Abstract
While the HIV-1-glycan shield is known to shelter Env from the humoral immune response, its quantitative impact on antibody elicitation has been unclear. Here we use targeted deglycosylation to measure the impact of the glycan shield on elicitation of antibodies against the CD4 supersite. We engineered diverse Env trimers with select glycans removed proximal to the CD4 supersite, characterized their structures and glycosylation, and immunized guinea pigs and rhesus macaques. Immunizations yielded little neutralization against wild-type viruses, but potent CD4-supersite neutralization (titers 1:>1,000,000 against 4-glycan-deleted autologous viruses with over 90% breadth against 4-glycan-deleted heterologous strains exhibiting tier-2 neutralization character). To a first approximation, the immunogenicity of the glycan-shielded protein surface was negligible, with Env-elicited neutralization (ID50) proportional to the exponential of the protein-surface area accessible to antibody. Based on these high titers and exponential relationship, we propose site-selective deglycosylated trimers as priming immunogens to increase the frequency of site-targeting antibodies.
eTOC
Zhou et al. engineer diverse HIV-Env trimers with selective glycans removed around the CD4 receptor-binding site. They demonstrate the impact of the glycan shield on immunogenicity, uncover an exponential relationship between exposed antibody-accessible protein surface and Env immunogenicity, and achieve vaccine-elicited neutralization of glycan-deleted viruses.
INTRODUCTION
The HIV-1-Env spike, the sole component of the virus that extends outside the protective viral membrane, is shielded from potentially neutralizing antibody by three primary mechanisms: sequence variation (Gaschen et al., 2002; Korber et al., 2001; Starcich et al., 1986), conformational masking (Kwong et al., 2002; Munro et al., 2014), and glycan shielding (Wei et al., 2003). A focus on conserved Env surfaces (Kwong et al., 1998; Zhou et al., 2015; Zhou et al., 2007) and stabilization of the functionally critical prefusion-closed conformation of the Env trimer (de Taeye et al., 2015; Kwon et al., 2015; Sanders et al., 2013) provide solutions, respectively, to evasion mechanisms of sequence variation and conformational masking. With respect to glycan-immune evasion, the HIV-1-glycan shield has been visualized (Gristick et al., 2016; Lee et al., 2016; Stewart-Jones et al., 2016), but the quantitative impact of the glycan shield on the elicitation of Env-directed antibodies remains unclear.
Atomic-level structures of fully glycosylated Env trimers reveal complex oliogosaccharides proximal to the viral membrane (Lee et al., 2016), which extend as prominent ridges of interlocking oligomannose to shield exposed surfaces of the viral spike including the trimer apex (Stewart-Jones et al., 2016). Analysis of known broadly neutralizing HIV-1 antibodies reveals that many of these rare antibodies require recognition of N-linked glycan for high affinity binding (McLellan et al., 2011; Pejchal et al., 2011; Walker et al., 2011; Walker et al., 2009). However, neutralizing antibodies that target the CD4 supersite, the site of Env vulnerability associated with binding to the CD4 receptor (Zhou et al., 2015; Zhou et al., 2007), are one of the few categories of broadly neutralizing antibodies that do not require glycan to bind to the Env trimer (Kwong and Mascola, 2012). In terms of sequence variation, the CD4 supersite has the lowest sequence entropy of epitopes on the closed spike (Pancera et al., 2014). And in terms of accessibility, the CD4 binding site appears to be accessible to antibody in the prefusion-closed conformation, with the exception of glycan occlusion (Chen et al., 2009; Pancera et al., 2014; Zhou et al., 2015). However, the CD4 supersite in the prefusion-closed state appears to be minimally immunogenic: antibodies elicited by Env trimer immunization in the prefusion-closed state do not target the CD4-binding site and display no neutralization breadth (Cheng et al., 2016; de Taeye et al., 2015; Sanders et al., 2015). Thus, the CD4 supersite in the prefusion-closed state appears to be primarily protected by the glycan shield.
