Abstract
Slight alterations in nanoparticles’ surface properties can significantly influence the corona composition which may alter their interaction with the biological milieu. Size and porosity of silica nanoparticles (SNPs) are known to be predominant factors influencing their dose-dependent toxicity. Little is known however about the extent and type of protein adsorption on SNPs as a function of physicochemical properties and the role this might play on mechanisms of cellular uptake and toxicity. In this work we investigated the influence of size and porosity of SNPs on protein adsorption, cellular uptake, and toxicity in RAW 264.7 macrophages. Toxicity of the SNPs was found to be concentration dependent, and the formation of the protein corona mitigated toxicity for all particles. Detailed analysis of the amount of proteins recovered from each nanoparticle revealed similarities in the protein adsorption profile as a function of size and porosity. The mechanism of uptake was highly dependent on size rather than porosity or the adsorbed proteins.
Keywords: silica nanoparticles, protein adsorption, protein corona, cytotoxicity, cellular uptake
Graphical Abstract
INTRODUCTION
Silica nanoparticles (SNPs) have shown potential for use in nanomedicine as drug carriers, cell markers, imaging agents, biosensors, and alike.1–5 Better understanding of how SNPs interact with cells and biological fluids is required to predict safety, biodistribution, pharmacokinetics, effective performance for the intended use, clearance, and possible adverse effects. Due to a lower loading capacity of the nonporous or mesoporous SNPs compared with other drug delivery carriers such as liposomes or hollow structures, a higher dosage of SNPs would be required to deliver the same amount of active drug moieties. This would lead to administering a higher amount of SNPs as carriers. The physicochemical properties of nanocarriers such as size, charge, surface functionality, shape, hydrophobicity, and aggregation state determine their biological fate.6–8 Once nanoparticles are in contact with the bloodstream, proteins bind to their surface almost immediately by a dynamic process and form a “protein corona”. Formation of protein corona changes the physicochemical properties of the nanoparticles such as size, charge, and hydrophilicity. Protein corona can further lower the surface free energy of particles and prevents the pristine surface interaction with biological fluids and cells, which changes cellular uptake, toxicity, and biocompatibility.9,10 Changes in the physicochemical properties such as shape, size, and surface chemistry may greatly influence the composition of the protein corona on SNPs. It has been shown, for example, that the size of SNPs quantitatively influences the type of identified proteins adsorbed to each particle.11
Previously we studied the influence of size, surface charge, geometry, and porosity of SNPs on mechanisms of cellular uptake, biodistribution, and toxicity in vivo. Our studies showed that the porosity of silica nanomaterials drastically influences in vivo toxicity and tissue biodistribution.12 SNPs were sequestered primarily in the liver and spleen. SNPs with a larger size, higher aspect ratio, and increased porosity had higher accumulations in the lung, while amine modifications reduced this accumulation.13 Additionally, our data showed that an increased surface charge density leads to the initiation of thrombotic and hemolytic events. Hemolysis was dependent on charge characteristics, with negatively charged Stöber particles (nonporous hydroxyl terminated) at the lowest level of hemolytic induction and positively charged amine-modified materials at the highest level of induction.14 However, the influence of nanoparticle physicochemical properties on protein adsorption and how such adsorption affect cellular uptake and toxicity is poorly understood.
Herein, we investigated the effect of nanoparticle size and porosity on protein adsorption and their subsequent toxicity and uptake in a macrophage cell line. RAW 264.7 was used to represent the physiological scavengers of foreign nanoparticles exposed to in vivo systems.15 Specifically, we chose Stöber silica nanoparticles (SNPs) of approximate diameters 50 and 500 nm to evaluate the effect of size, mesoporous SNPs of approximate diameter 500 nm for evaluating the effect of porosity on protein adsorption, toxicity, and mechanism of cellular uptake in a RAW 264.7 macrophage cell line. Toxicity studies were conducted in the presence and absence of serum. Pharmacological inhibitors of cellular uptake were used to elucidate the uptake mechanisms.
RESULT AND DISCUSSION
Synthesis and Characterization of SNPs
The goal of this research was to investigate the impact of the physicochemical properties of silica nanoparticles (SNPs) on protein adsorption, cellular uptake, and toxicity. To compare the effect of size, spherical nonporous SNPs of the two different diameters, namely, 46 ± 4.9 and 432 ± 18.7 nm, were synthesized using a modified Stöber method. To evaluate the effect of porosity, mesoporous spherical SNPs were produced with a diameter of 466 ± 86 nm, comparable to 432 ± 18.7 nm for the nonporous Stöber particle. All particles with engineered physicochemical features were evaluated for toxicity on RAW 264.7 cell lines in the presence and absence of serum protein. These cell lines were used as a model macrophage to represent the physiological scavengers of foreign nanoparticles exposed to in vivo systems. Synthesized particles were characterized using transmission electron microscopy (TEM), dynamic light scattering (DLS), scanning electron microscopy (SEM), X-ray diffraction (XRD) techniques, nitrogen adsorption–desorption analysis, and zeta potential measurement for size, morphology, mesoporous arrangement, surface area, pore size measurement, and surface charge.
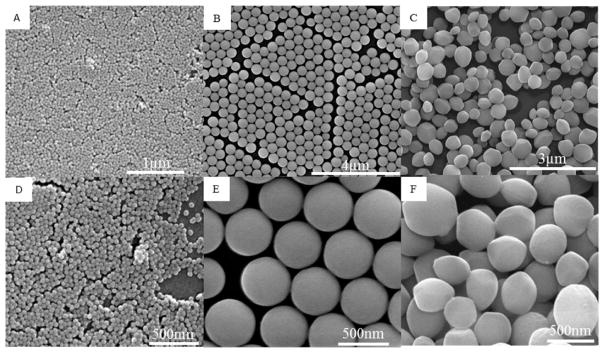
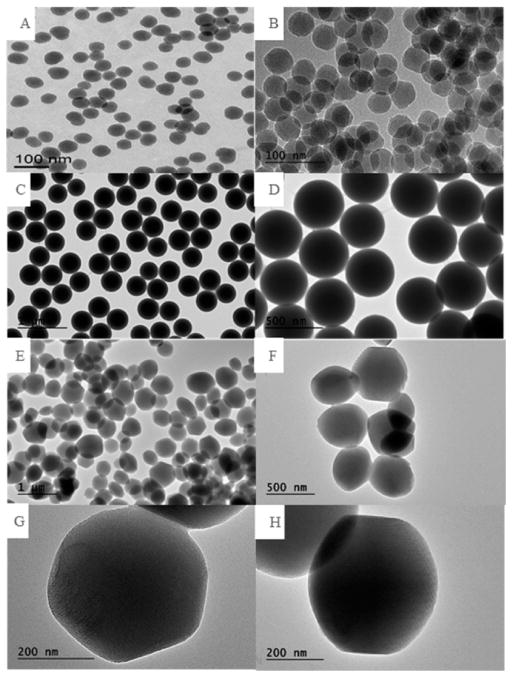
SEM (Figure 1) and TEM (Figure 2) image analyses of the nonporous silica nanoparticles (Stöber) and mesoporous silica nanoparticles (MSN) represent the morphology and size of each particle. Evident from TEM image analyses (Figure 2), Stöber particles were 46 ± 4.9 and 432 ± 18.7 nm in diameter, respectively, and the average diameter of mesoporous silica nanoparticles was 466 ± 86 nm.
Figure 1.
Scanning electron microscopy images of (A, D) Stöber SNPs with an average diameter of 46 nm (Stöber50), (B, E) Stöber SNPs with an average diameter of 432 nm (Stöber500), and (C, F) mesoporous SNPs with an average diameter of 466 nm (MSN500).
Figure 2.
Transmission electron microscopy images of (A, B) Stöber SNPs with an average diameter of 46 ± 4.9 nm, (C, D) Stöber SNPs with an average diameter of 432 ± 18.7 nm, (E, F) mesoporous SNPs with an average diameter of 466 ± 86 nm, and (G, H) high-resolution image of a single mesoporous particle.
Increasing the size of the Stöber particle from 46 to 432 nm shows an increase in the smoothness on the surface of the particle as observed previously.16 Comparing Stöber particles with mesoporous ones, the mesoporous particles showed higher polydispersity due to the micelle-based production method used for the synthesis of the particles. The synthetic process for mesoporous particles depends on micelle formation as a template in low-surfactant concentration. This is a very sensitive process toward temperature and concentrations, and slight changes in these parameters influence the morphology, size, and porosity of the particles.
Uniformly aligned mesopores along and perpendicular to the axes of mesoporous SNPs were observed (Figure 2G and 2H). High-resolution TEM images showed 2D-hexagonal mesopores in the close-packing structure for mesoporous SNPs.
