diversity. Such capabilities will also open new ways to look at metabolomics data.
Materials and Methods
Pseudomonad Culture and Extract Conditions
Frozen stocks of Pseudomonas spp. were inoculated into 600 µL of liquid Tryptic Soy Broth (TSB, Bacto Soybean-Casein Digest Medium, 30 g / liter) in 2.0 mL 96 deep well plates (Thermo Scientific, Nunc 2.0 mL DeepWell Plate). Cultures were grown overnight at 30°C and 200 rpm and then diluted 500x into a second 2.0 mL 96 deep well plate containing fresh TSB liquid. 5 µL of the 500x dilution was inoculated into a third 2.0 mL 96 deep well plate containing 600 µL TSB agar (15 g agar / liter), sealed with 96 Well-Cap Mats (Thermo Scientific, Nunc 96 Well-Cap Mats), and incubated at 30°C for 72 hours. The cultures were extracted with 300 µL 50/50 v/v ethyl acetate (Fisher Scientific, HPLC grade)/methanol (Fisher Scientific, HPLC grade). The plates were resealed with the same 96 Well-Cap Mats, sonicated for 10 minutes, and extracted for an additional 50 minutes. 250 µL of these crude extracts were transferred into a pre-washed 96 well plate (Agilent Technologies, 96 well plates, 0.5 mL, polypropylene) and lyophilized to dryness. The extract protocol was repeated once more for a total extract volume of 500 µL.
LC-MS/MS Analysis
Dried samples were redissolved in 200 µL of methanol and centrifuged for 5 minutes at 1000 rpm. 150 µL of material was transferred into a new 96 well plate containing 50 µL of 400 µM glycocholic acid (Calbiochem, sodium salt) to serve as an injection standard and quality control for the chromatography (final concentration 100 µM), and then sealed with Zone-Free Sealing Film (Excel Scientific, Inc.). MS analysis was performed on a micrOTOF-Q II (Bruker Daltonics) mass spectrometer with ESI source, controlled by OTOF control and Hystar. MS Spectra were acquired in positive ion mode over a mass range of 100–2000 m/z. An external calibration with ESI-L Low Concentration Tuning Mix (Agilent Technologies) was performed prior to data acquisition and hexakis(1H,1H,3H-tetrafluoropropoxy)phosphazene (Synquest Laboratories) m/z 922.009798 was used as a lock mass internal calibrant during data acquisition. The following instrument settings were used for data acquisition: capillary voltage of 4500 V, nebulizer gas (nitrogen) pressure of 3 bar, ion source temperature of 200 °C, dry gas flow of 9 L/min, source temperature, and spectra acquisition rate of 3 Hz for MS1 and MS2. Minutes 0–0.5 were sent to waste. Minutes 0.5–10 were recorded with Auto MS/MS turned on. The 10 most intense ions per MS1 scan were selected and subjected to collision induced dissociation according to the following fragmentation and isolation list (values are m/z, isolation width, and collision energy, respectively): 100, 4, 16; 300, 5, 24; 500, 6, 30; 1000, 8, 40; 1500, 10, 50; 2000, 12, 70. In addition, the basic stepping function was used to fragment ions at 100% and 160% of the CID calculated for each m/z from the above fragmentation and isolation list with a timing of 50% for each step. Similarly, basic stepping of collision RF of 198 and 480 Vpp with a timing of 50% for each step and transfer time stepping of 75 and 92 µs with a timing of 50% for each step. MS/MS active exclusion parameter was set to 5 and released after 0.5 min. The injected samples were chromatographically separated using an Agilent 1290 Infinity Binary LC System (Agilent Technologies) controlled by Hystar software (Bruker Daltonics), using a 50×2.1 mm Kinetex 1.7 µM, C18, 100 Å chromatography column (Phenomenex), 30°C column temperature, 0.5 mL/min flow rate, mobile phase A 99.9% water (J.T.Baker, LC-MS grade) 0.1% formic acid (Fisher Scientific, Optima LC/MS), mobile phase B 99.9% acetonitrile (J.T.Baker, LC-MS grade) 0.1% formic acid (Fisher Scientific, Optima LC/MS), with the following gradient: 0–0.5 min 10% B, 0.5–1 min 50% B, 1–6 min 100% B, 6–9 min 100% B, 9–9.5 min 10% B, 9.5–10 min 10% B. 0 µL (blank) injections, methanol injections containing glycocholic acid, and agar treated and extracted under the same conditions as the culture conditions, were used as controls.