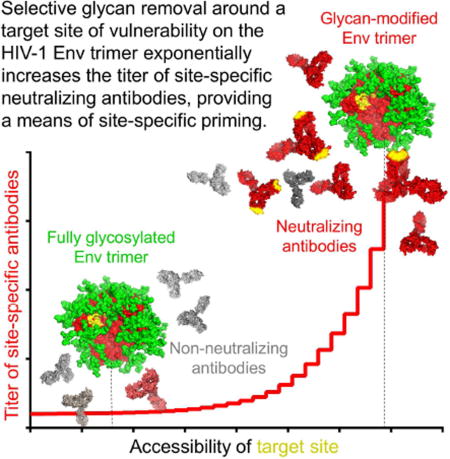
To measure the impact of targeted deglycosylation on the elicitation of antibodies against the CD4 supersite, we used molecular dynamics (MD) simulations to select four N-linked glycans proximal to the CD4 supersite for removal. We determined the structure of a ‘4-glycan-deleted’ Env trimer and engineered multiple additional Env trimers with the same four N-linked glycans removed. We characterized the antigenicity, structures, and glycan content of these engineered trimers, used them to immunize guinea pigs and rhesus macaques, and analyzed week 18 sera for neutralization of wild-type and 4-glycan-deleted viruses, as well as a panel of 3-glycan-deleted viruses. The results reveal N-linked glycosylation proximal to the CD4 supersite to dampen the titers of antibodies targeting the CD4 supersite by more than 1000-fold. We relate the elicited neutralization to a physical property of the immunogens, the antibody-accessible protein surface area. Overall, the increased site-selective immunogenicity of the deglycosylated trimers suggests their potential utility as priming immunogens to increase the polyclonal frequency of site-targeting antibody lineages.
RESULTS
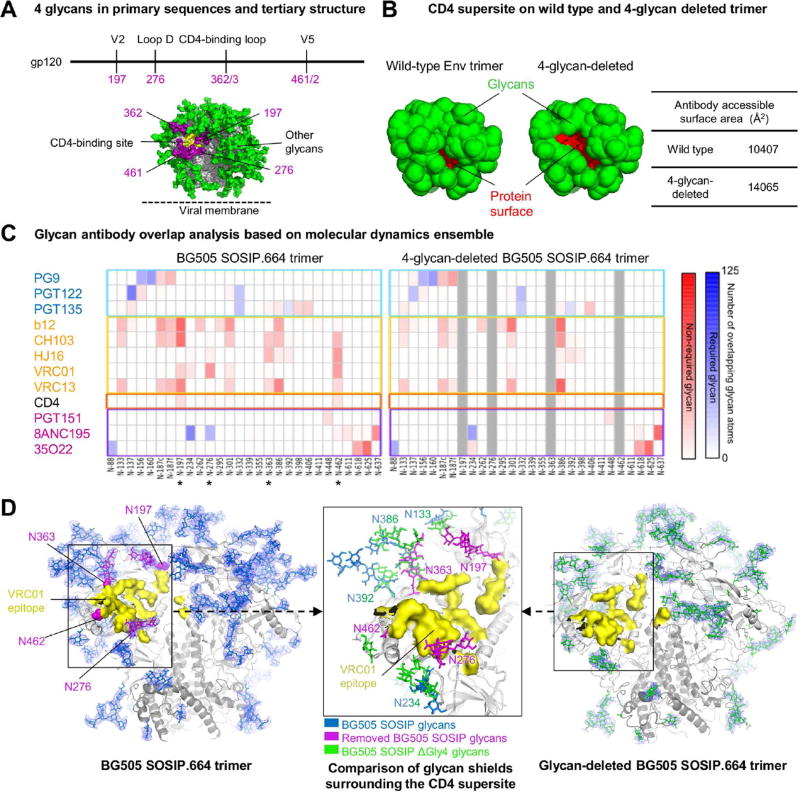
Identification of proximal glycans for targeted removal
MD simulations of fully glycosylated Env trimers coupled to antibody-overlap analysis have provided insight into the impact of the glycan shield on elicitation of neutralizing antibodies (Stewart-Jones et al., 2016). Antibody VRC01 (Wu et al., 2010; Zhou et al., 2010) is a member of a multidonor class that achieves the highest neutralization breadth of CD4 supersite-directed antibodies, with select members neutralizing over 95% of HIV-1 strains (Rudicell et al., 2014; Zhou et al., 2015). In an MD simulation of the BG505 SOSIP.664 Env trimer with all N-linked sites modeled with Man5GlcNAc2, the smallest high mannose N-linked glycan commonly found, N197, N276, N362, and N462 each occluded access of antibody VRC01 to the CD4 supersite (Stewart-Jones et al., 2016); we therefore analyzed the removal of these four N-linked glycans (Figure 1A), and their effect on shielding the protein component of the CD4 supersite (Figure 1B). 500 ns of MD simulation of 4-glycan-deleted BG505 SOSIP confirmed the loss of overlap of glycans with antibody VRC01, except for minor interactions with glycans N302 and N386 (Figure 1C, Table S1A). In general, CD4 supersite-directed antibodies showed less glycan interaction, although some glycans, e.g., N386, displayed increased glycan-antibody overlap in simulations with the 4-glycan-deleted Env (Figure 1C).