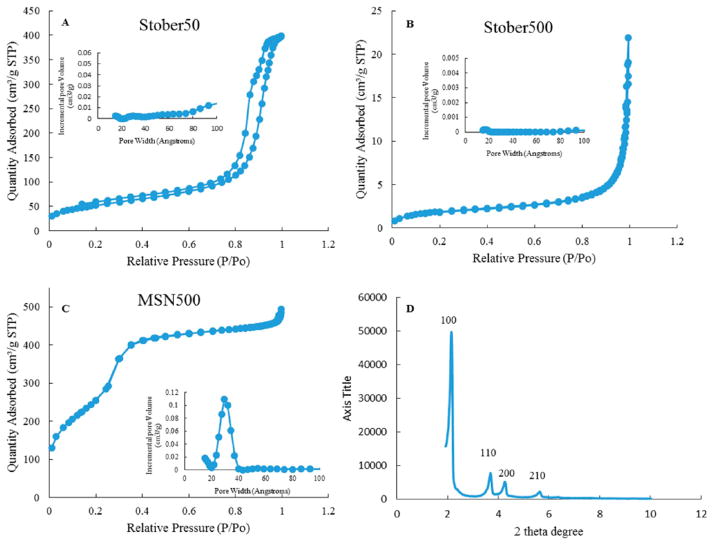
Nitrogen adsorption/desorption isotherms for nonporous Stöber particles and mesoporous SNPs were investigated to evaluate the porosity of the particles (Figure 3). The adsorption isotherms for Stöber particles were type II (according to IUPAC classification), indicating nonporous structure, while mesoporous SNPs exhibited type IV isotherms, typical for mesopore structures. Although the Stöber 46 nm particles are classified as nonporous, interparticle porosity is observed for these particles, which is indicated by the hysteresis at high relative pressure due to N2 condensation.17 The interparticle porosity of these particles can be attributed to their bumpy surface comparing to larger Stöber particles with a smoother surface as shown in TEM images (Figure 2B).
Figure 3.
Nitrogen adsorption–desorption isotherms of (A) Stöber 42 nm, (B) Stöber 432 nm, and (C) MSN 466 nm, and (D) X-ray diffraction patterns of MSN 466 nm. (Insets) Pore size distribution plots for each type.
Table 1 shows the results of characterization of surface area, external surface area, pore size, and pore volume of nonporous and mesoporous SNPs based on nitrogen adsorption–desorption full isotherms. The Brunauer–Emmet–Teller method18,19 was used to calculate the surface area. t-Plots were used to obtain the external surface area,20 and the Barrett–Joyner–Halenda method18,19 was used to determine the pore size distribution of each particle based on nitrogen adsorption isotherms. As expected, by increasing the size of the nonporous Stöber particles from 46 to 432 nm, the surface area decreased, respectively, from 186 to 6.8 m2 g−1. Mesoporous SNPs with an average 466 nm diameter possessed a relatively high surface area of 950 m2/g. Comparing MSN500 and Stöber500 particles, which have an almost similar size, the surface area increased 140-fold by incorporating mesoporosity in MSN500 particles. The external surface area of the particle, which is referred to as cell-contactable surface area, determines protein adsorption and cell cytotoxicity. MSN500 particles showed a narrow distribution with a pore size of 3 nm (Figure 3C, insert).
Table 1.
Hydrodynamic Size and Zeta Potential of Particles Dispersed in Water, Media, and Media with Serum Proteins
| hydrodynamic size (nm)
|
zeta potential (mV)
|
TEM | surface area (m2 g−1) | external surface area (m2 g−1) | pore size (nm) | pore volume (cm3 g−1) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| water | RPMI | RPMI and 10% serum | water | RPMI | RPMI and 10% serum | ||||||
| Stöber50 | 108.1 ± 6.3 | 95.52 ± 2.5 | 190.5 ± 4.8 | −40.3 ± 1.9 | −22.7 ± 1.4 | −9.57 ± 0.4 | 46 ± 4.9 | 186.03 | 178.84 | 13.16 | 0.0024 |
| Stöber500 | 520.3 ± 4.8 | 497.7 ± 5.3 | 539.7 ± 6.2 | −53.1 ± 1.5 | −32.1 ± 1.1 | −9.48 ± 0.5 | 432 ± 18.7 | 6.83 | 6.83 | N/A | N/A |
| MSN500 | 799.8 ± 22.9 | 999 ± 23 | 915 ± 72 | −44 ± 0.8 | −20.4 ± 0.7 | −8.03 ± 0.5 | 466 ± 86 | 950.09 | 121.86 | 3.05 | 0.5729 |
XRD measurement of MSN500 particles revealed distinct peaks (100, 110, 200, 210), which are typical of MCM-41-type mesopore arrangement (Figure 3D). The 2D-hexagonal mesopore arrangement in the close-packing structure was confirmed by TEM images (Figure 2G and 2H). The FT-IR spectrum of MSN500 particles before and after the washing step by acidic ethanol to extract all CTAB molecules is shown in Figure S1. The elimination of a carbon chain band in wavenumber range 3000–2800 cm−1 confirms complete removal of CTAB surfactant, which has the template role in the process of mesopore formation in MSN500 particles.
The hydrodynamic size of the particles was measured by dynamic light scattering in DI water, media, and media with 10% serum and is presented in Table 1. Comparison of the hydrodynamic diameter of MSN500 with diameter measured by TEM showed a higher tendency of these particles to agglomerate in solution, which could be due to a high surface area and higher surface energy that is confirmed by nitrogen adsorption–desorption isotherm measurements. Zeta potential measurements showed that Stöber nanoparticles dispersed in water were highly negatively charged, rendering colloidal stability in aqueous medium. Zeta potential values decreased for all particles suspended in the cell culture media (RPMI) due to higher ionic strength (RPMI 10 800 μS),21 screening effect of counterions, and shorter Debye length compared to water. Adsorption of proteins on the surface of particles reduces the zeta potential values to a neutral range (from −10 to 10 mV) in the media containing serum. This could reduce the stability of the particles and increase the tendency for agglomerations.
Protein Corona and Its Effect on Cell Toxicity
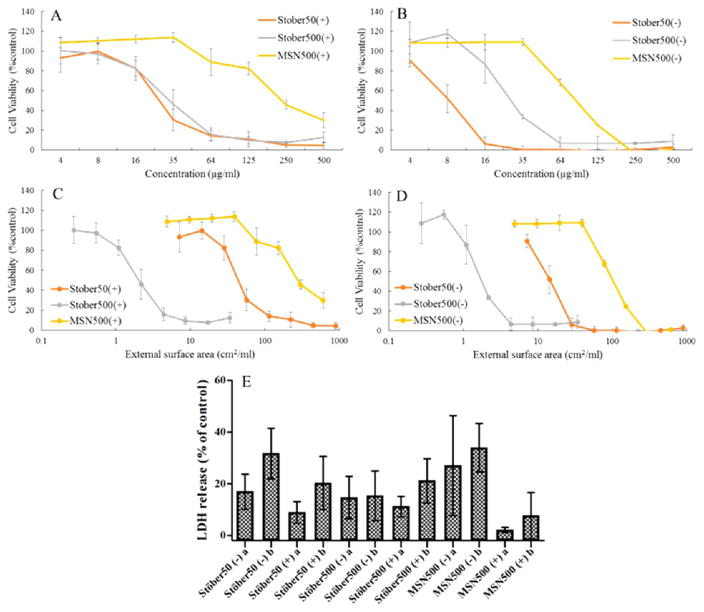
It has been well established that the presence of the protein shield (corona) around the nanoparticles alters the physiochemical properties of the nanoparticle surface and can contribute to changes in zeta potential, aggregation state, and cellular uptake. We investigated the effect of protein corona on SNPs based on the size and porosity and its subsequent effect on the cytotoxicity of nanoparticles toward RAW 264.7 macrophages. Figure 4 shows the toxicity profile of different SNPs in the presence and absence of serum protein. The toxicity of SNPs is associated with the surface silanol (≡SiOH) groups that can interact with membrane components by hydrogen bonding or SNPs can dissociate to form SiO− above the isoelectric point of silica at pH 2–3 and electrostatically interact with tetraalkylammonium-containing phospholipids causing cell membrane lysis.22,23 Another possible reason may be the generation of reactive oxygen species (ROS) by the reaction of a surface radical of SNPs with water. Amorphous silica contains a significant amount of strained 3-membered siloxane rings that give rises to surface-associated radicals that react with water to generate the most reactive hydroxyl radical HO•. These radicals react with the cells to initiate inflammatory responses and can cause cell death. The Stöber (50 and 500 nm) particles were found to be more toxic compared to the MSN500 in vitro. The reduced solid fraction in MSN particles leads to the decrease in availability of accessible surface silanol groups for hydrogen bonding and electrostatic interactions with the cell membrane leading to reduced toxicity.22 Also, the ROS production for the surfactant-extracted MSN500 is less due to the negligible concentration of strained siloxane ring which also contributes to the reduced cell toxicity of the MSN500.24 The presence of the protein corona reduces the toxicity of SNPs that ameliorates cell viability for all SNP particles compared to the serum-free condition. The probable reason for this decrease in toxicity is due to the reduced uptake efficiency of the SNPs. The surface energy of bare SNPs reduces upon formation of the corona, which screens the electrostatic interactions with surface silanol and inhibits SNP adhesion onto the cell surface and decreases subsequent internalization.25,26 Furthermore, the protein corona formed around the particles initiates or limits specific interaction with receptors on the cell surface and may result in altered intracellular localization reducing toxicity.9 Also, the adsorption of serum protein reduces the competitive adsorption of other important constitutional components for the cells and improving the bioavailability of those molecules for the cells reducing toxicity.27 These adsorbed proteins on the particles after digestion in the lysosome can provide a source of nutrition to the cells, leading to the increase of cell proliferation.28 These factors increase cell viability for SNPs in the presence of serum compared to the absence of serum.
