Abstract
To decipher neural circuits underlying brain functions, viral tracers are widely applied to map input and output connectivity of specific neuronal populations. Despite the successful application of retrograde transsynaptic viruses for identifying presynaptic neurons of transduced neurons, analogous anterograde transsynaptic tools for tagging postsynaptically targeted neurons remain under development. Here, we report that adeno-associated virus (AAV1 and AAV9) exhibit anterograde transsynaptic spread properties. AAV1-Cre from transduced presynaptic neurons effectively and specifically drove Cre-dependent transgene expression in selected postsynaptic neuronal targets, and thus allowed the tracing and functional manipulation of axonal projections from the latter input-defined neuronal population. Application of this tool in superior colliculus (SC) revealed that SC neuron subpopulations receiving corticocollicular projections from auditory and visual cortex specifically drove flight and freezing, two different types of defense behavior, respectively. Such anterograde transsynaptic tagging is thus useful for forward screening of distinct functional neural pathways embedded in complex brain circuits.
Introduction
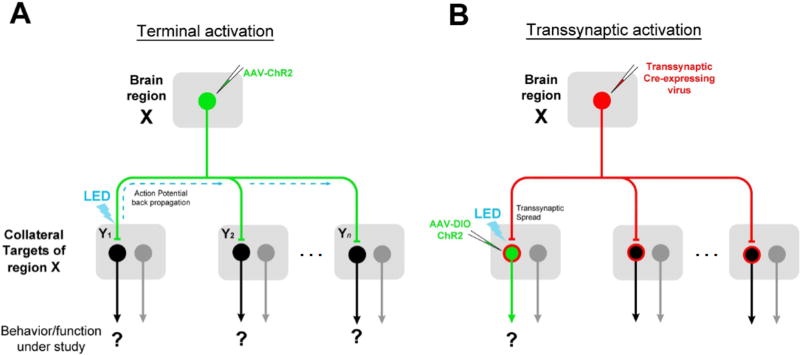
A key for deciphering complex brain circuits is the ability to identify neural pathways underlying distinct behaviors or brain functions. Given that a brain region (“X”, Figure 1) is known to be involved in a specific behavior/function, how to screen the neural pathways downstream of X that mediate this behavior/function remains challenging. Traditional anterograde tracing methods allow the identification of all regions targeted by neurons in X. However, for each of these regions (“Y1” to “Yn”, Figure 1), possibility exists that there are neurons that do not receive direct input from X and are unrelated to the X-dependent function under study. These neurons are not necessarily molecularly or genetically different from those directly involved in the function of interest based on our current knowledge. Therefore, to precisely trace the neural pathway underlying the X-dependent function under study, it is necessary to identify in a relevant downstream nucleus (Y) the neuronal subpopulation that receives direct input from X. This requires afferent-dependent tagging of postsynaptic neurons only in a selected target region, which to our knowledge has not been achieved with current approaches (Nassi et al., 2015). A straightforward strategy for labeling those input-defined postsynaptic neurons is to apply an anterograde transsynaptic tracer in the source region coupled with tracer-dependent transgene expression in the selected target region. This would allow functional manipulations of a specific second-order downstream pathway starting from the source nucleus.
Figure 1. Advantages of anterograde transsynaptic mapping of functional circuits.
(A) A brain region “X” is known to mediate a behavior/function of interest. Neurons in X project to multiple target nuclei (“Y”). To map the relevant downstream circuit, conventional method relies on activation of ChR2-expressing X axon terminals in a given target nucleus. This may result in unwanted activation of collateral targets via antidromic stimulation (marked by dash lines).
(B) A virus capable of anterograde transsynaptic spread would allow direct activation of postsynaptic cells in a target region that specifically receive input from region X, by enabling Cre-dependent transgene expression (green) in a Y nucleus.
Transcellular/transsynaptic viral tracers can be potentially powerful tools for mapping functional circuits in the above described manner (Figure 1B). However, while retrograde transsynaptic viral tracers, e.g. rabies virus and pseudorabies virus (PRV), have been widely used for mapping presynaptic inputs to transduced neurons (Wickersham et al., 2007; Wall et al., 2010; Defalco et al., 2001; Ekstrand et al., 2008), analogous anterograde transsynaptic tools for tracing neural pathways immediately downstream of targeted postsynaptic neurons remain under development. Although several viruses, e.g. herpes simplex virus (HSV) and vesicular stomatitis virus (VSV), have been found previously to exhibit transneuronal/transsynaptic spread, the neurotoxicity of these viruses and their uncontrollable spread across multiple serial synapses largely limit their applications in mapping circuits in a more precise manner (Lo and Anderson, 2011; Beier et al., 2011). Recent studies suggest that AAV1 can be transported anterogradely down the axon (Castle et al., 2013; Castle et al., 2014). However, evidence for its potential transneuronal/transsynaptic transduction remain largely unclear and controversial (Oh et al., 2014; Salegio et al., 2013; Hutson et al., 2015; Harris et al., 2012; Aschauer et al., 2013). For example, from all the images in the Allen Institute database (Oh et al., 2014), no evidence for anterograde transneuronal spread of AAV1 was obtained. Here, we thoroughly investigated the elusive transneuronal/transsynaptic transport of AAV from presynaptic to postsynaptic cell populations. Together with the development of an intersectional approach, we have established AAV1 as an effective anterograde transsynaptic tracer for mapping input-defined functional neural pathways.
Results
Anterograde transneuronal spread of AAV1-Cre
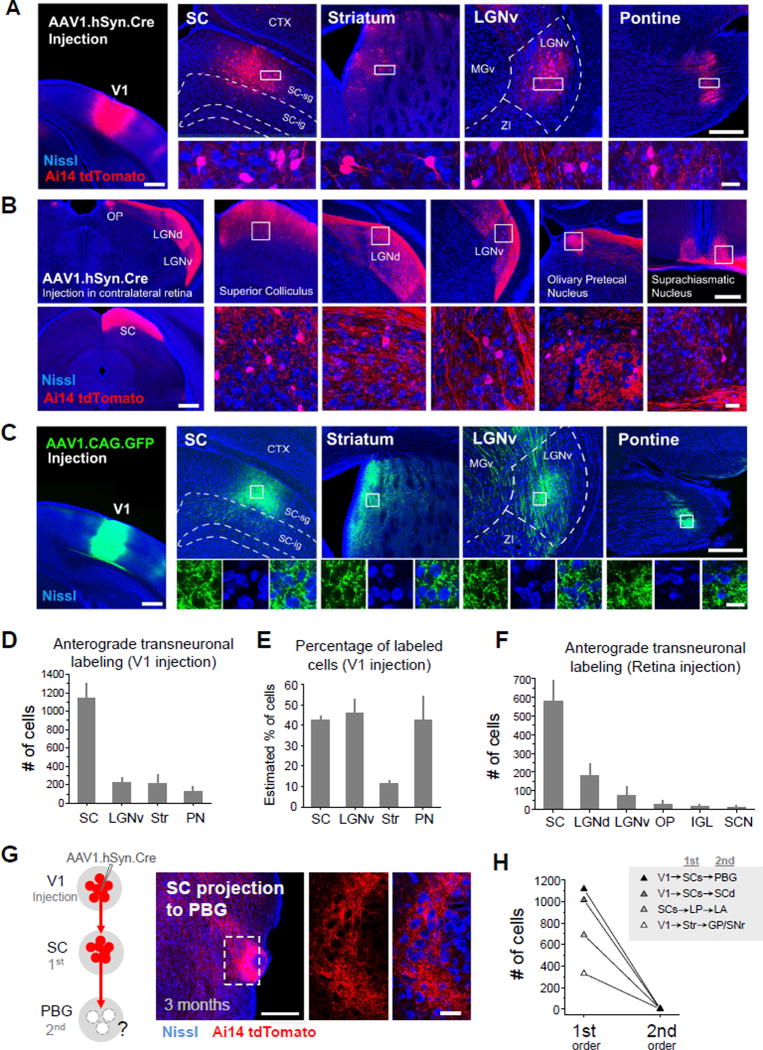
To test for any anterograde transneuronal transport of AAV, we injected AAV2/1-hSyn-Cre into the primary visual cortex (V1) of Ai14 (Cre-dependent tdTomato reporter) mice (Madisen et al. 2010). Following a 4 week post-injection survival time, we observed tdTomato-expressing cell bodies intermingled with terminal fields of tdTomato-labeled axons in all regions known to be directly targeted by V1, including the superior colliculus (SC), striatum (Str), ventral lateral geniculate nucleus (LGNv), and the pontine nucleus (PN) (Figure 2A). As these four regions do not project back to V1 (Simmons et al., 1982; Oh et al., 2014; Zingg et al., 2014), the presence of tdTomato+ cell bodies in these areas may only be explained by transneuronal spread of the Cre virus from V1 axons to neurons in their targeted structures. AAV1-Cre was also able to retrogradely spread to presynaptic neurons, although with a low efficiency, resulting in Cre-dependent transgene expression in presynaptic neurons (Figure S1, also see Tervo, et al., 2016; Rothermel, et al., 2013; Aschauer et al., 2013). Therefore in brain regions that are reciprocally connected with V1 (e.g. dorsal lateral geniculate nucleus, LGNd), tdTomato labeling of cell bodies can result from both anterograde and retrograde transneuronal transport of the Cre virus. These regions are not considered in the current study because of the ambiguity.