Molecular Networking
All LC-MS/MS data was converted to mzXML format using Compass Data Analysis (Bruker Daltonics) and uploaded to the Global Natural Products Social Molecular Networking webserver (http://gnps.ucsd.edu). The LC-MS/MS data for the 260 Pseudomonas isolates was analyzed using the Molecular Networking workflow with the following settings: Parent Mass Tolerance 0.9 Da, Ion Tolerance 0.45 Da, Min Pairs Cos 0.6, Min Matched Peaks 6, Network TopK 10, Minimum Cluster Size 2, and Maximum Connected Component Size 100. Molecular networking will merge all identical MS and MS/MS spectra, including identical MS/MS spectra of isomers. The molecular network was visualized using Cytoscape version 2.8.3 and displayed using an unweighted force directed layout. The data is publicly accessible at http://gnps.ucsd.edu under the MassIVE Accession number MSV000079450 and the networking results and parameters can be found at the following link: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c50e46ab24bc4e31a914c69e1df63b4e. Upon addition of the 370 wheat-associated Pseudomonas samples, Parent Mass Tolerance and Ion Tolerance were set to 1 Da and 0.5 Da, respectively, while Maximum Connected Component Size was increased to 100, in order to accommodate the additional 569,800 MS/MS scans used for the network. The remaining network settings were unchanged: Min Pairs Cos 0.6, Min Matched Peaks 6, Network TopK 10, and Minimum Cluster Size 2. At these settings, the GNPS community has determined that 1% of the annotations are incorrect, 4% not enough information to tell, 4% could be isomers or correct and 91% was determined to be correct.17 The data is publicly accessible under MassIVE Accession number MSV000079619 and the networking results and parameters can be found at the following link: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=b28463fdb3ce4d6cbc4bc6ea0129fdf3. The index itself can be comprised of MS/MS spectra from intact molecules but also products of in-source fragments, different adducts (Na+,K+,NH4+, Al3+, Fe3+, etc.)28, biosynthetic intermediates, biosynthetic diversification, isotopes (13C, 34S, halogenated compounds) or shunt products and system impurities.51, 52 Since molecular networking is a map of the diversity of MS/MS spectra, the index contains all such possibilities. System impurities can be readily identified by ensuring one has the proper blanks and are color coded grey in the network (Figure 1). The majority of the time, in-source fragments are spotted in the following ways within the molecular network: 1) MS/MS output of in-source fragments have similar pattern as the parent, therefore such artifacts usually become a sub-cluster of a molecular family and 2) Retention times (RT) of in-source fragments are always identical to the parent because are generated from the parent ion eluting from the column at the certain point of time. We export all the RT for all ions in GNPS so it can be verified during the analysis. 3) Since these are lipopeptides, as one would expect that if they are source fragments that one would see the loss of the lipid chain or the amino acids within the same sample not between samples, such mass differences within a parent mass is not observed 4) by observing the masses of the masses of the molecules produced by a known producer strain. For the peptides analyzed in the networks, no source fragment ions were observed.
Isolation of Poaeamide B
An overnight culture of Pseudomonas synxantha CR32, isolated from the bat species Myotis mystacinus found in the Hranice Abyss of the Czech Republic, was prepared from frozen stock in 7 mL of TSB liquid medium in a 14 mL round bottom culture tube (Corning) and shaken at 200 RPM and 30°C. 20 µL of overnight culture was used to inoculate 25 lawns of Pseudomonas synxantha CR32 on 10 mL TSB agar plates. The lawns were incubated for 72 hours at 30°C, transferred into a 500 mL Erlenmeyer flask, and extracted three times with 200 mL of 50/50 v/v ethyl acetate/methanol (Fischer). The extract supernatant was filtered away from the media and dried in vacuo. Dried crude material was dissolved in 2 mL of methanol and separated on an Agilent 1260 HPLC equipped with a 250 × 10 mm Discovery 5 µM, C18, 180 Å chromatography column (Supelco). LC conditions were as follows: 30°C column temperature, 2.0 mL/min flow rate, mobile phase A 99.9% water and 0.1% formic acid, mobile phase B 99.9% acetonitrile and 0.1% formic acid with the following isocratic gradient: 0–30 min 80% B, 31–33 min 100% B, 34–35 min 80% B. Poaeamide B was collected at 26–28 minutes on MS-based fraction collection. Molecules were verified simultaneously by MS/MS fragmentation.