Figure 4.
Toxicity of silica nanoparticles toward RAW264.7 macrophages after 24 h incubation. Cell viability curves of RAW264.7 with different concentration of SNPs as a function of mass in the presence (A) and absence (B) of protein. Cell viability curves as a function of total external surface area of particles in the presence (C) and absence (D) of protein. (E) Integrity of plasma membrane after 24 h incubation with LC50 concentrations of SNPs investigated by lactate dehydrogenase (LDH) release assay. [(−) = absence of serum and (+) = presence of serum; (a) = LC50 concentration (lower) in serum-free medium for each SNPs and (b) = LC50 concentration (higher) in media with serum for each SNPs.]
Size-dependent toxicity (Figure 4A and 4B) was observed in this study in accordance to previous reports.29 Toxicity increased by an increase in the surface area upon a decrease Table 2 shows decreased toxicity comparing the LC50 of in size. Stöber500 to Stöber50 by increasing the size of the particle. The decrease in toxicity by increasing surface area in the case of mesoporous particles seems counterintuitive; however, this can be explained from the data on size, zeta potential, and density of silanol groups per square nanometer on the surface of particles. Table 1 shows a decrease of the zeta potential and increase of the hydrodynamic diameter in the media containing FBS for mesoporous particles. Taking into consideration the far lower increase of the hydrodynamic diameter for Stöber500 particles, aggregation of mesoporous particles to a higher extent is expected. Increased aggregation leads to a decrease in the effective cell contactable surface area that causes less cell uptake, cell association, and toxicity as compared to similar sized Stöber500 particles. In addition to the aggregation effect, the influence of the density of the silanol groups per square nanometer surface of particles on protein adsorption and toxicity needs to be taken into account. The reduced solid fraction in MSN particle due to porosity leads to the decrease in availability of accessible surface silanol groups for hydrogen bonding and electrostatic interactions with cell membrane potentially resulting in reduced toxicity. This would also explain the previously reported decreased amount of cell association for mesoporous particles compared to nonporous SNPs.14
Table 2.
IC50 Values of Synthesized and Characterized SNPs for RAW264.7 Macrophages
| LC50 (μg mL−1)
|
||
|---|---|---|
| serum (+) | without serum (−) | |
| Stöber50 | 26.2 ± 2.7 | 8.3 ± 1.2 |
| Stöber500 | 33.7 ± 0.6 | 27.4 ± 1.5 |
| MSN500 | 223.6 ± 15.6 | 96.8 ± 12.0 |
Cellular toxicity of inorganic nanoparticles could be due to changing any metabolic activity, vital cellular functions for cell growth, DNA damage, cell cycle arrest, or disruption of plasma membrane integrity. The integrity of RAW264.7 plasma membranes treated with LC50 SNPs concentration for 24 h was evaluated by lactate dehydrogenase (LDH) release assay and is presented in Figure 4E. LC50 values of each SNP in the presence and absence of proteins were used as a measure of cell membrane integrity. For Stöber50, the LC50 concentration values were 8.3 (lower) and 26.2 μg mL−1 (higher) in the absence and presence of protein, respectively. Therefore, we treated the cells with both concentrations in the presence and absence of protein for comparison. For Stöber500 particles, 27.4 (lower) and 33.7 μg mL−1 (higher) concentrations were used, whereas for MSN500, 96.8 (lower) and 223.6 μg mL−1 (higher) concentrations were used. The LDH release values are plotted in Figure 4E. The bare particles in the absence of proteins had higher membrane damage compared to the particles in the presence of protein for all IC50 concentrations tested (except Stöber500 in higher concentration). The surface-adsorbed serum provides some degree of protection to the cells and reduces membrane damage.28 The LDH release was also found to be concentration dependent for each particle. In respective media, all SNPs exhibited higher membrane damage at higher LC50 concentration.
Identifying the Protein Corona
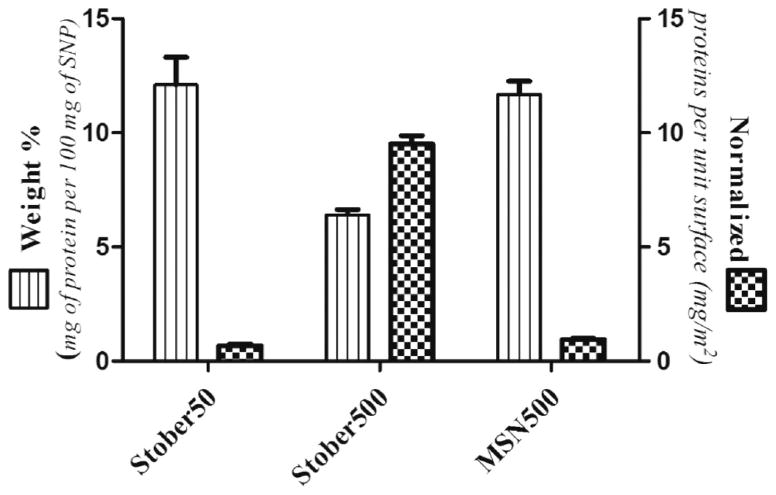
To further investigate the interaction of the SNPs with cells we sought to gain insight into the types and the abundance of proteins that adsorb on the corona upon contact with the particles. The changes in size (DLS) and zeta potential values upon interaction with serum proteins are evidence of corona formation on SNPs (Table 1). The drop in the zeta potential values to from around −8 to −10 mV (Table 1) also suggests the adsorption of proteins around SNPs that shield and alter the surface charge of SNPs.30,31 To evaluate the effect of particle diameter and porosity on protein adsorption, quantitative and qualitative analyses of protein adsorption were performed. The amount of the total proteins adsorbed onto different SNPs was estimated using the bicinchoninic acid assay (BCA) (Figure 5) and TGA analysis (Supporting Information, Figure S2). The Stöber50 SNPs adsorbed the greatest amount of proteins followed by MSN500 and Stöber500, respectively. The amount of protein adsorption was in accordance with the surface area of the particles (Table 1). Stöber50 SNP with an external surface area of 178.84 m2 g−1 was found to adsorb around ~12 wt % of protein from serum, whereas MSN500 with a protein-accessible external surface area of 121.8 m2 g−1 also adsorbed a comparable amount of protein. On the other hand, a decrease in the surface area for Stöber500 to around 6.83 m2 g−1 resulted in a decrease in protein adsorption. A similar trend of protein adsorption value onto SNPs was also reflected from the TGA data (Supporting Information, Figure S2), although normalizing the total amount of protein to the total surface area showed a reverse trend of protein adsorption (Figure 5 and Table S1). The Stöber500 with a higher particle diameter and decreased curvature adsorbed more proteins per unit surface area compared to Stöber50 with a higher curvature. As compared to the MSN500 the smoother surface of Stöber500 favors more protein binding. The decrease in the surface curvature and smoother surface of Stöber500 favor more protein binding as proteins are able to pack more closely on a smoother less curved surface compared to a highly curved and porous surface.32,33 In addition, Si–OH/nm2 could also influence protein adsorption. The roughness of Stöber50 and porosity of MSN500 lead to a decrease in the effective silanol density presented to proteins, which can in turn lead to reduced protein adsorption, reflected in the fact that the Stöber500 particles have the greatest protein adsorption on a mg m−2 basis.
Figure 5.
Amount of protein recovered from each nanoparticle by BCA assay.
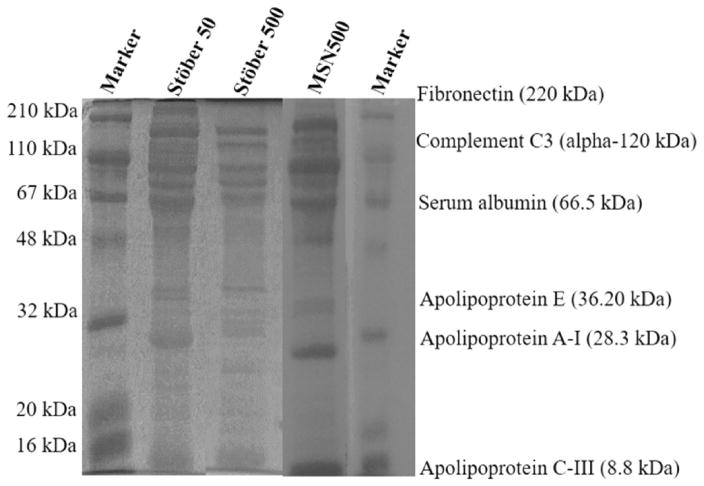
The adsorbed plasma proteins from the hard corona were identified with one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and liquid chromatography mass spectroscopy (LS-MS/MS). From the SDS-PAGE analysis it is apparent that substantial amounts of proteins were extracted for each SNPs (Figure 6). The intensity of each band reflects the amount of protein recovered from each SNP. The band for Stöber500 had less amount of proteins (less intense bands) compared to Stöber50 and MSN500 particles as the surface area of 500 nm SNPs was lower. This is in accordance with the adsorption values obtained by the BCA assay. MSN500 and Stöber50 adsorb a higher number of proteins as evident from the SDS-PAGE analysis. A very limited difference was observed in the elution profiles of proteins extracted, suggesting that most of the adsorbed proteins were of similar molecular weights.