Figure 2. Anterograde transneuronal transport of AAV1-Cre.
(A) Left, tdTomato (red) expression in the injection site following injection of AAV1-hSyn-Cre into V1 of an Ai14 reporter mouse. Blue is Nissl staining. Right four panels, tdTomato fluorescence in regions downstream of V1 (SC, striatum, LGNv, pontine nucleus). High-magnification images (bottom) show tdTomato-labeled cell bodies in the region indicated by the white box. Scale bars: 500 µm, left panel; 250 µm, right top panels; 25 µm, right bottom panels. The same scales apply to (B) and (C) below correspondingly.
(B) Anterograde transneuronal labeling in the SC, LGNd, LGNv, olivary pretectal nucleus (OP), and suprachiasmatic nucleus (SCN) following injection of AAV1-hSyn-Cre into the contralateral retina. High-magnification images (bottom panels) show tdTomato-labeled cell bodies in boxed regions.
(C) Similar experiment as in (A) except that AAV1-CAG-GFP was injected. Note that no GFP-labeled cell bodies were found in regions downstream of V1 (4 week post-injection survival time).
(D) Quantification of total number of tdTomato-labeled cells in selected structures downstream of V1 following injection of AAV1-hSyn-Cre (4 week post-injection survival time, n = 4 mice, Error bar = SD).
(E) Estimated percentage of tdTomato-labeled cells as compared with the total number of cells within a defined local region in each downstream structure (D) that includes the majority of transneuronally labeled neurons (see Figure S2 and Experimental Procedures). N = 4. Error bar = SD.
(F) Quantification of total number of labeled cells found in structures downstream of the contralateral retina following injection of AAV1-hSyn-Cre (4 week post-injection survival, n = 4 mice, Error bar = SD).
(G) Left, injection of AAV1-hSyn-Cre in V1 of Ai14 reporter mice labels neurons in SC (1st order connection), which in turn project strongly to PBG (2nd order connection). Middle and right, dense tdTomato+ axons (red) were observed in PBG, but no tdTomato+ cell bodies (right panels) after 3 month post-injection survival time. Scale bar: 250 µm, middle panel; 25 µm, right panels.
(H) Quantification of number of labeled cells in the first and second order downstream regions in four different mice. No second order spread was observed after 3 month post-injection survival time. Abbreviations: GP, globus pallidus; LA, lateral amygdala; LP, lateral posterior nucleus of thalamus; PBG, parabigeminal nucleus; SCs, superior colliculus, superficial layer; SCd, superior colliculus, deep layer; SNr, substantia nigra reticulate.
To further confirm the anterograde transneuronal spread of AAV1, we injected AAV1-hSyn-Cre unilaterally into the retina of Ai14 mice, and examined tdTomato expression in brain regions known to receive retinal ganglion cell (RGC) inputs. In all regions examined, including the SC, LGNd, LGNv, olivary pretectal nucleus (OP) and the suprachiasmatic nucleus (SCN), we observed tdTomato+ cell bodies (Figure 2B). Again, since none of the above central structures project back to the retina (Morin and Studholme, 2014), the tdTomato labeling of cell bodies can only be a result of anterograde transneuronal spread of the virus from RGC axons to target cells.
It is interesting to note that anterograde transneuronal transport of AAV has not been widely reported, despite the extensive use of AAVs expressing GFP or channelrhodopsin2 (ChR2) (Harris et al., 2012; Aschauer et al., 2013; Yizhar et al., 2011; Oh et al., 2014). Consistent with previous results, at 4 weeks following injections of AAV1-CAG-GFP into V1, we did not find any GFP+ cell bodies in regions directly targeted by V1, despite the presence of strongly labeled axonal terminals (Figure 2C). This result indicates that the efficiency of the transneuronal spread is relatively low. That is, only a small number of viral particles might be transported transneuronally from the initial host cell, leading to extremely weak GFP expression in the downstream neurons, which may be below the detection threshold. Thus, in any application of AAV-mediated anterograde transneuronal tagging, an amplification step is required, such as utilizing Cre to unlock robust transgene expression.
We counted the total number of anterogradely labeled neurons in each of the target structures of interest. For V1 injections, SC contained the largest number of labeled neurons, followed by LGNv, striatum (Str) and the pontine nucleus (PN) (Figure 2D). The number of tdTomato-labeled neurons was compared with that of Nissl-stained cell somata within the same local area (Figure S2). We found that in the SC, LGNv, and PN, the percentage of anterogradely labeled cells reached about 40% (Figure 2E), suggesting that the number of neurons that receive the transneuronally transported Cre virus is relatively high. For retina injections, SC also contained the largest number of anterogradely labeled neurons, followed by LGNd and LGNv (Figure 2F). The OP, intergeniculate leaflet (IGL), and SCN also contained a small number of labeled neurons.
Outside of the regions known to be directly innervated by V1 or retinal axons, we did not observe any tdTomato+ cell bodies, suggesting that the virus does not further spread to second-order downstream structures. A good example is shown in Figure 2G. While SC neurons were labeled by tdTomato expression unlocked by anterogradely transported AAV1-Cre from V1, in the parabigeminal nucleus (PBG) which is directly targeted by SC (Huerta and Harting, 1984; Shang, et al., 2015), no tdTomato+ cell bodies were found, even at 3 months post injection (Figure 2G, right). In several other second-order nuclei examined, we did not find tdTomato-labeled cell bodies either (Figure 2H).
Dependence on viral type, serotype and other factors
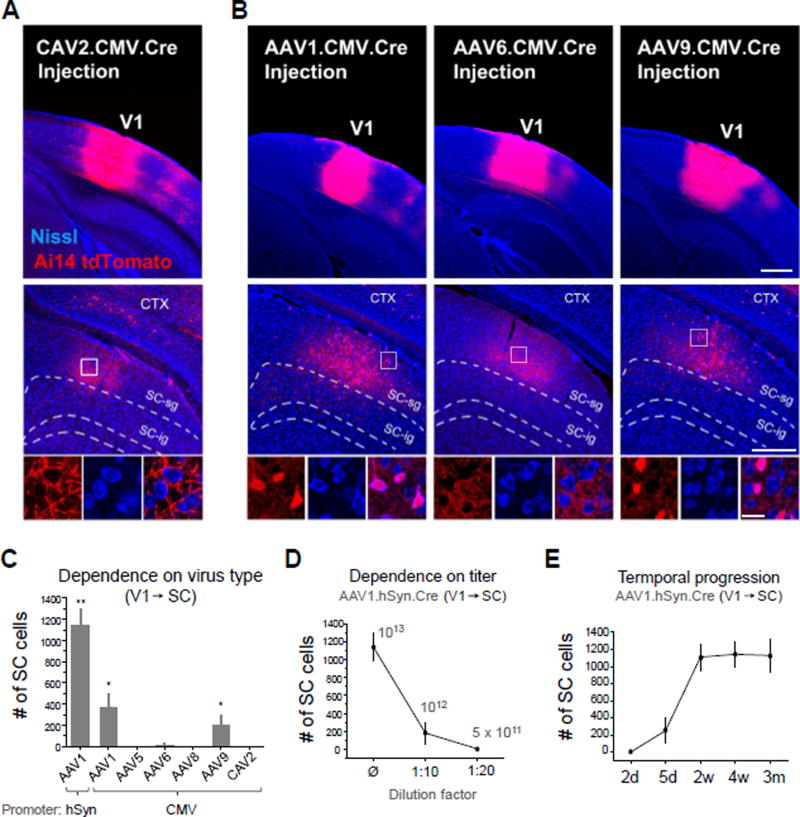
We next asked whether the observed anterograde transneuronal spread is a general property common to other viruses. To test this, we first injected Cre-expressing canine adenovirus (CAV2-CMV-Cre; Soudais et al., 2001) in a similar way as AAV1. At 4 weeks following V1 injections in Ai14 mice, no tdTomato+ cell bodies were observed in any regions exclusively downstream of V1 (Figure 3A). This suggests that anterograde transneuronal transport might be a unique property of AAV (and possibly some other viruses), due to some specific interactions between the virus and host cells. To test if this property varies with serotype, we compared transneuronal labeling with injections of AAV1, AAV6, AAV9, AAV5, and AAV8 encoding CMV-driven Cre into V1 (Figure 3B, 3C). AAV6, AAV5 and AAV8 did not exhibit anterograde transneuronal transport, unlike AAV1 and AAV9 (Figure 3B, 3C), despite similarly strong local transduction of neurons at the injection site (Figure 3B). These results further demonstrate that the observed anterograde transneuronal transport of AAV may reflect specific interactions between viral capsid proteins and host cells. It is worth noting that due to the higher titer of the injected AAV9 than AAV1 (see Experimental Procedures), the actual efficiency of AAV9 might be considerably lower than AAV1. Moreover, as both CAV2 and AAV6 drove high levels of Cre expression at the injection site (Figure 3A, 3B), yet no neurons were labeled in downstream targets, this suggests that Cre protein itself is incapable of transneuronal spread. The observed transneuronal labeling thus results from the spread of the Cre-encoding virus, rather than the Cre protein per se.
Figure 3. Serotype specificity of transneuronal transport.
(A) CAV2-CMV-Cre injection in V1 (upper panel). No anterograde transneuronal labeling was observed in regions downstream of V1 such as the SC (middle and bottom panels). High-magnification images (bottom) of the boxed region are shown (4 week post-injection survival).