Isolation of Bananamides 1, 2, and 3
An overnight culture of Pseudomonas fluorescens BW11P2, isolated from the banana rhizoplane in Galgadera, Sri Lanka, was prepared from frozen stock in 7 mL of TSB liquid medium in a 14 mL round bottom culture tube (Corning) and shaken at 200 RPM and 30°C. 5 mL of the overnight culture was added to 50 mL of liquid TSB containing 1 g of sterile glass beads (3 mm diameter Kimble Chase LLC) and 1 g of sterile Amberlite XAD-16 resin (Sigma) and incubated for 10 days at 30°C and 200 rpm in a 250 mL erlenmeyer flask. XAD-16 resin and cells were collected by vacuum filtration and extracted three times with 25 mL of 50/50 v/v ethyl acetate/methanol (Fischer) shaking for 1 hour at 200 rpm. The resin and glass beads were filtered and the crude extract supernatant dried in vacuo. To separate bananamides 1, 2, and 3, the dried crude material was dissolved in 5 mL of methanol and separated on an Agilent 1260 HPLC equipped with a 250 × 10 mm Discovery 5 µM, C18, 180 Å chromatography column (Supelco). LC conditions were as follows: 30°C column temperature, 2.0 mL/min flow rate, mobile phase A 99.9% water and 0.1% formic acid, mobile phase B 99.9% acetonitrile and 0.1% formic acid with the following isocratic gradient: 0–30 min 85% B, 31–33 min 100% B, 34–35 min 85% B. Bananamides 1, 2, and 3 were collected between 24.6–25.2, 20.6–21.2, and 16.1–16.7 minutes, respectively, on MS-based fraction collection. Molecules were verified simultaneously by MS/MS fragmentation.
NMR Measurements of poaeamide B and bananamides 1, 2, and 3
1D 1H-NMR, 2D 1H-1H double quantum filtered correlation spectroscopy (DQF-COSY), 2D 1H-13C heteronuclear single quantum coherence (HSQC), and 2D 1H-13C heteronuclear multiple bond correlation (HMBC) spectra of purified poaeamide B and bananamides 1, 2, and 3 were acquired at 25°C using a 600 MHz NMR (Magnex superconducting magnet, 14.1 T) fitted with a 1.7 mm cryoprobe and Bruker Avance II console operated using Bruker TopSpin 2.1 software. For NMR acquisition, 10–100 µg of poaeamide B and bananamides 1, 2, and 3 were dissolved in 50 µL of CD3OD (Cambridge Isotope Laboratories, Inc.).