Figure 6.
SDS–PAGE of adsorbed proteins recovered from hard corona; intensity of the band indicates the abundance of protein recovered from the particles. Proteins listed are for reference.
To get a better insight into the proteins recovered from the corona of each SNP, we used label-free liquid chromatography mass spectroscopy (LC-MS). The relative abundance of different proteins recovered was analyzed. The 22 most abundant proteins recovered from each particle are listed in Table 3. Proteins such as lipoproteins, coagulation factors, complement, and cellular components dominate the profile. A comparison of the proteins recovered from each SNPs reveals the commonality of certain proteins in all of them. Dutta et al. reported that plasma protein adsorption profiles remained uniform for differentially sized silica NPs.34 Clemments et al. did a corona analysis on silica NPs based on particle diameter and porosity.35 They found that the smallest particles adsorb higher amounts of proteins compared to larger particles irrespective of porosity, whereas the adsorption profile varied for porous and nonporous particles.35 In our study, we found that the adsorption amount was surface area dependent, whereas the profile of proteins adsorbed was independent of the surface area or porosity. Among the most abundant (relative) proteins from each particle, a similarity in adsorption profile is observed. Hemoglobin is the major protein recovered from all particles irrespective of size or porosity. Other proteins such as apolipoprotein, gelsolin, and thrombospondin were common in all coronas.
Table 3.
Twenty-Two Most Abundant Proteins Associated with Each SNPa
| Stöber50 | Stöber500 | MSN500 |
|---|---|---|
| hemoglobin subunit alpha-1 | hemoglobin subunit alpha-1 | hemoglobin subunit beta |
| hemoglobin subunit beta | gelsolin isoform a precursor | beta-globin |
| serum albumin | gelsolin chain A | hemoglobin chain A |
| hemoglobin fetal subunit beta | hemoglobin subunit beta | apolipoprotein A-I |
| hemoglobin chain B | thrombospondin-1 precursor | hemoglobin subunit alpha |
| gelsolin isoform a precursor | thrombospondin-1 | alpha-2-HS-glycoprotein precursor |
| unnamed protein | alpha-2-HS-glycoprotein precursor | alpha-2-HS-glycoprotein |
| complement C4 precursor | thrombospondin-1 isoform X4 | apolipoprotein A-I |
| vitronectin precursor | apolipoprotein E | complement C4 precursor |
| gelsolin | apolipoprotein E precursor | complement C4 isoform X2 |
| apolipoprotein E precursor | pigment epithelium-derived factor precursor | serum albumin |
| pigment epithelium-derived factor precursor | complement C4 precursor | vitronectin precursor |
| complement C4 isoform X2 | apolipoprotein A-I precursor | plasma serine protease inhibitor precursor |
| apolipoprotein A-I | hypothetical protein | interalpha (globulin) inhibitor H4 |
| thrombospondin-1 precursor | keratin 1 | gelsolin isoform a precursor |
| apolipoprotein A-II precursor | factor V | pigment epithelium-derived factor precursor |
| apolipoprotein E | factor XIIa inhibitor precursor | thrombospondin-1 precursor |
| thrombospondin-1 | alpha-2-macroglobulin precursor | complement factor I precursor |
| antithrombin-III precursor | coagulation factor V | complement factor H precursor |
| complement factor B precursor | alpha-1-antiproteinase precursor | complement factor B precursor |
| alpha-2-macroglobulin precursor | PKM2 protein | tetranectin precursor |
| alpha-1-antiproteinase precursor | pyruvate kinase PKM | fibrinogen alpha chain isoform X1 |
Additional details of database match, accession number, molecular weight, and average intensity values for each SNPs are detailed in the Supporting Information.
The toxicity of the nanoparticles can depend on the corona formed around them.36,37 The role of some proteins in biological systems can be classified as “opsonins” and “dysopsonins”.38,39 The opsonins promote uptake by macrophages, whereas the dysopsonins lower it. It is the abundance and availability of these two classes of proteins that determine the uptake and in turn toxicity of nanoparticles. The toxicity of the SNPs in the absence of protein in a “serum-free” condition was found to be higher for all SNPs. The surface energy of bare particles is the driving force for prominent adhesion of SNPs onto the surface of the cells and subsequent uptake.9 Apart from this, direct surface recognition of the SNPs by cell–surface receptors are also presumed for increased uptake. For example, scavenger receptors recognize certain patterns of nanomaterials and can initiate interactions.40,41 However, in the presence of serum proteins the nanoparticles may have increased or decreased interaction with the cell surface receptors depending on the accessibility of the opsonins or dysopsonins present in the corona. From the LCMS/MS data we found that opsonins such as complement proteins are found in the corona of these nanoparticles that may influence the uptake of SNPs in RAW 264.7 macrophages.42,43 On the other hand, dysopsonins such as apolipoproteins and serum albumin (except for Stöber500) are also found on the surface of the particles which may inhibit uptake. Dysopsonization may also result from adsorbed protein blocking the interaction of cellular response with nanomaterial surface.44 Therefore, the uptake and toxicity of NPs depend mainly on the proximal availability of specific proteins to receptors rather than being concealed in the corona. Thus, the decrease in the toxicity of SNPs upon adsorption of the proteins may be attributed to the presence of the dysopsonins on the surface of the SNPs that effectively inhibits the interaction with the RAW 264.7 and reduces the uptake efficiency of the particles.
Cell Uptake and Localization of Nanoparticles by TEM
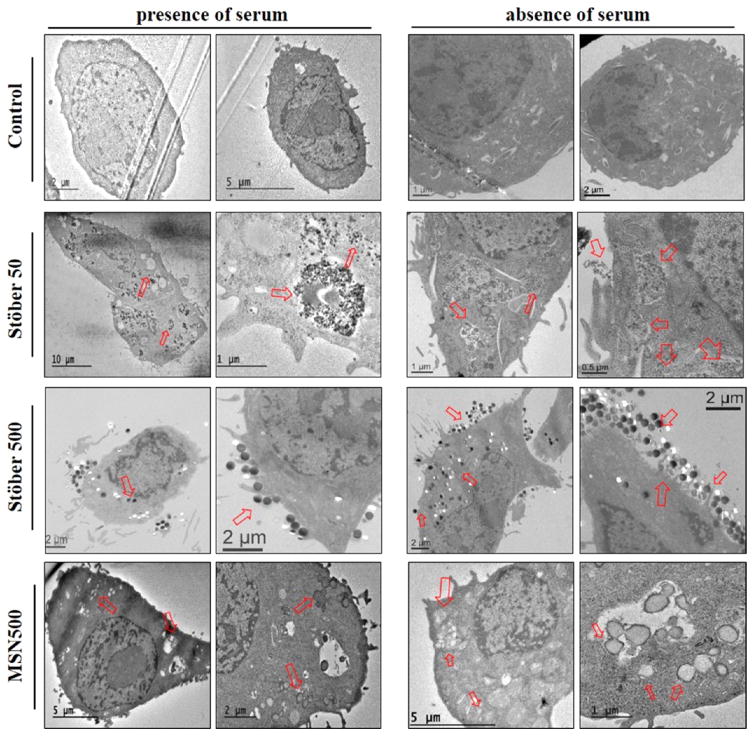
Proteins adsorbed onto the nanoparticle surface can also modulate the mode of uptake and subcellular localization.45–47 We evaluated cellular internalization by transmission electron microscopy (TEM) in the presence and absence of protein. For the Stöber50 SNPs, it was evident from the TEM images that uptake was higher in serum-free condition than in the presence of serum. The 50 nm particles were observed in membrane-bound organelles and also free in the cytoplasm (Figure 7) for both serum and serum-free media. Silica nanoparticles are directed to the lysosomal compartments and remain within these compartments through late-stage lysosomal digestion and then release into the cytoplasm.48 The availability of 50 nm particles in the vacuoles as well as in the cytosol (near membrane) favors the possibility of multiple endocytic pathways.9 The probability of the interplay of multiple mechanistic pathways for uptake of the 50 nm particles has also been favored by the IC-PMS data discussed in a later section of the manuscript. For the Stober500 without protein, more particles were seen inside the cells which correspond to an increased cellular uptake. Less uptake was seen for the Stober500 in the presence of protein. A large cluster of nanoparticles was found adjacent to the cell membrane for the Stober500 SNPs in the absence of serum even after washing three times with PBS. This sort of clustering of nanoparticles in the proximity of the cell membrane in the absence of serum was also reported by Lesniak et al.9 The cellular protrusions interact with these particles, resulting in increased accumulation near the cell surface. Also, the particles were all observed in the cytosol, which might be due to late-stage lysosomal degradation and escape.49 For the MSN500, limited differences in particle localization were noticed. In the presence and absence of proteins, the particles were mostly found in larger vacuole-like structures. In the absence of protein some MSN500 particles were also observed near the membrane in free cytosol, suggesting phagocytosis. Altogether the TEM images suggest more cellular association and enhanced uptake of SNPs in the absence of serum compared to the presence of serum.
Figure 7.