(B) Comparison of AAV1, AAV6, and AAV9. Injection into V1 resulted in robust anterograde transneuronal labeling in SC with AAV1 (left) and AAV9 (right), but not AAV6 (middle) (4 week post-injection survival, 60 nl injection each). Scale bars: 500 µm, top panels; 250 µm, middle panels; 25 µm, bottom panels. The same scales also apply to (A) correspondingly.
(C) Quantification of total number of cells labeled in SC for different Cre-expressing viruses injected into V1 of Ai14 mice (4 week post-injection survival, 60 nl injection, n = 4 mice each). Error bar = SD. **, p < 0.01, different from all other groups; *, p < 0.05, different from AAV5, AAV6, AAV8 and CAV2 groups (one-way ANOVA and post hoc test).
(D) Dependence of transneuronal labeling on viral concentration. Number of labeled cells in SC following injection of undiluted AAV1-hSyn-Cre in V1, 1:10 dilution, or 1:20 dilution (4 week post-injection survival, 60 nl injection, n = 4 each). Error bar = SD.
(E) Quantification of transneuronal labeling in SC following different post-injection survival times: 2 days, 5 days, 2 weeks, 4 weeks, and 3 months (60 nl injections of AAV1-hSyn-Cre in V1, n = 4 each). Error bar = SD.
Relatively high titers of the virus were needed for the anterograde transneuronal spread, as the number of labeled neurons rapidly reduced with decreasing titers of AAV1 (Figure 3D). The anterograde transneuronal labeling could be observed at as early as 5 days post injection, was fully expressed at 2 weeks post injection and then remained stable for as long as 3 months (Figure 3E).
Synaptic specificity of anterograde transneuronal spread
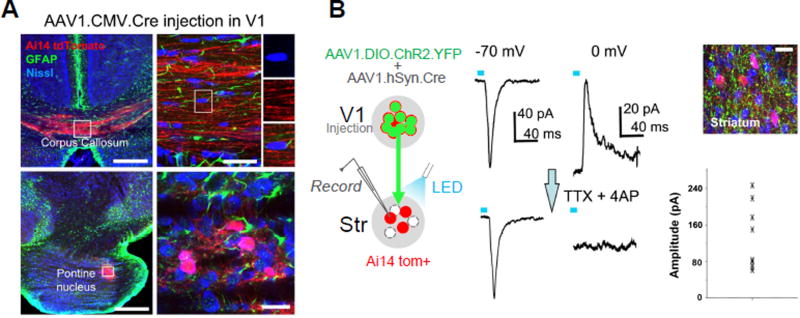
Does AAV spread specifically to synaptically connected target neurons or is it just released from axons locally capable of transducing all the neighboring cells? To address this issue, we first examined whether AAV is capable of transducing glial cells surrounding axonal fibers of passage, which might be expected if the virus was released non-specifically along the axon. We injected AAV1-CMV-Cre, which is capable of expressing Cre in both neurons and glia, in V1 of Ai14 mice. In the corpus callosum, which is comprised almost entirely of glia and axons (Sturrock, 1976; Aboitiz & Montiel, 2003), no tdTomato+ cell bodies were identified (Figure 4A, top panels), whereas strong transneuronal cell body labeling still occurred in the target structures of V1 (e.g. pontine nucleus, Figure 4A, bottom panels). These results suggest that the virus does not “leak” from fibers of passage. Additionally, in the sections containing transneuronal labeling, we co-stained the tissue with a marker for astrocytes, GFAP (Xu et al., 1999). In all sections examined, we did not observe co-labeling of tdTomato+ cell bodies with GFAP antibody (e.g. Figure 4A, bottom panels). Given that astrocytes can participate in a tripartite synaptic structure (Perea et al., 2009), the failure of their transduction by the virus suggests that the viral spread is highly restricted to neuronal structures in closest proximity to presynaptic terminals.
Figure 4. Synaptic specificity of anterograde viral spread.
(A) TdTomato expression (red) and GFAP labeling (green) in the corpus callosum (upper) and pontine nucleus (lower) following injection of AAV1-CMV-Cre into V1 of an Ai14 mouse. High-magnification images (right panels) show that Nissl stained cell bodies (blue) within the corpus callosum were negative for tdTomato (upper), and that tdTomato-positive cell bodies within PN were negative for GFAP staining (lower). Scale bars: 250 µm, left panels; 25 µm, right panels.
(B) Slice recording from transneuronally labeled neurons in the striatum (red) following co-injection of AAV1-hSyn-Cre and AAV1-EF1a-DIO-ChR2-YFP into V1 (left panel). Top right image shows ChR2-expressing axons (green) surrounding tdTomato-labeled striatal neurons. Scale: 25 µm. Middle panel, average LED-evoked excitatory (−70 mV) and inhibitory (0 mV) currents in an example tdTomato+ striatal neuron before and after perfusing in TTX and 4AP. LED stimulation is marked by a blue bar. Right bottom, a summary of amplitudes of average monosynaptic excitatory currents evoked by LED in 9 recorded striatal cells.
We next asked whether transneuronally labeled cells received functional synaptic input from neurons within the starter population. To test this, we injected a 1:1 mixture of AAV1-hSyn-Cre and a Cre-dependent ChR2 virus, AAV1-EF1α-DIO-ChR2-EYFP, into V1 of Ai14 mice (Figure 4B, left panel). Only neurons co-transduced with the two viruses could express ChR2. In slice preparations, we examined monosynaptic connectivity between ChR2-labeled V1 axons and tdTomato+ striatal neurons with whole-cell recording in the presence of TTX and 4-AP (Petreanu et al., 2009). In all recorded cells, blue LED pulses evoked excitatory synaptic currents which remained in the presence of TTX and 4-AP, while the evoked inhibitory currents were abolished by TTX and 4-AP (Figure 4B, middle and right panels). Since the ChR2-labeled V1 axons contained the Cre virus, the functional coupling between the former and the transneuronally labeled striatal neurons provides strong evidence for transsynaptic spread of the Cre virus to synaptically connected target neurons. To further support this notion, our control experiments (Castro-Alamancos and Favero, 2016) demonstrated that the LED-evoked current was only synaptic in nature, as antagonists of glutamate receptors completely blocked the current (Figure S3).
Mapping outputs of input-defined SC neuron subpopulations
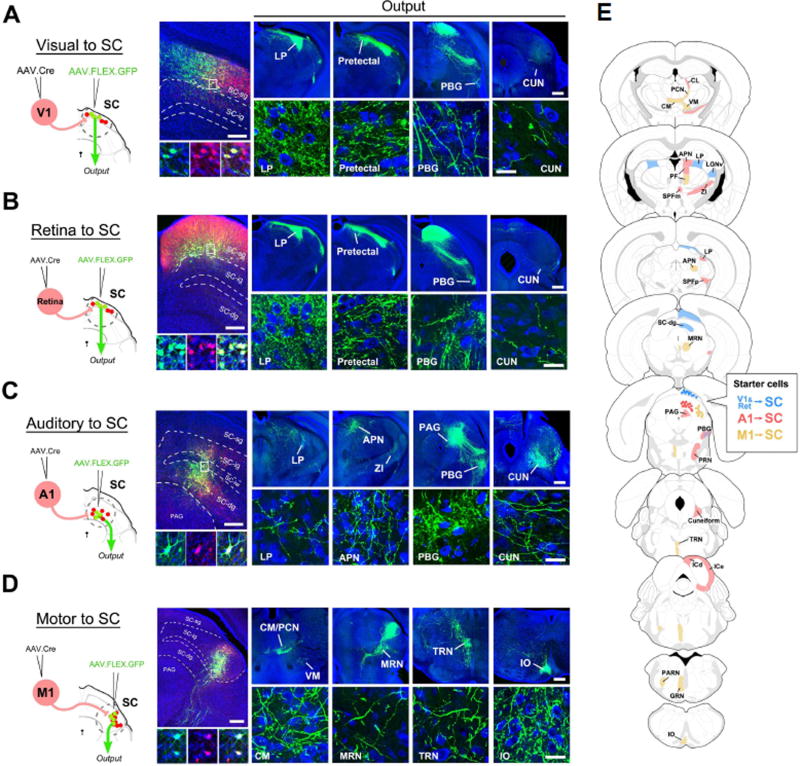
The anterograde transsynaptic property of AAV-Cre allows its application in mapping the axonal projections of neuronal populations with a specific presynaptic input source (i.e. input-defined). To test this idea, we used a two-step viral injection procedure and selected SC as the target structure, as it receives inputs from the retina as well as a variety of cortical areas, which terminate within distinct layers of SC (Huerta and Harting, 1984; Comoli et al., 2012). We first injected AAV1-hSyn-Cre in V1 of Ai14 mice, to label SC neurons that receive V1 input. Several days later, a second injection of AAV1-CAG-FLEX-GFP was made in SC (Figure 5A, left panel). After 4 weeks post-injection survival time, we observed robust GFP expression in tdTomato+ neurons specifically within the superficial gray layer of SC (i.e. SC-sg; Figure 5A, middle panel). 100% of GFP+ neurons were also tdTomato+, consistent with a result of co-transduction of Cre-dependent GFP and transneuronally transported Cre viruses. We examined the long-range axonal projections of this specific set of SC-sg neurons to various target structures. The most prominent axonal labeling was found in the lateral posterior nucleus (LP) of thalamus, with additional labeling seen in pretectal regions and PBG (Figure 5A, right panel). Next we examined SC neurons receiving RGC projections, which also target the superficial layer of SC. As with V1, paired injections in the retina and SC yielded a group of GFP+ neurons specifically within SC-sg (Figure 5B). Their axonal projection targets were found to closely match those of the V1 input-defined population (compare Figure 5A and Figure 5B), suggesting that the retina and V1 may target a similar group of SC neurons.