Genome Sequencing, Assembly, and Analysis
Genomic DNA from Pseudomonas synxantha CR32 (poaeamide B producer [m/z 1253], accession number KU936045 and KU936046) and Pseudomonas fluorescens BW11P2 (bananamides producer [m/z 1108, 1106, and 1080], accession number LRUN00000000, and KX437753 for the bananamide BGC) was isolated using a Wizard Genomic DNA Purification Kit (Promega) in n=3 biological replicates. Sequencing libraries were constructed from 1 µg of genomic DNA using the Ion Xpress™ Plus Fragment Library Kit (ThermoFisher). DNA was sheared using the Covaris S2 (Covaris) to an average of 400 bp. After nick-repair and adapter ligation, the Pippin Prep instrument (Sage Science) was used to size select for 475 bp fragments using a 2% agarose gel DF cassette with Marker L, following the standard protocol. The library was quantified using a DNA High Sensitivity kit on the BioAnalyzer 2100 system (Agilent). The Ion PGM™ Template OT2 Kit (ThermoFisher) was used for sample preparation with the Ion OneTouchTM 2 System with a modified thermoprofile. Changes to the thermoprofile included an increase in melting temperature to 97°C and extended cycling parameters. Sequencing was performed using an Ion Torrent Personal Genome Machine (ThermoFisher) with an Ion PGM™ Hi-Q Sequencing Kit (ThermoFisher), according to the standard protocol, on a 318v2 sequencing chip (ThermoFisher). De novo genome assembly was performed using CLC Genomics Workbench software v5.01 (CLC bio); the full bananamide BGC was reconstructed by combining this with a second assembly using SPAdes.53 Sequencing of the Pseudomonas synxantha CR32 poaeamide B gene cluster resulted in two contigs of 9.9 kb and 31.3 kb. The Pseudomonas fluorescens BW11P2 genome assembled into 6.0 Mb of 130 contigs with an N50 of 87 kb. BGCs in the genomes were analyzed with antiSMASH and processed with custom Python scripts. BGC annotations were submitted to MIBiG50 with accession numbers BGC0001346 (bananamides) and BGC0001347 (poaeamide B). Phylogenetic analysis was performed using MEGA 7.0.54
To obtain an overview of the evolutionary relationships of biosynthetic gene clusters (BGCs) of different types of Pseudomonas cyclic lipopeptides, we compiled a list of 18 different biosynthetic gene clusters of cyclic lipopeptides. The phylogenetic tree of the adenylation domains contained distinct functional clades in which the adenylation domains share the same amino acid substrate specificity (Supplementary Figure 12)36, 52 Several sub-groups of cyclic lipopeptides are identified that are more distantly related to poaeamide B and the bananamides. The BGCs encoding larger assembly line structures, such as the syringopeptin BGC, have lower overall sequence and architectural similarity. Poaeamide B and bananamide BGCs are closely related to six other BGCs (arthrofactin, orfamide, massetolide, poaeamide A, viscosin, and WLIP). Using this tree, we constructed pseudo-sequences of adenylation domain clades for all BGCs that represent the functional architectures of the encoded assembly-lines to estimate the evolutionary distances between the gene clusters (Supplementary Figure 12). We used the distance metric with domain types defined as the adenylation domain clades from Supplementary Figure 12, and with weights of the Jaccard index, Goodman-Kruskal gamma index, and domain duplication index at 0.5, 0.25 and 0.25. Such analysis revealed many interesting aspects of evolutionary relationships that manifest themselves in the MS/MS data of the molecules found in the index. Overall, four other subfamilies of cyclic lipopeptide BGCs can be distinguished: sessilin/tolaasin, syringopeptin/nunapeptin/chicopeptin, putisolvin/entolysin/xantholysin and cichofactin/syringafactin. Some pathways are encoded on two separate genomic loci, while others are encoded in a single BGC configuration; the distribution of these two architectural configurations is notably discontinuous, also when plotted onto a phylogeny of C-starter domains, which constitute the most conserved part of the assembly lines. This suggests that multiple independent split/join events might have taken place during the evolution of this BGC family (Supplementary Figure 13). To understand specific evolutionary events on the domain level, such as duplications, deletions and insertions, a 2D-clustered heatmap was constructed (Supplementary Figure 14). The BGC of poaeamide B is related to BGCs which encode the production of poaeamide A, massetolides, and