TEM images of the intracellular uptake and localization of SNPs in RAW 264.7 macrophages in the presence and absence of proteins. TEM images of ultrathin sections of pure RAW 267.4 cells in the presence and absence of proteins were taken as control for each case. Arrows indicate localization of particles within the cells and also extracellular adhesions.
Particle Uptake
The cellular uptake pathway of nanoparticles can have direct consequences on intracellular localization and cytotoxicity. The surface properties, protein adsorption, shape, size, and geometry of nanoparticles are among factors that govern the quantity and mechanism of uptake.12,14,50,51
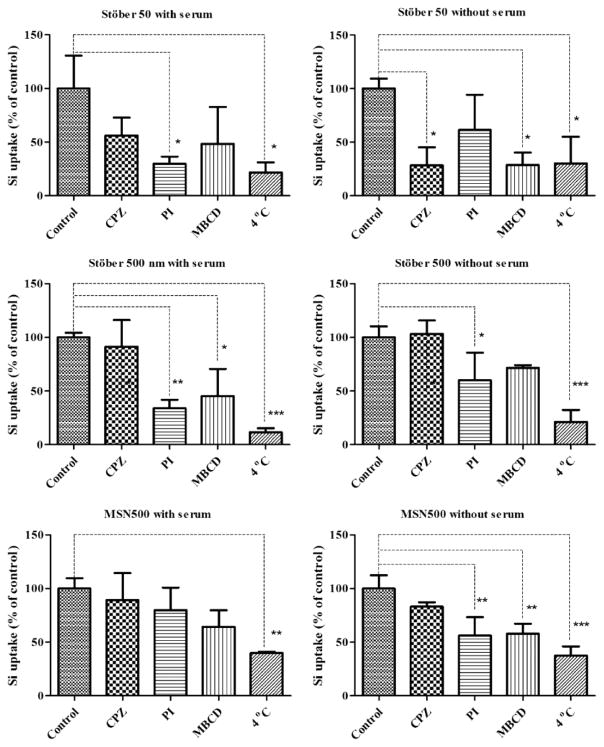
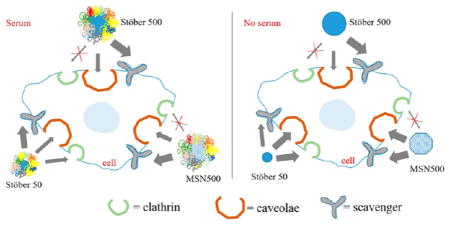
The protein corona formed onto these particles plays a prominent role in the uptake phenomenon. The protein corona inhibits recognition by scavenger receptors but promotes clathrin-dependent uptake.52 In contrast, the corona may also initiate scavenger receptors-mediated cellular toxicity.53 The formation of the corona can also shield to reduce the targeting ability of particles and alter the uptake pathway.54 To evaluate the effect of size and protein corona on the uptake mechanism we used inductively coupled plasma mass spectrometry (ICPMS) to quantify the uptake of nanoparticles. All nanoparticles were sonicated for 15 min to prevent aggregation prior to addition to media with and without serum for the uptake studies. Energy-dependent endocytosis can be inhibited by treating cells at lower temperature and also by inhibitors.55 We incubated the cells at 4 °C to evaluate the energy-dependent uptake of the nanoparticles. Compared to control, a drastic decrease in uptake of all SNPs was observed at 4 °C, indicating the energy dependence of the internalization process. For the energy-dependent cellular endocytotic uptake mechanism of NPs, specific pharmacological inhibitors were used. Chlorpromazine hydrochloride (CPZ) inhibits clathrin-mediated endocytosis by inducing a loss of clathrin and adaptor protein complex 2 from the surface of the cell. Methyl-β-cyclodextrin (MBCD) inhibits caveolae-mediated endocytosis by forming inclusion complexes with cell membrane cholesterol, and polyinosinic acid (PI) was used for scavenger-mediated endocytosis. Figure 8 represents the relative cellular uptake of the SNPs with various inhibitors and at 4 °C in the presence and absence of serum. For the 50 nm SNPs, uptake was inhibited by CPZ (~40% with protein and ~71% without protein), MBCD (~52% with protein and ~71% without protein), and PI (70% with protein and 39% without protein). The presence of serum had some influence on the internalization of the 50 nm SNPs that altered the dispersity of the particles in media. PI inhibition increased in the presence of serum followed by MBCD and CPZ, whereas in the absence of serum CPZ inhibited the most followed by MBCD and PI. The inhibition of uptake in the presence of PI (scavenger) is contradictory for a particle size of 50 nm. Generally, macrophages internalize particles larger than 500 nm by phagocytosis.56 The reason for this observed behavior of the 50 nm is probably due to the aggregation of the particles in the cell’s vicinity. The apparent “secondary size” formed upon aggregation of the particles acts as larger particles that are exposed to the cells. The probability of the aggregation of these particles in media is also suggested by the DLS data. These apparently larger particles are then engulfed by the scavenger-mediated process.
Figure 8.
Cellular uptake pathway of SNP in RAW 264.7 using specific endocytosis inhibitors in the presence and absence of serum. Values represent the mean ± SD (n = 3). *Significantly different from control. *p < 0.05, **p < 0.01, and ***p < 0.001 are marked with an asterisk.
The presence or absence of protein does not alter the uptake pathway for Stober500. For Stober500 nm SNPs, the scavenger receptor-mediated endocytosis (inhibition is ~67% with protein and ~40% without protein) dominated the uptake. However, inhibition of MBCD was also seen for 500 nm SNPs in the presence of protein, demonstrating the caveolae-mediated uptake of particles. Although evidence for caveolin expression in macrophages is scarce and conflicting,57 there are reports in the literature that show the predominance of caveolin proteins in RAW 264.7 macrophages.52,58 Two proteins mediate the caveolin-mediated internalization, caveolin-1 and caveolin-2. Caveolin-1, present primarily on the cell surface, does not colocalize significantly with caveolin-2, which is present primarily in the Golgi compartment in all macrophages studied.57 Caveolae-mediated uptake is observed for larger particles ≈ 500 nm.59 For the 500 nm particles, in the presence and absence of protein, negligible inhibition with the CPZ was found as clathrin pits are small enough to accommodate these larger particles.60 For the MSN 500 nm particle MBCD and PI inhibited the uptake irrespective of the presence or absence of proteins. In the absence of protein the uptake of MSN500 was inhibited by MBCD followed by PI, whereas in the presence of protein the scavenger-mediated pathway was more prominent than the caveolae-mediated uptake. From these results it can be concluded that the size of the nanoparticle has a predominant effect on modulating the uptake phenomenon, whereas we did not observe any prominent effect of surface porosity on the uptake mechanism. The presence or absence of protein may affect the rate of uptake, but no direct correlation from our data can be drawn for changes in the mechanism of uptake. It must be noted that the use of a macropinocytosis-specific inhibitor would help to distinguish if caveolae-mediated internalization is occurring or if extraction of cholesterol is altering macropinocytosis. This phenomenon needs to be further examined in future studies.
CONCLUSION
In summary, we investigated the effect of serum protein adsorption onto a series of SNPs based on the size and porosity and their subsequent cellular impact. The toxicity for all particles in the presence of serum was lowered compared to serum-free condition, which was in agreement with previously reported literature.28 The protein adsorption amount varied directly with the available surface area of the particles. The presence and absence of corona had a limited effect on the uptake mechanism of SNPs. The uptake mechanism was predominantly governed by size. The small sized 50 nm SNPs followed multiple endocytosis pathways, whereas for the larger particles the scavenger-mediated pathway regulated the uptake along with caveolae-mediated pathway. The uptake of the particles was inhibited in the presence of serum, but no significant difference was observed for the uptake mechanisms.
EXPERIMENTAL PROCEDURES
Materials
Cetyltrimethylammonium bromide (CTAB, ≥99.0%) and tetraethyl orthosilicate (TEOS, 98%) were obtained from Sigma-Aldrich. Absolute ethanol (200 proof) and ammonium hydroxide were from Acros Organics and EMD Millipore Corp., respectively. Hydrochloric acid was purchased from BDH Aristar. All chemicals were used as received.
Synthesis of Nonporous and Mesoporous Silica Nanoparticles
A modified Stöber method was used to produce nonporous spherical silica nanoparticles (Stöber).61 First, 900 mL of 200 proof ethanol was mixed with 25.2 mL of DI water and 22.5 mL of ammonium hydroxide (29.7%) at room temperature in a 1.5 L glass flask. Then tetraethyl orthosilicate (TEOS) (33.3 mL) was added dropwise under stirring at 500 rpm, and the reaction was left under stirring for 24 h. The product was washed twice by centrifugation at 15 000 rpm for 20 min and stored in ethanol. By changing the concentration of TEOS and aqueous ammonia the targeted size of the particles was obtained.
Mesoporous silica nanoparticles were produced by methods previously described through a one-step condensation under dilute silica source and low surfactant concentration conditions.14,62 Briefly, 2 g of CTAB was dissolved in 700 mL of DI water, and 50 mL of ammonium hydroxide was added to the mixture. The mixture was stirred at room temperature for 1 h to stabilize the solution and form aligned CTAB micelles. TEOS was added dropwise under stirring at 300 rpm and the stirring continued for 4 h, after which the mixture was autoclaved at 100 °C for 24 h. The product was washed twice by centrifugation, and particles were suspended in ethanolic HCl (1.5 mL of HCl in 150 mL of ethanol) and heated at 60 °C for 6 h to remove the surfactant.