Figure 5. Mapping of axonal outputs of input-defined neuronal populations in SC.
(A) Left, AAV1-hSyn-Cre was injected into V1 of Ai14 mice, followed by a second injection of AAV1-CAG-FLEX-GFP into SC. Middle, GFP-labeled neurons in SC-sg were also tdTomato+. Right four panels, GFP-labeled axons in various regions (LP, pretectal area, PBG, cuneiform nucleus (CUN)) downstream of SC. High-magnification images (bottom) reveal ramified axons and their terminal and bouton structures. Blue, Nissl staining. Scale bars: 250 µm, middle top panel; 500 µm, right top panels; 25 µm, bottom panels. The scales also apply to (B), (C), (D) correspondingly.
(B) Axonal outputs of SC-sg neurons that receive input from the contralateral retina. Data are displayed in a similar way as in (A).
(C) Axonal outputs of SC neurons that receive input from A1, which are located mainly in SC-dg and sparsely in SC-ig.
(D) Axonal outputs of SC neurons that receive input from M1, which are located mainly in the lateral aspect of SC-ig.
(E) Summary of observed target regions for SC neuron subpopulations receiving input from V1/retina (blue), from A1 (red), and from M1 (yellow) respectively. Abbreviations: CL, central lateral nucleus of thalamus; PCN, paracentral nucleus; VM, ventral medial nucleus of thalamus; CM, central medial nucleus of thalamus; APN, anterior pretectal nucleus; PF, parafascicular nucleus; SPFm and SPFp, subparafascicular nucleus, magnocellular and parvicellular; ZI, zona incerta; MRN, midbrain reticular nucleus; PRN, pontine reticular nucleus; TRN, tegmental reticular nucleus; ICd and ICe, inferior colliculus, dorsal and external; PARN, parvicellular reticular nucleus; GRN, gigantocellular reticular nucleus; IO inferior olivary complex.
We further examined SC neuron groups specifically receiving inputs from the primary auditory cortex (A1) and primary motor cortex (M1), respectively. Following paired injections into A1 and SC, we observed numerous GFP+ cells within the medial aspect of deep grey layer (SC-dg), with a few scattered in overlying intermediate grey layer (SC-ig) (Figure 5C). This labeling pattern is consistent with the known distribution of A1 projections to SC (Xiong et al., 2015). Axonal outputs of A1 input-defined SC neurons were apparently different from those of superficial SC neurons, as prominent projections were seen in the PBG, periaqueductal gray (PAG), cuneiform nucleus (CUN), rostral pontine reticular nucleus (PRNr), and among other regions (Figure 5C, 5E). Only very sparse input to LP was observed as compared with the strong projection to LP from superficial SC neurons. In comparison, paired injections in M1 and SC labeled a population of neurons restricted to the lateral aspect of SC-ig (Figure 5D). Axonal outputs from this group of SC neurons differed further from those defined by A1 and V1/retina inputs (Figure 5D).
To summarize the axonal output profiles of input-defined SC neuron subpopulations, we examined all brain sections containing GFP+ axons and plotted those regions containing observable synaptic boutons on corresponding atlas sections (Figure 5E, with original images shown in Figure S4). SC neurons receiving V1 and retinal inputs exhibited very similar output profiles (Figure 5A–5B; Figure 5E, blue; Figure S4), while those defined by A1 and M1 inputs showed profiles distinct from each other and from the V1/retina-defined population (Figure 5C–5D; Figure 5E, red and yellow respectively; Figure S4). For example, unlike V1- and A1-defined populations, M1-defined SC neurons exhibited prominent projections to some contralateral targets, especially in the posterior part of the brainstem, such as the tegmental reticular nucleus (TRN), gigantocellular reticular nucleus (GRN), and inferior olive (IO) (Figure 5D; Figure 5E; Figure S4). Together these results demonstrate that AAV-Cre mediated transsynaptic labeling can be utilized to map specific classes of projection neurons within a given brain region, each with distinct input and output patterns.
Distinct behavioral functions for input-defined SC neuron subpopulations
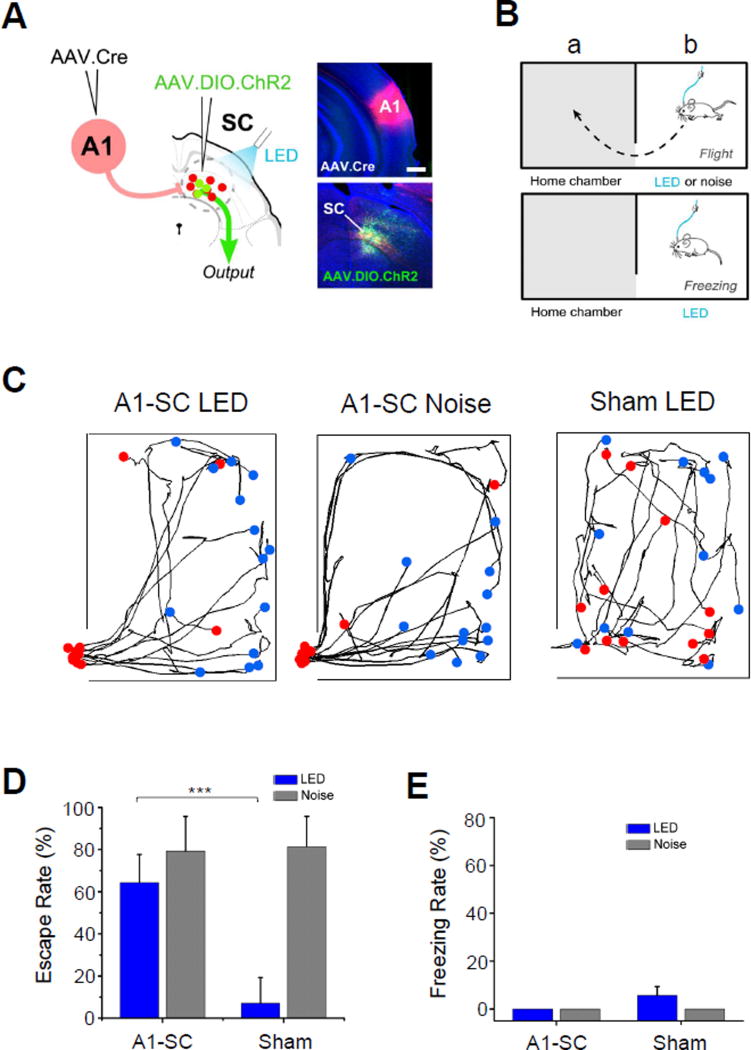
The superior colliculus integrates diverse sensory and motor information, and has been implicated in controlling orienting behaviors as well as innate defense behaviors, such as freezing and escape (Schenberg et al., 2005; Dean et al., 1988; Liang et al., 2015; Wei et al., 2015; Shang et al., 2015). Since SC neuron subpopulations receiving V1 and A1 inputs have different downstream target profiles, we speculated that each might participate in driving a distinct SC-mediated behavior. To test this, we performed paired injections of AAV1-Cre in either A1 or V1 and AAV1-EF1α-DIO-ChR2-EYFP in SC to enable Cre-dependent expression of ChR2 in either deep or superficial layer SC neurons, respectively (Figure 6A, Figure 7A). We could then activate the SC neurons in awake, freely moving mice using pulses of blue LED light (at 20 Hz for 5 sec) delivered through an implanted optical fiber (see Experimental Procedures). We tested both freezing and escape responses using a two-chamber set up, in which the mouse was acclimated to a “home” chamber on one side for 10 minutes, upon which a door was then removed to allow its exploration of the adjoining, “novel” chamber (Figure 6B). As the mouse was exploring the novel chamber, SC was optically activated and subsequent escape to home chamber or freezing was monitored (Figure 6B). Mice expressing ChR2 in A1-recipient, deep layer SC neurons demonstrated a robust escape response following LED light activation (Figure 6C, 6D, see Supplementary Video 1). Such LED-induced escape behavior was not observed in sham mice receiving injection of only GFP expressing virus (Figure 6C, 6D). The effect of optically activating A1-recipient SC neurons was similar as applying a loud noise sound (5 s, 70 dB sound pressure level), presentation of which alone robustly drove an escape response in most of trials (Figure 6C, 6D, noise), consistent with our previous study (Xiong et al., 2015). No freezing response was observed for these mice on any trials (Figure 6E). In our control slice recording experiments, we demonstrated that in these injected mice, A1 corticofugal neurons transduced with AAV1-Cre did not express ChR2 themselves (Figure S5). Therefore the observed escape behavior could not be attributed to activation of some other A1 targets.
Figure 6. A1-recipient SC neurons drive an innate escape behavior.
(A) Schematic illustration of paired injections in A1 and SC, as well as LED illumination applied (A1-SC). Right panels, images of injection sites (red for tdTomato; green for ChR2) in an example animal. Scale bar: 500 µm.
(B) Schematic illustration of two-chamber behavior setup for testing freezing or escape.