orfamides (Figure 4, and Supplementary Figure 14 and 15). While almost all A-domain sequences of poaeamide A and poaeamide B show similarity, the poaeamide B BGC distinguishes itself from the poaeamide A BGC through the presence of a distinct A-domain substituting an Ile for a Leu residue in the 4th position (Supplementary Figure 15). In addition, poaeamide B biosynthesis shows similarity to the massetolide gene cluster with an duplication of the seventh A-domain that activates leucine. The bananamides, however, are more of a molecular and evolutionary outlier. As reflected in the comparisons of the biosynthetic machineries, the first five modules of the bananamide NRPS assembly line are similar and co-linear with those of the arthrofactin gene cluster. The observed evolutionary relationships between these BGCs corroborate the structural relationships of the molecules visualized by molecular networking (Figure 2, 4, and Supplementary Figure 14 and 15). Conservation on the domain level, however, allows for the identification of evolutionary modularity underlying their structures.55 For example, the substructure synthesized by modules 5–6–7 in the poaeamide A, poaeamide B, massetolide, orfamide, WLIP, viscosin, and arthrofactin assembly-lines is shared between all of these molecules (Figure 5). The module conservation indicates a possible key role of this Leu-Ser-Leu substructure in mediating the biological activity of this group. All data and Python scripts used for generating each of the figures are available at https://git.wageningenur.nl/Xiaowen/pseudomonas/tree/master
Supplementary Material
Acknowledgments
Financial support was provided by the National Institute of Health (NIH) grants GM097509 (B.S.M. and P.C.D.). M.H.M. was supported by Rubicon (825.13.001) and Veni (863.15.002) grants from the Netherlands Organization for Scientific Research (NWO). J.R. and V.J.C were supported by a grant from the Netherlands BEBasic Foundation (project F07.003.01) R.D.M. was supported by KU Leuven grant GOA/011/2008. M.G.K.G. is the recipient of a post-doctoral fellowship from FWO Vlaanderen (12M4615N). A.M.K. and T.L.C. were supported by NSF grants DEB-1115895 and DEB-1336290 and US FWS grant F12AP01081. L.M.S. was supported by National Institutes of Health IRACDA K12 GM068524 grant award. J.G.M. was supported by BBSRC Institute Strategic Program (ISPG) Grant BB/J004553/1, and University of East Anglia start-up funding. T.H.M. was supported by by the BBSRC Institute Strategic Program (ISPG): Optimization of nutrients in soil-plant systems (BBS/E/C/00005196). We further acknowledge Bruker and NIH Grant GMS10RR029121, P41-GM103484 for the support of the shared instrumentation and the computational infrastructure that enabled this work. NMR data was acquired at the University of California, San Diego Skaggs School of Pharmacy and Pharmaceutical Sciences NMR Facility. We acknowledge Dr. Vanessa V. Phelan for a review of this manuscript and Mingxun Wang, Andrew T. Nelson, and Louis-Félix Nothias-Scaglia for their contributions.
Footnotes
Accession numbers
LC-MS/MS data is publicly accessible under the MassIVE accession number MSV000079450 or can be accessed by following this link: https://gnps.ucsd.edu/ProteoSAFe/result.jsp?task=5728ca4b0dfd4c058e0ef6151a31f9c4&view=advanced_view Molecular networking results and parameters can be found by follow this link: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c50e46ab24bc4e31a914c69e1df63b4e
The full genome sequencing data for Pseudomonas fluorescens strain BW11P2 (bananamides producer) can be found under NCBI accession number LRUN00000000. The genome sequencing data for the bananamide BGC can be found under NCBI accession number KX437753 and under MiBIG accession BGC0001346.
The genome sequencing data for the poaeamide B BGC from Pseudomonas synxantha strain CR32 (poaeamide B producer) can be found under NCBI accession numbers KU936045 and KU936046. In addition, the poaeamide B BGC can be found under MiBIG accession BGC0001347
Contributions:
D.D.N., A.M., X.L., M.H.M., and P.C.D. designed research.
D.D.N., A.M., N.K., X.L., M.S., J.F., K.A., T.L.L., B.M.D., B.S.M., M.H.M., and P.C.D. performed research.
D.D.N., A.M., N.K., X.L., M.S., M.G.K.G., J.F., B.M.D., R.D.M, M.H.M., and P.C.D. analyzed data.
M.G.K.G, V.J.C, T.C., J.G.M., T.H.M., L.M.S., A.M.K, J.R., and R.D.M. contributed microbial strains or extracts.
D.D.N., A.M., and P.C.D. wrote the paper.
The authors declare no conflict of interest.