Nanoparticle Characterization
The shape and size of the particles were investigated by electron microscopy techniques. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images were obtained with a FEI Quanta 650 FE-SEM operating at 20 kV and a JEOL JEM 1400 microscope operating at 120 kV, respectively. X-ray diffraction patterns of mesoporous particles were analyzed to confirm 2D-hexagonal mesopore structure in the close-packing structure on a Bruker D2 Phaser X-ray diffractometer using Cu Ka radiation (λ = 0.1542 nm) at 45 kV and 40 mA. The XRD spectra were recorded at a scanning speed of 0.01 deg/s, with a step size of 0.02° in a 2θ scattering angle and in the 2θ range of 2–10. FT-IR spectra were recorded on a Varian 3100 FT-IR spectrometer with attenuated total reflectance mode to confirm washing the CTAB out of pores in mesoporous particles. Hydrodynamic diameter and zeta potential measurements were performed by dynamic light scattering (DLS) in a Malvern Instruments Zetasizer Nano ZS. Nitrogen adsorption–desorption isotherm analysis was conducted on a Micromeritics ASAP 2020 (Norcross, GA) surface area and porosity analyzer at −196 °C to measure internal and external surface area and pore size. All samples were dried at 100 °C overnight prior to analysis. Pore volume and pore size distributions were obtained from an adsorption branch by using the Barrett, Joyner, and Halenda method. The external surface areas of all samples were calculated from the t-plots of their N2 adsorption isotherms. The Brunauer–Emmett–Teller specific surface areas were calculated by using adsorption data at P/P0 = 0.05–0.20.18,20,63,64
Cell Culture and Cytotoxicity Assay
The cytotoxicity of the assynthesized SNPs was tested on RAW 264.7 macrophages. Cells were cultured at 37 °C in 5% CO2 in RPMI with 10% fetal bovine serum (FBS). For the cytotoxicity assays cells were seeded onto 96-well plates with 6000 cells/well and allowed to grow for 24 h. After 24 h the media was replaced and the cells were washed with PBS. Fresh media was then added without (control) and with varying concentrations of 4 μg/mL to 800 μg/mL of SNPs in the presence and absence of 10% fetal bovine serum. The cells were then incubated for 24 h, after which the media was aspirated and the cells were washed twice with PBS. Cell viability was measured with CCK-8 from Dojindo according to an established protocol (Dojindo, Rockville, MD). The absorbance of the plate was measured at 450 nm. For membrane integrity the LDH Cytotoxicity Assay Kit (Thermo Scientific Pierce) was used according to the manufacturer’s protocol.
Protein Adsorption Experiments
A stock solution of 8 mg/mL of SNPs was used for all adsorption studies. A 2 mL amount of RPMI containing 10% FBS was preincubated at 37 °C for 30 min in a shaker incubator. An aliquot containing 1 mg/mL of particles was added to the media and incubated for 1 h. After incubation the particles were centrifuged at 13 000 rpm for 10 min and washed with PBS (7.4) three times to remove the unbound or loosely bound proteins. To make sure that we are not losing any particles (at 13 000 rpm and 10 min time) in the solution we centrifuged a known concentration of each Stober50, Stober500, and MSN500 particles (500 and 1 mL) and dried the pellets formed and measured the amount (mass) of nanoparticles recovered. No observable difference was seen, and thus, the centrifuging speed of 13 000 rpm and for 10 min was optimum to separate the SNPs from solution.
Protein Quantification by Bicinchoninic Acid (BCA) Assay
The hard corona recovered after washing and centrifugation was removed by sonicating the nanoparticles in extraction buffer (63 mM Tris-HCl, pH 6.8, 40 mM DTT, 0.01% (w/v) bromophenol blue, 10% glycerol, 2% (w/v) SDS) as described previously.65 The suspension was placed in a thermal cycler (Applied Biosystems, 2720 Thermal Cycler) to boil at 95 °C for 5 min and then cooled. The suspension was then centrifuged at 13 000 rpm for 10 min, and the supernatant was used for quantification using the BCA assay kit (G-Biosciences) according to manufacturer’s protocol.
SDS PAGE
One-dimensional sodium dodecyl sulfate-poly-(acrylamide gel electrophoresis) (SDS-PAGE) was utilized to analyze the proteins recovered after incubation with serum. After the final (3×) washing and centrifugation step, the proteins (hard corona) were removed by eluting the particle in Laemmli buffer (Bio-Rad). The suspension was placed in a thermal cycler (Applied Biosystems, 2720 Thermal Cycler) to boil at 95 °C for 5 min and then cooled. It was subsequently centrifuged at 13 000 rpm for 10 min, and the supernatant was used for the SDS-PAGE analysis. A 10 μL amount of each supernatant was run on a Bio-Rad Mini-PROTEAN electrophoresis system (120 V, 1.5 h) alongside a standard protein marker (PAGEMark Tricolor, G-Biosciences). The gels were washed with deionized water three times prior to staining for 2 h using Coomassie Brilliant Blue R-250 reagent (Bio-Rad), followed by destaining with constant shaking overnight.
Proteomic Analysis
For detailed identification of the proteins recovered from the SNPs, a similar method of preparation was used as described above. After loading the samples (stained with Coomassie blue) in gel it was allowed to run for 3 min so that the proteins enter from the stacking gel section to the separating gel section. The band was precisely cut and digested with Trypsin and Lys-C (Promega). The digested samples were dissolved in 0.1% formic acid, and 5 μL was injected into an electrospray liquid chromatography mass spectrometry (LC-MS/MS), Bruker Maxis II ETD. Peptides were separated by applying a gradient of 5–60% acetonitrile and water at a flow rate of 400 nL/min. The acquired data from the LC-MSMS were searched using the MASCOT search engine against the NCBInr database using the taxonomy filter of mammalian database. The label-free relative quantification of the proteins was done using the MASCOT Distiller interface (www.matrixscience.com). After chromatogram alignment and peptide retention time determination, a weighted mean m/z of each peptide was calculated and a tab delimited file was created to extract peptide intensity using MASCOT Distiller interface. Protein abundance (intensity) was calculated from all qualified peptides corresponding to a particular protein. An average of two repeats was used for the calculation. Data were modified to remove duplicates of each representation, and the higher average intensity values were considered. The proteins were then listed in order of decreasing abundance (relative). Protein abundance/quantity calculated by this method was represented by unitless numerical values.
Cell Uptake by TEM
The interaction and uptake of the nanoparticles with RAW 264.7 were studied using TEM. Cells were grown on ACLAR sheets in 6-well plates and treated with desired concentration of SNPs in presence and absence of proteins. Controls were treated by media with and without proteins. After incubation for 4 h, cells were washed 3 times with PBS. 1 mL of fixing solution (2.5% glutaraldehyde + 1.0% paraformaldehyde) was added to the cells, and the mixture was incubated at room temperature for 30 min followed by 4 °C overnight. Ultrathin sections of control cells and treated cells were imaged by a JOEL JEM 1400 microscope.
Cellular Uptake Pathway Determination
Different endocytic inhibitors were employed to determine the endocytic pathways responsible for cellular uptake. Chlorpromazine hydrochloride (CPZ), methyl-β-cyclodextrin (MBCD), and polyinosinic acid (PI) were used to inhibit the clathrin-mediated, caveolin-mediate, and scavenger-mediated endocytosis pathways, respectively. RAW 264.7 cells were preincubated with optimized doses (in terms of CCK-8 toxicity assay) for 30 min at 37 °C. Inhibitors were then removed; cells washed with PBS and SNPs (50 μg mL−1) suspended in media with and without serum were added to the cells and further incubated for 2 h for the uptake study. Control experiments were prepared in the same way without preincubation with inhibitors. SNP internalization was quantified using inductive coupled plasma-mass spectroscopy (ICP-MS).
Inductive Coupled Plasma-Mass Spectroscopy (ICP-MS)
The cell pellets were transferred from centrifuging tubes to PTFE (Savillex) vial and the liquid evaporated. A 0.5 mL amount of trace metal grade HNO3 was added and allowed to digest at 150 °C for 2 h to complete digestion. After digestion the HNO3 was allowed to dry down on the hot plate. The oxidized product was dissolved in 2 mL of water and transferred into a PS autosampler tube. An internal standard of Cs was added to each sample. A known standard Si reference was taken to generate the standard curve. Measurements were made using an Agilent 7500ce ICPMS instrument under operating conditions suitable for routine multielement analysis. All chemicals used were of trace metal grade.
Statistical Analysis
Data are expressed as mean ± SDs for at least three separate experiments. Statistical analyses were performed by one-way ANOVA. The Tukey post-test was used where a difference was detected. The difference compared to control was considered significant when p < 0.05.
Acknowledgments
Support from the National Institute of Environmental Health Sciences of the NIH (R01ES024681) and the Skaggs Graduate Research Fellowship (Mostafa Yazdimamaghani) is acknowledged. This work made use of the University of Utah shared facilities of the Micron Microscopy Suite and the University of Utah USTAR shared facilities supported in part by the MRSEC Program of the NSF under Award No. DMR-1121252.