Figure 1. Identification of glycans for targeted removal, with molecular dynamics and crystal structure of a 4-glycan-deleted HIV-1 Env trimer.
(A) Location of 4 glycans (purple) proximal to the HIV-1 CD4-binding site (yellow) in primary sequences and tertiary structure.
(B) Antibody accessible- protein surface area on wild type and glycan-deleted BG505 SOSIP based on molecular dynamics simulation of BG505 SOSIP with Man5 glycans. The antibody accessible-protein surface area was calculated using a 10Å probe with Naccess. Protein and glycan surfaces are shown red and green, respectively.
(C) Glycan-antibody overlap analysis derived from a 500 ns molecular dynamics simulations of Man-5- and 4-glycan-deleted models of the HIV-1 glycan shield. The extent of steric overlap with antibody is shown in blue for those glycans that are known to be required for recognition and in red for glycans not known to be required for recognition.
(D) Comparison of crystal structures BG505 SOSIP (left) and 4-glycan-deleted BG505 SOSIP (right) trimer indicates selective removal of 4 glycans (purple) around the VRC01-binding site (yellow) reduces glycan shielding. Center inset shows the superposition of glycan shield around the VRC01-binding site in wild type and 4-glycan-deleted BG505 SOSIP trimer. Ordered glycan residues are shown in stick representation with 2Fo-Fc electron density for glycans shown at 0.8 σ on gp140 trimer glycans and colored in blue and green for wild type and glycan-deleted BG505 SOSIP trimer, respectively.
See also Figure S1 and Table S1.
Structure of a fully glycosylated ‘4-glycan-deleted’ Env trimer from clade A
To provide structural insight, we crystallized a 4-glycan-deleted BG505 N137A SOSIP.664 Env trimer in complex with antibody 35O22 and an antibody from the PGT121-lineage (3H and 109L) (Garces et al., 2015; Pancera et al., 2014). Diffraction data extended to a nominal resolution of 3.65 Å, and refinement indicated ~40% of the N-linked glycan to be ordered (Table S1B, Supplemental Experimental Procedures). Comparison of the N-glycan-deleted structure with the previously determined fully glycosylated BG505 SOSIP.N332 structure (PDB 5FYL) revealed changes in the organization of the glycan shield (Figure S1).
As expected, electron density for the 4-deleted glycans (N197, N276, N363 and N463) was entirely lacking. Glycans proximal to glycan-deleted sequons, for instance N386, which we previously observed to form extensive stem-stem interactions with glycan N363 in the fully glycosylated structure (Stewart-Jones et al., 2016), partially occupied the region of the now-deleted glycan N363 (Figure 1D). More distal differences were also observed, for example, with glycan N160 at the membrane-distal trimer apex: while retaining a similar NAG-stem orientation, N160 in the 4-glycan-deleted structure adopted an alternative oligomannose branch conformation involving interactions with N160 glycans on neighboring protomers, forming a cage over the trimer apex (Figure 1D). Interestingly, the order induced by glycan crowding now correlated with the number of glycans within a 20 Å radius (Figure S1C, D), reduced from 50 Å in the fully-glycosylated Env trimer (Stewart-Jones et al., 2016), perhaps a reflection of decreased long-range order in the shield of the glycan-deleted Env trimers.
We also assessed the conformational impact of glycan deletions on the protein structure; removal of the four CD4-binding-site proximal glycans caused only limited changes in protein structure, most pronounced in loop D. The structure of the 4-glycan-deleted Env trimer thus revealed the deglycosylated CD4 supersite to retain a mostly unaltered protein conformation, with substantially reduced glycan shielding around the CD4 supersite, and altered conformations of retained glycans.