(C) Movement tracking for an example A1-SC mouse under LED stimulation (left), A1-SC mouse under noise stimulation (middle) and sham mouse under LED stimulation (right) in the novel chamber during 5 s LED activation or 5 s noise stimulation. Each curve represents one trial. Blue dot indicates the starting location at the initiation of LED or noise stimulus, and red dot indicates the location at the end of the stimulus. Red dot beyond the novel chamber boundary indicates that the animal has returned to the adjacent home chamber within 5 sec (bottom left). For “sham”, AAV1-FLEX-GFP was injected in SC.
(D) Summary of percentage of trials that induced escape behavior (n = 7 mice for A1-SC group, n = 5 mice for sham). Error bar = SD. ***, p < 0.001, t test.
(E) Percentage of trials that induced freezing behavior. Error bar = SD.
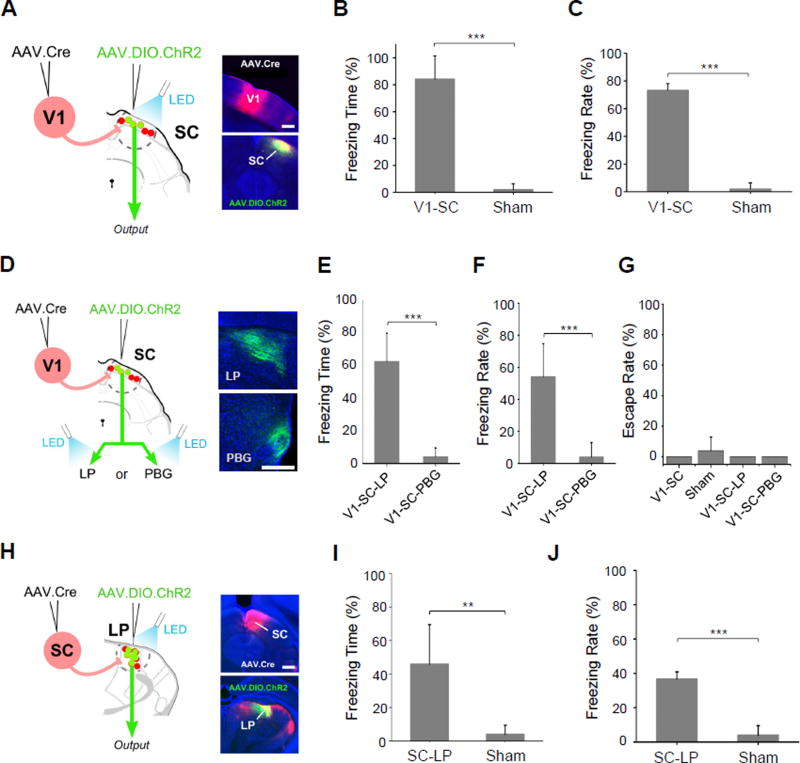
Figure 7. V1-recipent SC neurons drive freezing behavior.
(A) Paired injections labeling SC neurons receiving V1 input. LED illumination was applied to cell bodies in SC (V1-SC). Scale bar: 500 µm.
(B) Percentage of time spent freezing within the time window of LED illumination (n = 5 mice for each group). Error bar = SD. ***, p < 0.001, t test.
(C) Percentage of trials that induced freezing. ***, p < 0.001, t test.
(D) LED illumination was applied to ChR2+ SC axon terminals in either LP (V1-SC-LP) or PBG (V1-SC-PBG). Right, images showing ChR2 labeled SC axons in LP or PBG. Scale bar: 250 µm.
(E) Percentage of time spent freezing within the time window of LED illumination (n = 5 mice for each group). ***, p < 0.001, t test.
(F) Percentage of trials that induced freezing. ***, p < 0.001, t test.
(G) Percentage of trials that induced escape behavior.
(H) Paired injections labeling LP neurons that receive input from SC. LED illumination was applied to LP (SC-LP). Right panel, images showing injection sites in SC and LP. Scale bar: 500 µm.
(I) Percentage of time spent freezing within the time window of LED illumination (n = 6 mice for SC-LP, n = 5 mice for sham). Error bar = SD. **, p < 0.05, t test.
(J) Percentage of trials that induced freezing behavior. Error bar = SD. ***, p < 0.001, t test.
Mice expressing ChR2 in V1-recipient, superficial layer SC neurons (Figure 7A), on the other hand, all demonstrated freezing response following LED light activation (Figure 7B, 7C, see Supplementary Video 2), which was not observed for sham mice (Figure 7B, 7C). None of the mice exhibited escape behavior following LED activation (Figure 7G). Our results thus reveal distinct functional roles of superficial versus deep layer SC neurons in controlling two different defense behaviors.
Our tracing result has demonstrated that LP is a major axonal target of V1-recipient SC neurons (Figure 5A). In addition, combined anterograde transneuronal labeling and retrograde labeling confirmed that a large fraction of V1-recipient SC neurons project to LP (Figure S6). We thus explored whether the freezing behavior evoked by SC-sg activation might be mediated through this structure. To test this, we first optogenetically activated the axon terminals of SC-sg neurons in LP (Figure 7D) in freely behaving mice, which resulted in similar, though somewhat weaker expression of freezing (Figure 7E, 7F, V1-SC-LP), implicating LP’s role in driving SC-mediated freezing behavior. In contrast, optically activating SC axon terminals in PBG did not produce any freezing behavior (Figure 7E, 7F, V1-SC-PBG), suggesting that PBG does not play a role in this behavior. None of these activations induced escape behavior (Figure 7G).
With terminal activation, it is possible however that nonspecific antidromic stimulation of collateral targets could occur, resulting in activation of undesired SC targets. Moreover, LED light from the optic fiber might also activate ChR2-labeled axons passing through the LP to other targets. Both scenarios would confound interpretation of results. To overcome these limitations, we used AAV-Cre transsynaptic labeling to express ChR2 specifically in LP neurons that receive input from SC (Figure 7H). To achieve this, AAV1-Cre was injected into SC and AAV1-DIO-ChR2 was injected into LP, which has unidirectional connectivity with SC (Huerta and Harting, 1984, Comoli et al., 2012). ChR2-expressing LP neurons were then selectively activated in freely moving animals. All mice demonstrated freezing behavior comparable to that elicited by activation of SC projections in LP (Figure 7I, 7J). This result provides strong support for the structure of LP as a downstream mediator of the freezing response generated by activation of superficial SC neurons.
Anterograde transsynaptic labeling in a cell-type specific manner
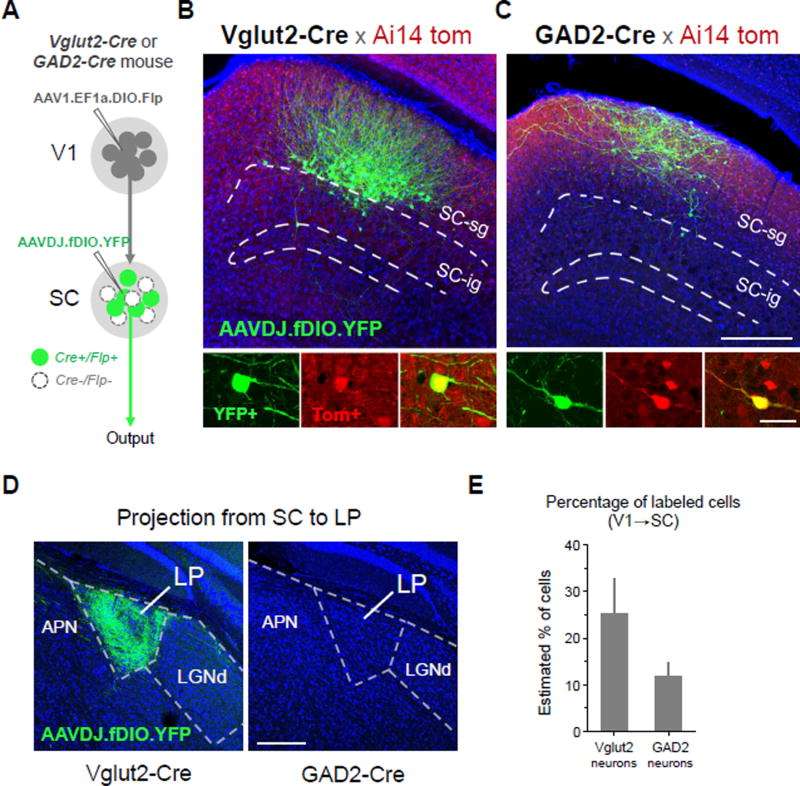
In the above experiments, the cell type of transsynaptically labeled postsynaptic neurons could not be selected. To achieve ability of selecting the postsynaptic cell type, we developed an intersectional approach which depends on both Cre and flippase (Flp) recombinases. A new AAV1 vector expressing Cre-dependent Flp (AAV1-EF1a-DIO-Flp) was made for this purpose. We injected this new virus into V1 of Vglut2-Cre or GAD2-Cre mice, and Flp-dependent YFP virus (AAVDJ-fDIO-YFP) into SC of the same animal. Only in those Cre-expressing SC neurons can Flp and thus Flp-dependent YFP be expressed (Figure 8A). YFP-labeled SC neurons were found in both Vglut2-Cre and GAD2-Cre mice (Figure 8B, 8C), indicating that both glutamatergic and GABAergic SC neurons can receive direct V1 input. YFP-labeled axons were found in LP (target of SC) of Vglut2-Cre but not GAD2-Cre mice (Figure 8D). Moreover, in the injected GAD2-Cre mice, YFP-labeled axons were not found in any structures outside SC (data not shown). These observations demonstrate that unlike glutamatergic neurons, GABAergic SC neurons are local interneurons without long-range projections. Quantification of the number of YFP-labeled SC neurons in comparison with tdTomato+ SC neurons within the same local area revealed that about 25% of tdTomato+ glutamatergic neurons were YFP+, while about 12% of tdTomato+ GABAergic neurons were YFP+ (Figure 8E). As tdTomato+ SC neurons might not all receive V1 input, these numbers gave an estimation of the lower bound of transsynaptic labeling efficiency for the two types of SC neuron respectively. Together, our results demonstrate that these AAV-based transsynaptic approaches have the capacity to specifically and robustly label diverse populations of neurons downstream of a given brain structure.