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.6b09950.
FTIR data, quantification of proteins by TGA, table for normalized protein adsorption amount to surface area, SDS PAGE, and details of the corona composition proteins (PDF)
References
- 1.Lu J, Liong M, Li Z, Zink JI, Tamanoi F. Biocompatibility, Biodistribution, and Drug-Delivery Efficiency of Mesoporous Silica Nanoparticles for Cancer Therapy in Animals. Small. 2010;6:1794–1805. doi: 10.1002/smll.201000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J, Kim HS, Lee N, Kim T, Kim H, Yu T, Song IC, Moon WK, Hyeon T. Multifunctional Uniform Nanoparticles Composed of a Magnetite Nanocrystal Core and a Mesoporous Silica Shell for Magnetic Resonance and Fluorescence Imaging and for Drug Delivery. Angew Chem, Int Ed. 2008;47:8438–8441. doi: 10.1002/anie.200802469. [DOI] [PubMed] [Google Scholar]
- 3.Rieter WJ, Kim JS, Taylor KM, An H, Lin W, Tarrant T, Lin W. Hybrid Silica Nanoparticles for Multimodal Imaging. Angew Chem, Int Ed. 2007;46:3680–3682. doi: 10.1002/anie.200604738. [DOI] [PubMed] [Google Scholar]
- 4.Tapec R, Zhao XJ, Tan W. Development of Organic Dye-Doped Silica Nanoparticles for Bioanalysis and Biosensors. J Nanosci Nanotechnol. 2002;2:405–409. doi: 10.1166/jnn.2002.114. [DOI] [PubMed] [Google Scholar]
- 5.Tu HL, Lin YS, Lin HY, Hung Y, Lo LW, Chen YF, Mou CY. In Vitro Studies of Functionalized Mesoporous Silica Nanoparticles for Photodynamic Therapy. Adv Mater. 2009;21:172–177. [Google Scholar]
- 6.Dobrovolskaia MA, Patri AK, Potter TM, Rodriguez JC, Hall JB, McNeil SE. Dendrimer-Induced Leukocyte Procoagulant Activity Depends on Particle Size and Surface Charge. Nanomedicine. 2012;7:245–256. doi: 10.2217/nnm.11.105. [DOI] [PubMed] [Google Scholar]
- 7.Dowding JM, Das S, Kumar A, Dosani T, McCormack R, Gupta A, Sayle TX, Sayle DC, von Kalm L, Seal S. Cellular Interaction and Toxicity Depend on Physicochemical Properties and Surface Modification of Redox-Active Nanomaterials. ACS Nano. 2013;7:4855–4868. doi: 10.1021/nn305872d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Townson JL, Lin YS, Agola JO, Carnes EC, Leong HS, Lewis JD, Haynes CL, Brinker CJ. Re-Examining the Size/Charge Paradigm: Differing in Vivo Characteristics of Size-and Charge-Matched Mesoporous Silica Nanoparticles. J Am Chem Soc. 2013;135:16030–16033. doi: 10.1021/ja4082414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells. ACS Nano. 2012;6:5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 10.Docter D, Bantz C, Westmeier D, Galla HJ, Wang Q, Kirkpatrick JC, Nielsen P, Maskos M, Stauber RH. The Protein Corona Protects against Size-and Dose-Dependent Toxicity of Amorphous Silica Nanoparticles. Beilstein J Nanotechnol. 2014;5:1380–1392. doi: 10.3762/bjnano.5.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C. Nanoparticle Size Is a Critical Physicochemical Determinant of the Human Blood Plasma Corona: A Comprehensive Quantitative Proteomic Analysis. ACS Nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 12.Yu T, Greish K, McGill LD, Ray A, Ghandehari H. Influence of Geometry, Porosity, and Surface Characteristics of Silica Nanoparticles on Acute Toxicity: Their Vasculature Effect and Tolerance Threshold. ACS Nano. 2012;6:2289–2301. doi: 10.1021/nn2043803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu T, Hubbard D, Ray A, Ghandehari H. In Vivo Biodistribution and Pharmacokinetics of Silica Nanoparticles as a Function of Geometry, Porosity and Surface Characteristics. J Controlled Release. 2012;163:46–54. doi: 10.1016/j.jconrel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu T, Malugin A, Ghandehari H. Impact of Silica Nanoparticle Design on Cellular Toxicity and Hemolytic Activity. ACS Nano. 2011;5:5717–5728. doi: 10.1021/nn2013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical Studies to Understand Nanoparticle Interaction with the Immune System and Its Potential Effects on Nanoparticle Biodistribution. Mol Pharmaceutics. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carcouët CC, van de Put MW, Mezari B, Magusin PC, Laven J, Bomans PH, Friedrich H, Esteves ACC, Sommerdijk NA, van Benthem RA. Nucleation and Growth of Monodisperse Silica Nanoparticles. Nano Lett. 2014;14:1433–1438. doi: 10.1021/nl404550d. [DOI] [PubMed] [Google Scholar]
- 17.Vacassy R, Flatt R, Hofmann H, Choi K, Singh R. Synthesis of Microporous Silica Spheres. J Colloid Interface Sci. 2000;227:302–315. doi: 10.1006/jcis.2000.6860. [DOI] [PubMed] [Google Scholar]
- 18.Lu F, Wu SH, Hung Y, Mou CY. Size Effect on Cell Uptake in Well-Suspended, Uniform Mesoporous Silica Nanoparticles. Small. 2009;5:1408–1413. doi: 10.1002/smll.200900005. [DOI] [PubMed] [Google Scholar]
- 19.Tsai CP, Hung Y, Chou YH, Huang DM, Hsiao JK, Chang C, Chen YC, Mou CY. High-Contrast Paramagnetic Fluorescent Mesoporous Silica Nanorods as a Multifunctional Cell-Imaging Probe. Small. 2008;4:186–191. doi: 10.1002/smll.200700457. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H, Zhao X, Lu G, Do D. Improved Comparison Plot Method for Pore Structure Characterization of Mcm-41. Langmuir. 1996;12:6513–6517. [Google Scholar]
- 21.Lee P, Knight R, Smit JM, Wilschut J, Griffin DE. A Single Mutation in the E2 Glycoprotein Important for Neurovirulence Influences Binding of Sindbis Virus to Neuroblastoma Cells. Journal of Virology. 2002;76:6302–6310. doi: 10.1128/JVI.76.12.6302-6310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slowing II, Wu CW, Vivero-Escoto JL, Lin VSY. Mesoporous Silica Nanoparticles for Reducing Hemolytic Activity Towards Mammalian Red Blood Cells. Small. 2009;5:57–62. doi: 10.1002/smll.200800926. [DOI] [PubMed] [Google Scholar]
- 23.Nash T, Allison A, Harington J. Physico-Chemical Properties of Silica in Relation to its Toxicity. Nature. 1966;210:259–261. doi: 10.1038/210259a0. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Dunphy DR, Jiang X, Meng H, Sun B, Tarn D, Xue M, Wang X, Lin S, Ji Z. Processing Pathway Dependence of Amorphous Silica Nanoparticle Toxicity: Colloidal Vs Pyrolytic. J Am Chem Soc. 2012;134:15790–15804. doi: 10.1021/ja304907c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesniak A, Salvati A, Santos-Martinez MJ, Radomski MW, Dawson KA, Åberg C. Nanoparticle Adhesion to the Cell Membrane and Its Effect on Nanoparticle Uptake Efficiency. J Am Chem Soc. 2013;135:1438–1444. doi: 10.1021/ja309812z. [DOI] [PubMed] [Google Scholar]
- 26.Tarn D, Ashley CE, Xue M, Carnes EC, Zink JI, Brinker CJ. Mesoporous Silica Nanoparticle Nanocarriers: Biofunctionality and Biocompatibility. Acc Chem Res. 2013;46:792–801. doi: 10.1021/ar3000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Li W, Li Q, Li Y, Li Y, Zhang X, Huang Q. Effects of Serum Proteins on Intracellular Uptake and Cytotoxicity of Carbon Nanoparticles. Carbon. 2009;47:1351–1358. [Google Scholar]
- 28.Tedja R, Lim M, Amal R, Marquis C. Effects of Serum Adsorption on Cellular Uptake Profile and Consequent Impact of Titanium Dioxide Nanoparticles on Human Lung Cell Lines. ACS Nano. 2012;6:4083–4093. doi: 10.1021/nn3004845. [DOI] [PubMed] [Google Scholar]
- 29.Malugin A, Herd H, Ghandehari H. Differential Toxicity of Amorphous Silica Nanoparticles toward Phagocytic and Epithelial Cells. J Nanopart Res. 2011;13:5381–5396. [Google Scholar]
- 30.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 31.Mortensen NP, Hurst GB, Wang W, Foster CM, Nallathamby PD, Retterer ST. Dynamic Development of the Protein Corona on Silica Nanoparticles: Composition and Role in Toxicity. Nanoscale. 2013;5:6372–6380. doi: 10.1039/c3nr33280b. [DOI] [PubMed] [Google Scholar]
- 32.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein– Nanoparticle Interactions: Opportunities and Challenges. Chem Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 33.Gagner JE, Lopez MD, Dordick JS, Siegel RW. Effect of Gold Nanoparticle Morphology on Adsorbed Protein Structure and Function. Biomaterials. 2011;32:7241–7252. doi: 10.1016/j.biomaterials.2011.05.091. [DOI] [PubMed] [Google Scholar]