Multiple 4-glycan-deleted Env trimers
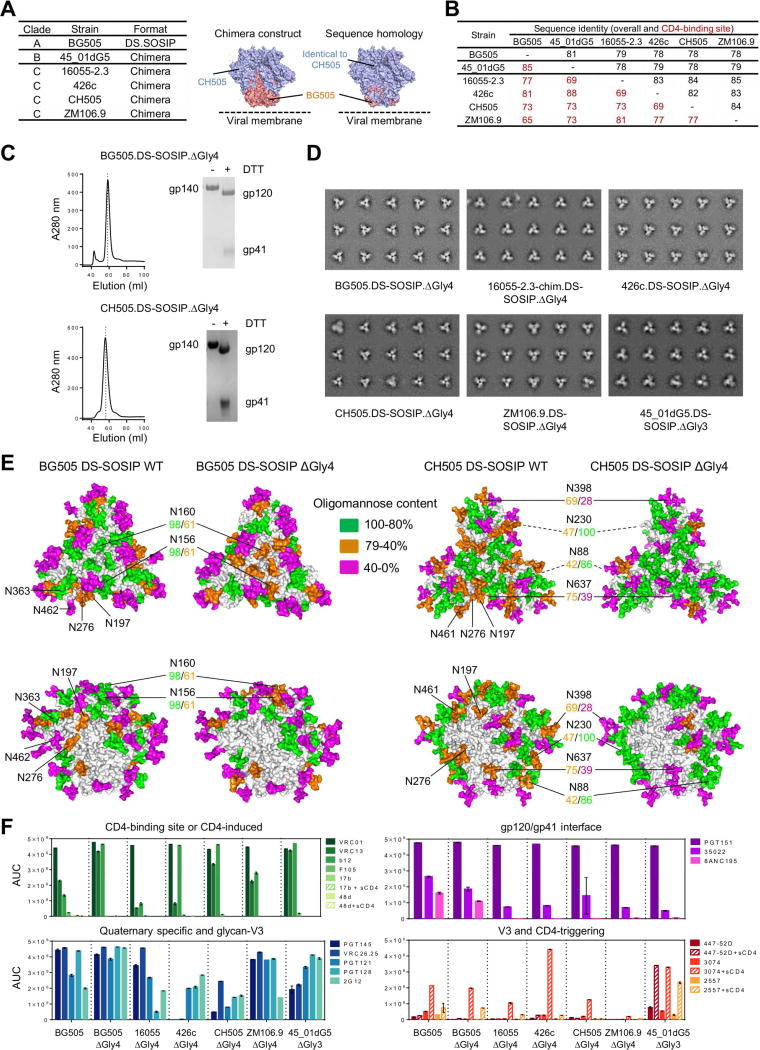
To provide insight into the properties and antigenicity of 4-glycan-deleted trimers, we used a chimeric strategy to create 4-glycan-deleted DS-SOSIP (named DS-SOSIP.ΔGly4) trimers from clade C strains, 16055, 426c, CH505, and ZM106.9 (Figures 2A, 2B). In all four of these clade C strains, the glycan at 362 was naturally absent, so creation of 4-glycan-deleted trimers required removal of only three glycans, at 197, at 276 and from the 460–463 region (Figure S2). We also created a 3-glycan-deleted DS-SOSIP (named DS-SOSIP.ΔGly3) from clade B strain, 45_01dG5, in which we retained the naturally occurring glycan at residue N463. Characteristics of the purified trimers indicated all to be fully cleaved, and negative-stain electron microscopy (EM) indicated the purified trimers to be primarily in the prefusion-closed state (Figures 2C, 2D, Figure S3). Glycan analysis (Behrens et al., 2016; Li et al., 2017) of 4-glycan-deleted BG505 and CH505 DS-SOSIP.ΔGly4 trimers confirmed the targeted glycans to be absent (Table S2). Moreover, removal of these four glycans did affect the processing for several of the remaining glycans, though estimated site occupancies were unchanged (Figure 2E, Table S2).
Figure 2. Construction and characteristics of diverse 4-glycan-deleted HIV-1 Env trimers.
(A) Construction of diverse glycan-deleted HIV-1 Env trimers. Chimeric soluble Env trimers of diverse HIV-1 Clade B and C strains were constructed using the gp41, and gp120 N- and C-termini regions of BG505 DS-SOSIP.664 prefusion stabilized molecule. Molecular surface of a model of the chimeric CH505.DS-SOSIP is shown with structural portions from CH505 and BG505 colored light blue and salmon, respectively (middle), the same model is shown with sequence identical to CH505 colored in light blue and sequence identical to BG505 colored in salmon (right).
(B) Overall and CD4-binding site sequence identity between diverse glycan-deleted HIV-1 Env trimers.
(C) Representative purification profiles of 4-glycan-deleted (ΔGly4) HIV Env DS-SOSIP trimers. Gel filtration and SDS-PAGE for clade A BG505 and chimeric clade C CH505 are shown.
(D) Negative stain EM of the purified glycan-deleted HIV-1 Env trimers. The proteins are highly homogenous and 2D particle class-averages are shown.