References
- 1.Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 2009;26:1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 2.Kloepper JW, Leong J, Teintze M, Schroth MN. Pseudomonas siderophores: A mechanism explaining disease-suppressive soils. Curr. Microbiol. 1980;4:317–320. [Google Scholar]
- 3.Ron EZ, Rosenberg E. Natural roles of biosurfactants. Environ. Microbiol. 2001;3:229–236. doi: 10.1046/j.1462-2920.2001.00190.x. [DOI] [PubMed] [Google Scholar]
- 4.Raaijmakers JM, de Bruijn I, de Kock MJD. Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant. Microbe. Interact. 2006;19:699–710. doi: 10.1094/MPMI-19-0699. [DOI] [PubMed] [Google Scholar]
- 5.Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 6.Davies J. Specialized microbial metabolites: functions and origins. J. Antibiot. 2013;66:361–364. doi: 10.1038/ja.2013.61. [DOI] [PubMed] [Google Scholar]
- 7.Ichinose Y, Taguchi F, Mukaihara T. Pathogenicity and virulence factors of Pseudomonas syringae. J. Gen. Plant Pathol. 2013;79:285–296. [Google Scholar]
- 8.Raaijmakers JM, Vlami M, de Souza JT. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek. 2002;81:537–547. doi: 10.1023/a:1020501420831. [DOI] [PubMed] [Google Scholar]
- 9.Mascuch SJ, et al. Direct detection of fungal siderophores on bats with white-nose syndrome via fluorescence microscopy-guided ambient ionization mass spectrometry. PLoS One. 2015;10:e0119668. doi: 10.1371/journal.pone.0119668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoyt JR, et al. Bacteria isolated from bats inhibit the growth of Pseudogymnoascus destructans, the causative agent of white-nose syndrome. PLoS One. 2015;10:e0121329. doi: 10.1371/journal.pone.0121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JY, et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laatsch H. AntiBase 2014: The Natural Compound Identifier. 2014 [Google Scholar]
- 13.Buckingham J. Dictionary of Natural Products, Supplement 4. Taylor & Francis; 1997. [Google Scholar]
- 14.Blunt JW, Munro M. MarinLit. A database of the literature on marine natural products. 2003 [Google Scholar]
- 15.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1743–52. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen DD, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2611–20. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jošić D, et al. Antifungal activities of indigenous plant growth promoting Pseudomonas spp. from alfalfa and clover rhizosphere. Frontiers in Life Science. 2015;8:131–138. [Google Scholar]
- 21.Pauwelyn E, et al. New linear lipopeptides produced by Pseudomonas cichorii SF1-54 are involved in virulence, swarming motility, and biofilm formation. Mol. Plant. Microbe. Interact. 2013;26:585–598. doi: 10.1094/MPMI-11-12-0258-R. [DOI] [PubMed] [Google Scholar]
- 22.Zachow C, et al. The Novel Lipopeptide Poaeamide of the Endophyte Pseudomonas poae RE*1-1-14 Is Involved in Pathogen Suppression and Root Colonization. Mol. Plant. Microbe. Interact. 2015;28:800–810. doi: 10.1094/MPMI-12-14-0406-R. [DOI] [PubMed] [Google Scholar]
- 23.Kersten RD, et al. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4407–16. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettit GR, et al. Isolation of labradorins 1 and 2 from Pseudomonas syringae pv. coronafaciens. J. Nat. Prod. 2002;65:1793–1797. doi: 10.1021/np020173x. [DOI] [PubMed] [Google Scholar]
- 26.Kong F, Singh MP, Carter GT. Pseudopyronines A and B, alpha-pyrones produced by a marine Pseudomonas sp. F92S91, and evidence for the conversion of 4-hydroxy-alpha-pyrone to 3-furanone. J. Nat. Prod. 2005;68:920–923. doi: 10.1021/np050038v. [DOI] [PubMed] [Google Scholar]
- 27.Chu M, et al. Structure of sch 419560, a novel alpha-pyrone antibiotic produced by Pseudomonas fluorescens. J. Antibiot. 2002;55:215–218. doi: 10.7164/antibiotics.55.215. [DOI] [PubMed] [Google Scholar]