- 34.Dutta D, Sundaram SK, Teeguarden JG, Riley BJ, Fifield LS, Jacobs JM, Addleman SR, Kaysen GA, Moudgil BM, Weber TJ. Adsorbed Proteins Influence the Biological Activity and Molecular Targeting of Nanomaterials. Toxicol Sci. 2007;100:303–315. doi: 10.1093/toxsci/kfm217. [DOI] [PubMed] [Google Scholar]
- 35.Clemments AM, Botella P, Landry CC. Protein Adsorption from Biofluids on Silica Nanoparticles: Corona Analysis as a Function of Particle Diameter and Porosity. ACS Appl Mater Interfaces. 2015;7:21682–21689. doi: 10.1021/acsami.5b07631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrenberg MS, Friedman AE, Finkelstein JN, Oberdörster G, McGrath JL. The Influence of Protein Adsorption on Nanoparticle Association with Cultured Endothelial Cells. Biomaterials. 2009;30:603–610. doi: 10.1016/j.biomaterials.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 37.Clift MJ, Bhattacharjee S, Brown DM, Stone V. The Effects of Serum on the Toxicity of Manufactured Nanoparticles. Toxicol Lett. 2010;198:358–365. doi: 10.1016/j.toxlet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Walkey CD, Chan WC. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 39.Moghimi SM, Muir I, Illum L, Davis SS, Kolb-Bachofen V. Coating Particles with a Block Co-Polymer (Poloxamine-908) Suppresses Opsonization but Permits the Activity of Dysopsonins in the Serum. Biochim Biophys Acta, Mol Cell Res. 1993;1179:157–165. doi: 10.1016/0167-4889(93)90137-e. [DOI] [PubMed] [Google Scholar]
- 40.Nishikawa K, Arai H, Inoue K. Scavenger Receptor-Mediated Uptake and Metabolism of Lipid Vesicles Containing Acidic Phospholipids by Mouse Peritoneal Macrophages. J Biol Chem. 1990;265:5226–5231. [PubMed] [Google Scholar]
- 41.Orr GA, Chrisler WB, Cassens KJ, Tan R, Tarasevich BJ, Markillie LM, Zangar RC, Thrall BD. Cellular Recognition and Trafficking of Amorphous Silica Nanoparticles by Macrophage Scavenger Receptor A. Nanotoxicology. 2011;5:296–311. doi: 10.3109/17435390.2010.513836. [DOI] [PubMed] [Google Scholar]
- 42.Hsu M, Juliano RL. Interactions of Liposomes with the Reticuloendothelial System: Ii. Nonspecific and Receptor-Mediated Uptake of Liposomes by Mouse Peritoneal Macrophages. Biochim Biophys Acta, Mol Cell Res. 1982;720:411–419. doi: 10.1016/0167-4889(82)90120-3. [DOI] [PubMed] [Google Scholar]
- 43.Moghimi SM, Hunter AC, Murray JC. Long-Circulating and Target-Specific Nanoparticles: Theory to Practice. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 44.Lee KD, Pitas RE, Papahadjopoulos D. Evidence That the Scavenger Receptor Is Not Involved in the Uptake of Negatively Charged Liposomes by Cells. Biochim Biophys Acta, Biomembr. 1992;1111:1–6. doi: 10.1016/0005-2736(92)90267-p. [DOI] [PubMed] [Google Scholar]
- 45.Serda RE, Gu J, Burks JK, Ferrari K, Ferrari C, Ferrari M. Quantitative Mechanics of Endothelial Phagocytosis of Silicon Microparticles. Cytometry, Part A. 2009;75A:752–760. doi: 10.1002/cyto.a.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng ZJ, Liang M, Monteiro M, Toth I, Minchin RF. Nanoparticle-Induced Unfolding of Fibrinogen Promotes Mac-1 Receptor Activation and Inflammation. Nat Nanotechnol. 2011;6:39–44. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- 47.Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O’Neil CP, Lee LK, Swartz MA, Hubbell JA. Exploiting Lymphatic Transport and Complement Activation in Nanoparticle Vaccines. Nat Biotechnol. 2007;25:1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 48.Huang DM, Hung Y, Ko BS, Hsu SC, Chen WH, Chien CL, Tsai CP, Kuo CT, Kang JC, Yang CS. Highly Efficient Cellular Labeling of Mesoporous Nanoparticles in Human Mesenchymal Stem Cells: Implication for Stem Cell Tracking. FASEB J. 2005;19:2014–2016. doi: 10.1096/fj.05-4288fje. [DOI] [PubMed] [Google Scholar]
- 49.Herd HL, Malugin A, Ghandehari H. Silica Nanoconstruct Cellular Toleration Threshold in Vitro. J Controlled Release. 2011;153:40–48. doi: 10.1016/j.jconrel.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herd H, Daum N, Jones AT, Huwer H, Ghandehari H, Lehr CM. Nanoparticle Geometry and Surface Orientation Influence Mode of Cellular Uptake. ACS Nano. 2013;7:1961–1973. doi: 10.1021/nn304439f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng H, Yang S, Li Z, Xia T, Chen J, Ji Z, Zhang H, Wang X, Lin S, Huang C. Aspect Ratio Determines the Quantity of Mesoporous Silica Nanoparticle Uptake by a Small Gtpase-Dependent Macropinocytosis Mechanism. ACS Nano. 2011;5:4434–4447. doi: 10.1021/nn103344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng X, Tian X, Wu A, Li J, Tian J, Chong Y, Chai Z, Zhao Y, Chen C, Ge C. Protein Corona Influences Cellular Uptake of Gold Nanoparticles by Phagocytic and Nonphagocytic Cells in a Size-Dependent Manner. ACS Appl Mater Interfaces. 2015;7:20568–20575. doi: 10.1021/acsami.5b04290. [DOI] [PubMed] [Google Scholar]
- 53.Shannahan JH, Podila R, Aldossari AA, Emerson H, Powell BA, Ke PC, Rao AM, Brown JM. Formation of a Protein Corona on Silver Nanoparticles Mediates Cellular Toxicity Via Scavenger Receptors. Toxicol Sci. 2015;143:136–146. doi: 10.1093/toxsci/kfu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Bombelli FB, Hristov DR, Kelly PM, Åberg C, Mahon E, Dawson KA. Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 55.Kou L, Sun J, Zhai Y, He Z. The Endocytosis and Intracellular Fate of Nanomedicines: Implication for Rational Design. Asian J Pharm Sci. 2013;8:1–10. [Google Scholar]
- 56.Clift MJ, Rothen-Rutishauser B, Brown DM, Duffin R, Donaldson K, Proudfoot L, Guy K, Stone V. The Impact of Different Nanoparticle Surface Chemistry and Size on Uptake and Toxicity in a Murine Macrophage Cell Line. Toxicol Appl Pharmacol. 2008;232:418–427. doi: 10.1016/j.taap.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Gargalovic P, Dory L. Caveolin-1 and Caveolin-2 Expression in Mouse Macrophages High Density Lipoprotein 3-Stimulated Secretion and a Lack of Significant Subcellular Co-Localization. J Biol Chem. 2001;276:26164–26170. doi: 10.1074/jbc.M011291200. [DOI] [PubMed] [Google Scholar]
- 58.Lei MG, Morrison DC. Differential Expression of Caveolin-1 in Lipopolysaccharide-Activated Murine Macrophages. Infection and immunity. 2000;68:5084–5089. doi: 10.1128/iai.68.9.5084-5089.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-Dependent Internalization of Particles Via the Pathways of Clathrin-and Caveolae-Mediated Endocytosis. Biochem J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahmoudi M, Azadmanesh K, Shokrgozar MA, Journeay WS, Laurent S. Effect of Nanoparticles on the Cell Life Cycle. Chem Rev. 2011;111:3407–3432. doi: 10.1021/cr1003166. [DOI] [PubMed] [Google Scholar]
- 61.Stöber W, Fink A, Bohn E. Controlled Growth of Monodisperse Silica Spheres in the Micron Size Range. J Colloid Interface Sci. 1968;26:62–69. [Google Scholar]
- 62.Huh S, Wiench JW, Yoo JC, Pruski M, Lin VSY. Organic Functionalization and Morphology Control of Mesoporous Silicas Via a Co-Condensation Synthesis Method. Chem Mater. 2003;15:4247–4256. [Google Scholar]
- 63.He Q, Zhang Z, Gao Y, Shi J, Li Y. Intracellular Localization and Cytotoxicity of Spherical Mesoporous Silica Nano-and Microparticles. Small. 2009;5:2722–2729. doi: 10.1002/smll.200900923. [DOI] [PubMed] [Google Scholar]
- 64.Szekeres M, Tóth J, Dékány I. Specific Surface Area of Stoeber Silica Determined by Various Experimental Methods. Langmuir. 2002;18:2678–2685. [Google Scholar]
- 65.Clemments AM, Muniesa C, Landry CC, Botella P. Effect of Surface Properties in Protein Corona Development on Mesoporous Silica Nanoparticles. RSC Adv. 2014;4:29134–29138. [Google Scholar]