Figure 8. Cell-type specific anterograde transneuronal labeling.
(A) Strategy for labeling glutamatergic (Vglut2-Cre+) or GABAergic (GAD2-Cre+) subpopulations of SC neurons that receive input from V1.
(B) Selective labeling of V1-recipient glutamatergic neurons in superficial layers of SC. Anterograde transneuronal transport of AAV1-EF1a-DIO-Flp enables Cre-dependent expression of Flp in glutamateric neurons (Ai14 tdTomato+) receiving input from V1. A second injection of AAVDJ-fDIO-YFP into SC enables Flp-dependent expression of YFP specifically in V1-recipient glutamatergic neurons (green), filling soma, dendrites, and axons. YFP+ neurons co-localize with Ai14 tdTomato expression in Vglut2-Cre+ neurons (bottom panels, enhanced with anti-RFP immunostaining).
(C) Selective labeling of V1-recipient GABAergic neurons using same strategy as in (B) but with injections in a GAD2-Cre mouse. Scale: 250 µm, top panel; 25 µm bottom panel. The scales also apply to (B) correspondingly.
(D) Long range axonal projection to LP from glutamatergic, but not GABAergic, V1-recipient SC neurons. Scale: 250 µm.
(E) Percentage of YFP+/Tomato+ cells out of total Tomato+ cells quantified for local regions expressing YFP in Vglut2-Cre and GAD2-Cre mice (4 week post-injection survival, 60 nl injections, n = 4 each). Error bar = SD.
Discussion
Significant progress has been made in developing viral tools for revealing the inputs to a given brain region or cell type (Callaway and Luo, 2015; Junyent et al., 2015; Oyibo et al., 2014; Defalco et al., 2001; Gradinaru et al., 2010; Schwarz et al., 2015; Ugolini, 2010). However, creation of a complementary set of tools for labeling transsynaptic output remains an active area of investigation. Here we show that AAV can be transported anterogradely and can transsynaptically transduce neuronal populations in a wide variety of brain regions postsynaptic to a given injection site. When combined with a conditional expression strategy (i.e. two-step viral injection approach), this transsynaptic transport property enabled us to reveal the outputs of distinct subpopulations of SC neurons that receive different corticocollicular inputs and demonstrate their unique functional roles in driving different types of defense behavior.
AAV vectors expressing various fluorescence reporter genes have been widely used for anterograde circuit mapping. However, anterograde transneuronal spread of this virus has remained controversial (Salegio et al., 2013; Hutson et al., 2015; Harris et al., 2012; Oh et al., 2014). Indeed, in this study, AAV1-CAG-GFP failed to result in any GFP+ cell bodies in regions postsynaptic to the injection site (Figure 2C). This result is consistent with the experimental data provided by the online database of the Allen Institute, where all neuronal projections were anterogradely traced with AAV1, but with no evidence of transneuronal transduction. Only when AAV1-Cre was injected into Ai14 mice, or was combined with a second virus injection to conditionally express a Cre-dependent fluorescence reporter, did we see robust labeling of neurons downstream of the injection location. In this case, the lack of transneuronal cell body labeling with AAV1-CAG-GFP injections may possibly be attributed to that only a small number of viral particles escape the initial host cell and enter downstream neurons, resulting in only extremely weak fluorescence expression which is below the detection threshold for confocal imaging. With AAV1-hSyn-Cre, however, only a small amount of Cre supplied by a small number of viral particles may be sufficient to unlock robust tdTomato expression in Ai14 mice or GFP expression supplied by a second Cre-dependent virus, thus revealing an otherwise hidden capacity of AAV for transneuronal spread.
Previous studies have examined the anterograde transneuronal properties of other viruses including vesicular stomatitis virus (VSV-G) and herpes simplex virus (HSV-1) (Breier et al, 2011; Lo et al, 2011). Though promising, these viruses suffer from toxicity issues which limit their use in physiological studies, and their uncontrollable spread to higher order neurons complicates interpretation of results from circuit mapping studies. AAV, therefore, offers several advantages over those viruses currently available for such a purpose. Namely, AAV has a well characterized lack of toxicity, which allows for the long term, robust expression of viral transgenes necessary for axonal mapping and functional manipulations. Additionally, as we demonstrate in this study, AAV appears not to spread beyond the first-order downstream neurons, thus facilitating a more conclusive interpretation of anatomical connectivity results. One limitation of using AAV1 for anterograde transneuronal studies, however, is the fact that AAV1-Cre can be transported also in the retrograde direction, as has been reported (Tervo, et al., 2016; Rothermel, et al., 2013; Aschauer et al., 2013; Aronoff et al., 2010; Masamizu, et al., 2014) and as we demonstrate in this study (Figure S1). Application should therefore be limited to pathways that do not contain reciprocal connections between targeted pre- and postsynaptic regions. Overall, its lack of toxicity and restricted spread to first-order downstream neurons suggest that AAV may be a valuable new alternative to currently available vectors for anterograde transneuronal circuit studies.
The exact mechanism by which AAV spreads transneuronally is unclear, though it appears to reflect a specific interaction between the virus and host cells, rather than a physical property such as passive diffusion, as injection of high titer Cre-expressing CAV2 and several other AAV serotypes failed to replicate the anterograde transneuronal labeling seen with AAV1 and AAV9. AAV is a small (~20 nm), replication-deficient virus that is normally endocytosed and transported to the nucleus through interactions between capsid proteins and cell surface receptors (Pillay et al., 2016; Kotterman et al., 2014). Evidence suggests that at high titers, some viral particles may be trafficked down axons where they may be released from host cell synaptic terminals, enabling local infection of adjacent neurons (Castle et al., 2013; Castle et al., 2014). Whether this always happens specifically through synaptically connected neurons is still uncertain. Here we provide several lines of evidence that are suggestive of a transsynaptic mechanism of spread. In particular, we show that (1) labeled cells are found only in regions that conform to known innervation patterns of a given upstream structure; (2) no labeling was found in cells surrounding fibers of passage; (3) labeled cells were never GFAP+ glial cells, suggesting specificity in the viral spread to closely apposed neurons; and (4) labeled cells were always functionally connected to their presynaptic starter population. These results support a transsynaptic, rather than a nonspecific or extrasynaptic mechanism of spread in downstream target regions.
Although the exact efficiency of the anterograde transsynaptic spread is unknown, our experiments could provide a rough estimation. First, 25% and 12% of local SC-sg neurons can be transneuronally labeled by V1 injections in Vglut2-Cre and GAD2-Cre mice respectively (Figure 8), suggesting a lower bound of transneuronal transport efficiency for glutamatergic and GABAergic SC neuron populations respectively. Second, for SC-sg neurons retrogradely labeled from CTB injection in LP, about 65% of the cells were transneuronally labeled from V1 injection (Figure S6), suggesting that for the group of V1-recipient, LP-projecting SC neurons the efficiency of transneuronal labeling is relatively high. Together, these data suggest that a reasonably good efficiency can be achieved under our experimental conditions, by controlling the titer and volume of viral injection.
When combined with secondary injections of a conditional virus, the anterograde transneuronal properties of AAV offer several unique opportunities for the study of neural circuits. In particular, this method provides experimental access to spatially restricted groups of neurons that would otherwise be impossible to target with conventional viral injections. These groups may then be targeted for mapping their axonal output or for manipulation or recording using genetically encoded opsins or Ca2+ indicators. In addition, the viral approach may be combined with transgenic mouse lines to gain access to specific subtypes of neurons receiving input from a given upstream region, as we demonstrated with injections of a novel Cre-dependent Flp expressing virus and a Flp-dependent YFP expressing virus in Vglut2-Cre and GAD2-Cre mice (Figure 8). Such approach enabled us to separate glutamateric and GABAergic neurons in SC that were innervated by V1.
The two-step injection strategy provides a new way to functionally categorize neurons according to their distinct input sources. As a proof of concept, we accessed discrete populations of neurons residing in superficial, deep, and lateral aspects of SC. This enabled us to reveal that each subpopulation, defined by its specific corticocollicular input, has a unique divergent axonal targeting profile. We further show that two of these subpopulations—those in superficial layer SC receiving input from V1, and those in deep layers of SC receiving input from A1—mediate distinct freezing and escape behaviors, respectively. Together these results reveal the potential for AAV-based anterograde transsynaptic labeling to access, map, and functionally manipulate specific groups of neurons that would otherwise be challenging to investigate using conventional approaches.