- 28.Moree WJ, et al. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdel-Mawgoud AM, Lépine F, Déziel E. Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010;86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonçalves-de-Albuquerque CF, et al. Possible mechanisms of Pseudomonas aeruginosa-associated lung disease. Int. J. Med. Microbiol. 2016;306:20–28. doi: 10.1016/j.ijmm.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Grundmann F, et al. Identification and isolation of insecticidal oxazoles from Pseudomonas spp. Beilstein J. Org. Chem. 2012;8:749–752. doi: 10.3762/bjoc.8.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer JS, et al. Biosynthetic Origin of the Antibiotic Pseudopyronines A and B in Pseudomonas putida BW11M1. Chembiochem. 2015;16:2491–2497. doi: 10.1002/cbic.201500413. [DOI] [PubMed] [Google Scholar]
- 33.Kersten RD, et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross H, et al. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 2007;14:53–63. doi: 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Kuiper I, et al. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 2004;51:97–113. doi: 10.1046/j.1365-2958.2003.03751.x. [DOI] [PubMed] [Google Scholar]
- 36.Li W, et al. The antimicrobial compound xantholysin defines a new group of Pseudomonas cyclic lipopeptides. PLoS One. 2013;8:e62946. doi: 10.1371/journal.pone.0062946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rainey PB, Brodey CL, Johnstone K. Biological Properties and Spectrum of Activity of Tolaasin, a Lipodepsipeptide Toxin Produced by the Mushroom Pathogen Pseudomonas-Tolaasii. Physiol. Mol. Plant Pathol. 1991;39:57–70. [Google Scholar]
- 38.Tran H, Kruijt M, Raaijmakers JM. Diversity and activity of biosurfactant-producing Pseudomonas in the rhizosphere of black pepper in Vietnam. J. Appl. Microbiol. 2008;104:839–851. doi: 10.1111/j.1365-2672.2007.03618.x. [DOI] [PubMed] [Google Scholar]
- 39.Kruijt M, Tran H, Raaijmakers JM. Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promoting Pseudomonas putida 267. J. Appl. Microbiol. 2009;107:546–556. doi: 10.1111/j.1365-2672.2009.04244.x. [DOI] [PubMed] [Google Scholar]
- 40.Rokni-Zadeh H, et al. Genetic and functional characterization of cyclic lipopeptide white-line-inducing principle (WLIP) production by rice rhizosphere isolate Pseudomonas putida RW10S2. Appl. Environ. Microbiol. 2012;78:4826–4834. doi: 10.1128/AEM.00335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vlassak K, Van Holm L, Duchateau L, Vanderleyden J, De Mot R. Isolation and characterization of fluorescent Pseudomonas associated with the roots of rice and banana grown in Sri Lanka. Plant Soil. 1992;145:51–63. [Google Scholar]
- 42.Emanuele MC, et al. Corceptins, new bioactive lipodepsipeptides from cultures of Pseudomonas corrugata. FEBS Lett. 1998;433:317–320. doi: 10.1016/s0014-5793(98)00933-8. [DOI] [PubMed] [Google Scholar]
- 43.Pauwelyn E. Epidemiology and pathogenicity mechanisms of Pseudomonas cichorii, the causal agent of midrib rot in greenhouse-grown butterhead lettuce (Lactuca sativa L.) Ghent University; 2012. [Google Scholar]
- 44.Morikawa M, et al. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J. Bacteriol. 1993;175:6459–6466. doi: 10.1128/jb.175.20.6459-6466.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D’aes J, et al. To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ. Microbiol. 2014;16:2282–2300. doi: 10.1111/1462-2920.12462. [DOI] [PubMed] [Google Scholar]
- 46.Huang C-J, et al. Characterization of Cichopeptins, New Phytotoxic Cyclic Lipodepsipeptides Produced by Pseudomonas cichorii SF1-54 and Their Role in Bacterial Midrib Rot Disease of Lettuce. Mol. Plant. Microbe. Interact. 2015;28:1009–1022. doi: 10.1094/MPMI-03-15-0061-R. [DOI] [PubMed] [Google Scholar]
- 47.Lange A, Sun H, Pilger J, Reinscheid UM, Gross H. Predicting the structure of cyclic lipopeptides by bioinformatics: structure revision of arthrofactin. Chembiochem. 2012;13:2671–2675. doi: 10.1002/cbic.201200532. [DOI] [PubMed] [Google Scholar]
- 48.Medema MH, et al. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput. Biol. 2014;10:e1003822. doi: 10.1371/journal.pcbi.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber T, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Medema MH, et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat. Chem. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput. Biol. 2014;10:e1004016. doi: 10.1371/journal.pcbi.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medema MH, Takano E, Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 2013;30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.