Additionally, application of this method provides a straightforward means for examining the potential role of downstream structures in mediating a function or behavior observed from activation of a given upstream structure. Using this approach, we implicate LP as an important downstream structure of SC-sg in mediating evoked freezing responses, as also suggested by a previous report (Wei et al., 2015, but see Shang et al., 2015). In addition to optogenetically activating ChR2-expressing SC axon terminals in LP, we went one step further and directly activated SC-recipient cell bodies in LP and found robust evoked freezing behavior. Employing an anterograde transsynaptic labeling approach to this functional test allowed us to bypass potential problems associated with optogenetic activation of axon terminals. These could include activation of axon collaterals of SC neurons, as illustrated in Figure 1A, and activation of fibers of passage in LP. Instead, directly activating the LP neurons that receive SC input provides more conclusive evidence implicating this structure’s role in mediating the freezing response evoked by SC-sg activation. Altogether, our results demonstrate that the AAV-based anterograde transsynaptic tagging approaches described in this study may be broadly applicable for use in the forward screening of functional circuits across multiple synaptic steps.
Experimental Procedures
Additional methods and more details are included in Supplemental Experimental Procedures.
Animal preparation and stereotaxic surgery
All experimental procedures used in this study were approved by the Animal Care and Use Committee at the University of Southern California. Male and female C57BL/6J and Ai14 (Cre-dependent tdTomato reporter) mice (Jackson Laboratories) aged 2–6 months were used in this study. Stereotaxic injection of viruses was carried out as we previously described (Ibrahim et al., 2016; Liang et al., 2015; Xiong et al. 2015). Injection coordinates were based on the Allen Reference Atlas (Dong, 2007).
For virus injection into the retina, 800 nl of virus was injected into the left eye at a depth of 0.7 mm from the surface of the lateral most curvature of the eye using a 40° angle of approach relative to the optic axis.
Injection of viruses for anterograde transneuronal labeling
To demonstrate the anterograde transneuronal properties of AAV, AAV2/1-hSyn-Cre-WPRE-hGH (UPenn Vector Core, 2.5 × 1013 GC/ml) was injected into either V1 (60 nl total volume; 3.9 mm posterior and 2.6 mm lateral to bregma and 0.5 mm ventral from the cortical surface) or retina (800 nl total volume) of Ai14 mice using the pressure injection method. Animals were usually euthanized 4 weeks following injection to allow time for viral transport and transgene expression.
To examine the potential anterograde transneuronal properties of other viruses, AAV2/1-CMV-Pl-Cre-rBG (UPenn Vector Core, 2.7 × 1013 GC/ml), AAV2/5-CMV-Pl-Cre-rBG (UPenn Vector Core, 2.8 × 1013 GC/ml), AAV2/6-CMV-Pl-Cre-rBG (UPenn Vector Core, 3.5 × 1013 GC/ml), AAV2/8-CMV-Pl-Cre-rBG (UPenn Vector Core, 4.4 × 1013 GC/ml), AAV2/9-CMV-Pl-Cre-rBG (UPenn Vector Core, 1.6 × 1014 GC/ml), AAV2/1-CB7-Cl-eGFP-WPRE-rBG (UPenn Vector Core, 4.2 × 1013 GC/ml), or CAV2-CMV-Cre (Montpellier Vector Core, 1.3 × 1012 GC/ml) was injected into V1 (60 nl total volume) of Ai14 mice (for Cre-expressing viruses) or wild-type C57BL/6J mice (for GFP-expressing virus). Original titers were used, while for testing dependence on titer AAV1-CMV-Cre was diluted accordingly. Note that the original titer was lowest for AAV1 among tested AAV serotypes all expressing CMV-driven Cre. Animals were euthanized 4 weeks following injection and postsynaptic structures were examined for the presence of cell body labeling.
Retrograde spread of AAV1
To test for potential retrograde transport of AAV, injections of either AAV2/1-hSyn-Cre-WPRE-hGH (60 nl) or AAV2/1-CB7-Cl-eGFP-WPRE-rBG (60 nl) were made into SC (3.9 mm posterior and 0.8 mm lateral to bregma and 1.5 mm ventral from the cortical surface) of either Ai14 or wild-type C57BL/6J mice, respectively. Animals were euthanized 4 weeks following injection.
Two-step viral injection
For mapping the axonal outputs of subpopulations of neurons in SC, AAV2/1-hSyn-Cre-WPRE-hGH was injected into V1, contralateral retina, A1, or M1 of Ai14 mice (60 nl). Following 2 to 7 days, a second injection of AAV2/1-CAG-FLEX-eGFP-WPRE-bGH (UPenn vector core, 1.7 × 1013 GC/ml) was made into the ipsilateral SC (60 nl). The spacing of the two injections over several days was selected to allow sufficient time for the clearance of any residual AAV-Cre virus that may have spread across the pial surface in an effort to eliminate any local contamination of the Cre-dependent virus injection site. Animals were allowed to recover for at least 4 weeks following the second injection.
For behavioral testing, AAV2/1-hSyn-Cre-WPRE-hGH was injected into V1, A1, or SC, as described above. Following 2 to 7 days, a second injection of AAV2/1-EF1a-DIO-hChR2-eYFP (UPenn vector core, 1.6 × 1013 GC/ml) was made into SC or LP. Animals were then prepared for optogenetic testing 4 weeks after the second injection. For injection of CTB-488 in LP, please see the Supplemental Experimental Procedure.
For labeling glutamatergic and GABAergic SC neuronal populations receiving V1 input, AAV1-EF1a-DIO-Flp (1.5 × 1014 GC/ml, custom design, ViGene Biosciences) was injected into V1 (60 nl) of Vglut2-Cre (Slc17a6tm2(cre)Lowl, Jackson Laboratories) or GAD2-Cre (Gad2tm2(cre)Zjh, Jackson Laboratories) mice crossed with Ai14. Following 2–7 days, a second injection of AAVDJ-EF1a-fDIO-YFP (UNC Vector Core, 1.6 × 1013 GC/ml) was injected into SC (60 nl). Animals were euthanized 4 weeks following the second injection.
Imaging and quantification
All images were generated using a confocal microscope (Olympus FluoView FV1000). The total number of tdTomato+ cells were counted across sections for each structure of interest. To estimate the percentage of tdTomato+ cells in innervated regions, 40× magnification images were taken across multiple sections for SC, LGNv, Str, and PN. A region of interest of fixed size was defined for each structure (Str = 150 × 200 µm, PN = 100 × 200 µm, and SC and LGNv = 200 × 300 µm area) and the total number of Tomato+ cells and Nissl+ cells were quantified within the region (see Figure S2). We chose to focus this quantification on restricted local regions with Tomato labeling, rather than the entire structure, as only a small portion of each target structure is innervated by a given V1 injection due to the topographic nature of its output. We defined the central regions of labeling in each structure to capture the majority of labeled neurons for this estimation. More details of the quantification were included in the Supplemental Experimental Procedure.
Slice preparation and recording
To confirm synaptic connectivity to anterogradely labeled cells in the striatum (Figure 4B) and SC (Figure S3), a 1:1 mixture of AAV2/1-hSyn-Cre-WPRE-hGH and AAV2/1-EF1a-DIO-hChR2-eYFP was injected into V1 of Ai14 mice (80 nl total volume). Following a 4 week post-injection survival time, acute brain slices containing striatum or SC were prepared. Whole-cell recordings were carried out as we previously described (Xiong et al., 2015; Ibrahim et al., 2016; Ji et al., 2016; also see Supplemental Experimental Procedure).
In vivo optogenetic preparation and stimulation
To examine whether SC neuron subgroups mediate different behavioral responses, mice were implanted with an optical fiber (200 µm diameter, Thorlabs) three weeks after secondary injection of AAV2/1-EF1a-DIO-hChR2-eYFP in SC, following our previous study (Xiong et al. 2015). Please see the Supplemental Experimental Procedures for details.
Behavioral testing and quantification
Escape behaviors were tested in a box containing two chambers connected by a small opening, as we previously described (Xiong, et al., 2015). Mice (n = 7 mice for A1-SC, n = 5 mice for V1-SC) were allowed to acclimate to one chamber for 10 minutes prior to behavioral testing. During this time, the opening connecting the two chambers was blocked with a removable door. After 10 mins the door was removed and the mouse was free to explore the adjacent, novel chamber. Following complete entry into the novel chamber, 5-s 20-Hz blue LED stimulation or 5-s 70 dB SPL white noise was applied. The animal behavior was recorded with a camera mounted above the box. If the mouse went back to the home chamber within 5 s of noise or LED stimulation, it was considered as a successful escape trial. Escape rate was calculated as the fraction of total trials for each animal that evoked escape.
Freezing behavior was tested in a single or double chamber box with a camera on top recording the whole process (n = 5 mice for V1-SC, n = 6 mice for SC-LP). After the animal explored the single chamber or the novel chamber for 5 minutes, 5-s 20-Hz blue LED stimulation was applied. Each session contained 4–6 trials, with an interval of at least 5 mins. Each animal was tested for 2 sessions separated by one day. If the animal became motionless for more than 1.5 s after the onset of stimulation, it was considered as a successful freezing response (Shang et al., 2015; Tovote et al., 2016; Wei et al., 2015; Wolff et al., 2014). Freezing time was quantified as the fraction of time spent freezing during the optogenetic stimulation. Freezing rate was the fraction of total trials that evoked freezing.
Supplementary Material
Acknowledgments
This work was supported by grants from the US National Institutes of Health (R01DC008983) and the David and Lucile Packard Foundation (Packard Fellowships for Science and Engineering) to L.I.Z., a NIH grant (R01EY019049) to H.W.T., and a NIH grant (F31DC015185) to B.Z. We thank Junxiang Huang and Leena A. Ibrahim for the help in viral vector construction.
References
- Aboitiz F, Montiel J. One hundred million years of interhemispheric communication: The history of the corpus callosum. Brazilian J. Med. Biol. Res. 2003;36:409–420. doi: 10.1590/s0100-879x2003000400002. [DOI] [PubMed] [Google Scholar]
- Aschauer DF, Kreuz S, Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier KT, Saunders A. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc. Natl. Acad. Sci. USA. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway XEM, Luo L. Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J. Neurosci. 2015;35:8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Gershenson ZT, Giles AR, Holzbaur ELF, Wolfe JH. Adeno-associated virus serotypes 1, 8, and 9 share conserved mechanisms for anterograde and retrograde axonal transport. Hum. Gene Ther. 2014;25:705–720. doi: 10.1089/hum.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Perlson E, Holzbaur EL, Wolfe JH. Long-distance Axonal Transport of AAV9 Is Driven by Dynein and Kinesin-2 and Is Trafficked in a Highly Motile Rab7-positive Compartment. Mol. Ther. 2013;22:1–13. doi: 10.1038/mt.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Alamancos MA, Favero M. Whisker-related afferents in superior colliculus. J. Neurophysiol. 2016;115:2265–2279. doi: 10.1152/jn.00028.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoli E, Das Neves Favaro P, Vautrelle N, Leriche M, Overton PG, Redgrave P. Segregated anatomical input to sub-regions of the rodent superior colliculus associated with approach and defense. Front. Neuroanat. 2012;6:1–19. doi: 10.3389/fnana.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P, Mitchell IJ, Redgrave P. Responses resembling defensive behaviour produced by microinjection of glutamate into superior colliculus of rats. Neuroscience. 1988;24:501–510. doi: 10.1016/0306-4522(88)90345-4. [DOI] [PubMed] [Google Scholar]
- Defalco J, Tomishima M, Liu H, Zhao C, Cai XL, Marth JD, Enquist L, Friedman JM. Virus-Assisted Mapping of Neural Inputs to a Feeding Center in the Hypothalamus. Science. 2010;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Dong H-W. The Allen Reference Atlas: A Digital Color Brain Atlas Of The C57BL/6J Male Mouse. Hoboken: John Wiley and Sons, Inc.; 2007. [Google Scholar]
- Ekstrand MI, Enquist LW, Pomeranz LE. The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends Mol. Med. 2008;14:134–140. doi: 10.1016/j.molmed.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski RM, Sadowski PD. The FLP recombinase of the Saccharomyces cerevisiae 2 microns plasmid attaches covalently to DNA via a phosphotyrosyl linkage. Mol. Cell. Biol. 1985;5:3274–3279. doi: 10.1128/mcb.5.11.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Wook Oh S, Zeng H. Adeno-associated viral vectors for anterograde axonal tracing with fluorescent proteins in nontransgenic and Cre driver mice. Curr. Protoc. Neurosci. 2012;1:1–18. doi: 10.1002/0471142301.ns0120s59. [DOI] [PubMed] [Google Scholar]
- Huerta MF, Harting JK. Connectional organization of the superior colliculus. Trends Neurosci. 1984;7:286–289. [Google Scholar]
- Hutson TH, Kathe C, Moon LDF. Trans-neuronal transduction of spinal neurons following cortical injection and anterograde axonal transport of a bicistronic AAV1 vector. Gene Ther. 2015;23:231–236. doi: 10.1038/gt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim LA, Mesik L, Ji XY, Fang Q, Li HF, Li YT, Zingg B, Zhang LI, Tao HW. Cross-Modality Sharpening of Visual Cortical Processing through Layer-1-Mediated Inhibition and Disinhibition. Neuron. 2016;89:1031–1045. doi: 10.1016/j.neuron.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji XY, Zingg B, Mesik L, Xiao Z, Zhang LI, Tao HW. Thalamocortical Innervation Pattern in Mouse Auditory and Visual Cortex: Laminar and Cell-Type Specificity. Cereb Cortex. 2016;26:2612–2625. doi: 10.1093/cercor/bhv099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junyent F, Kremer EJ. CAV-2 - Why a canine virus is a neurobiologist’s best friend. Curr. Opin. Pharmacol. 2015;24:86–93. doi: 10.1016/j.coph.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Xiong XR, Zingg B, Ji X, Zhang LI, Tao HW. Sensory Cortical Control of a Visually Induced Arrest Behavior via Corticotectal Projections. Neuron. 2015;86:1–13. doi: 10.1016/j.neuron.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Anderson DJ. A cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2009;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP, Studholme KM. Retinofugal Projections in the Mouse. J. Comp. Neurol. 2014;00:1–21. doi: 10.1002/cne.23635. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front. Neuroanat. 2015;9:80. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front. Neuroanat. 2015;9:80. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyibo HK, Znamenskiy P, Oviedo HV, Enquist LW, Zador AM. Long-term Cre-mediated retrograde tagging of neurons using a novel recombinant pseudorabies virus. Front. Neuroanat. 2014;8:86. doi: 10.3389/fnana.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea G, Navarrete M, Araque A. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 2009;32:421–431. doi: 10.1016/j.tins.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, et al. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel M, Brunert D, Zabawa C, Diaz-Quesada M, Wachowiak M. Transgene Expression in Target-Defined Neuron Populations Mediated by Retrograde Infection with Adeno-Associated Viral Vectors. J. Neurosci. 2013;33:15195–15206. doi: 10.1523/JNEUROSCI.1618-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenberg LC, Povoa RMF, Costa ALP, Caldellas AV, Tufik S, Bittencourt AS. Functional specializations within the tectum defense systems of the rat. Neurosci. Biobehav. Rev. 2005;29:1279–1298. doi: 10.1016/j.neubiorev.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, et al. Viral-genetic tracing of the input–output organization of a central noradrenaline circuit. Nature. 2015;524:88–92. doi: 10.1038/nature14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahibzada N, Dean P, Redgrave P. Movements resembling orientation or avoidance elicited by electrical stimulation of the superior colliculus in rats. J Neurosci. 1986;6:723–733. doi: 10.1523/JNEUROSCI.06-03-00723.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salegio EA, Samaranch L, Kells AP, Mittermeyer G, San Sebastian W, Zhou S, Beyer J, Forsayeth J, Bankiewicz KS. Axonal transport of adeno-associated viral vectors is serotype-dependent. Gene Ther. 2013;20:348–352. doi: 10.1038/gt.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang C, Liu Z, Chen Z, Shi Y, Wang Q, Liu S, Li D, Cao P. A parvalbumin-positive excitatory visual pathway to trigger fear responses in mice. Science. 2015;348:1472–1477. doi: 10.1126/science.aaa8694. [DOI] [PubMed] [Google Scholar]
- Simmons PA, Lemmon V, Pearlman AL. Afferent and efferent connections of the striate and extrastriate visual cortex of the normal and reeler mouse. J. Comp. Neurol. 1982;211:295–308. doi: 10.1002/cne.902110308. [DOI] [PubMed] [Google Scholar]
- Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 2001;15:2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- Sturrock RR. Light microscopic identification of immature glial cells in semithin sections of the developing mouse corpus callosum. J. Anat. 1976;122:521–537. [PMC free article] [PubMed] [Google Scholar]
- Tervo DGR, Hwang BY, Viswanathan S, Gaj T, Lavzin M, et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016:1–11. doi: 10.1016/j.neuron.2016.09.021. http://doi.org/10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed]
- Tovote P, Esposito MS, Botta P, Chaudun F, Jonathan P, et al. Midbrain circuits for defensive behaviour. Nature. 2016;534:206–212. doi: 10.1038/nature17996. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Advances in viral transneuronal tracing. J. Neurosci. Methods. 2010;194:2–20. doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Liu N, Zhang Z, Liu X, Tang Y, He X, Wu B, Zhou Z, Liu Y, Li J, et al. Processing of visually evoked innate fear by a non-canonical thalamic pathway. Nat. Commun. 2015;6:6756. doi: 10.1038/ncomms7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, Young JAT, Callaway EM. Monosynaptic Restriction of Transsynaptic Tracing from Single, Genetically Targeted Neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Gründemann J, Tovote P, Krabbe S, Jacobson GA, Müller C, Herry C, et al. Amygdala interneuron subtypes control fear learning through disinhibition. Nature. 2014;509:453–458. doi: 10.1038/nature13258. [DOI] [PubMed] [Google Scholar]
- Xiong XR, Liang F, Zingg B, Ji X, Ibrahim LA, Tao HW, Zhang LI. Auditory cortex controls sound-driven innate defense behaviour through corticofugal projections to inferior colliculus. Nat. Commun. 2015;6:7224. doi: 10.1038/ncomms8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Malouf AT, Messing A, Silver J. Glial fibrillary acidic protein is necessary for mature astrocytes to react to B-amyloid. Glia. 1999;25:390–403. doi: 10.1002/(sici)1098-1136(19990215)25:4<390::aid-glia8>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zingg B, Hintiryan H, Gou L, Song MY, Bay M, Bienkowski MS, Foster NN, Yamashita S, Bowman I, Toga AW, et al. Neural Networks of the Mouse Neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.