(E) BG505 (left) and CH505 (right) DS-SOSIP Man-5 models showing relative proportions of oligomannose versus complex glycosylation at each PNGS. Glycans removed in ΔGly4 proteins are indicated and glycans with >30% change in oligomannose content between wild type and ΔGly4 versions are highlighted between BG505 and CH505 SOSIP paired models and oligomannose proportions are shown and colored according to the key. Concealed glycans are indicated with dashed lines.
(F) Antigenic analysis of purified glycan-deleted HIV-1 Env trimers. Antibodies that target different HIV-1 sites of vulnerability and difference conformational states and the CD4-receptor were used to evaluate the antigenicity and conformational states.
See also Figure S2, S3 and Table S2.
Antigenic analysis of the purified trimers showed that all maintained binding to the cleavage-dependent antibody PGT151. Variable binding was observed with the quaternary conformation-specific antibodies PGT145 and CAP256-VRC26.25, with full binding only retained for BG505 and ZM106 (no binding was observed for strain 426c, the parent strain of which is resistant to neutralization by both of these antibodies). The glycan-deleted trimers showed increased binding to CD4-binding-site-directed antibodies (especially for CH103, b12, and VRC01). Notably, none of the glycan-deleted trimers showed substantial binding to antibodies such as 17b, 447-52d, and F105, which interact with open forms of HIV-1 Env, indicating retention of their prefusion-closed conformation (Figure 2F).
Potent neutralization of glycan-deleted viruses elicited by 4-glycan-deleted Env trimers
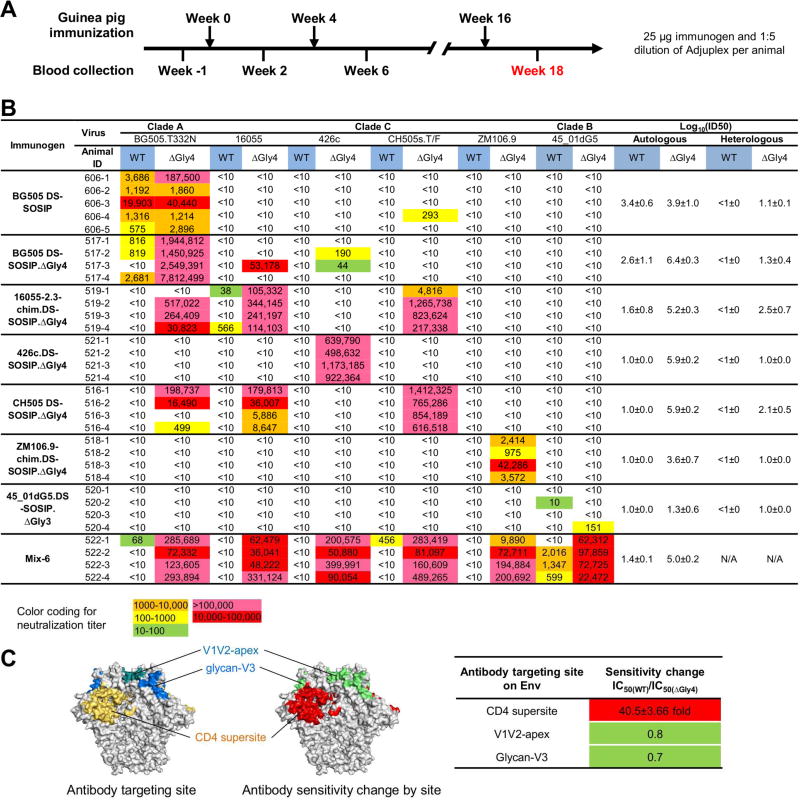
With a panel of 4-glycan-deleted Env trimers characterized, we proceeded to test their immunogenicity. We first needed to determine an appropriate immunization scheme, which we assessed with the fully glycosylated BG505 DS-SOSIP Env trimer, as it elicits autologous neutralizing antibodies (Sanders et al., 2015). We used an immunization schedule of 0, 4, and 16 weeks (Cheng et al., 2016) with alum, Adjuplex, and ISCO Matrix as adjuvants, and observed the highest titer of autologous neutralizing antibodies with Adjuplex (Figure S4). We therefore used Adjuplex and this regimen (Figure 3A) with seven additional experimental groups: five groups comprising each of the 4-glycan-deleted DS-SOSIP.ΔGly4 trimers separately, one group comprising the 3-glycan deleted DS-SOSIP.ΔGly3 trimer from 45_01dG5, and a “Mix-6” group comprising all six glycan-deleted trimers (Figure 3B).
Figure 3. High titers of autologous and heterologous neutralization achieved in guinea pigs.
(A) Immunization scheme for guinea pigs. Week 18 sera were used for neutralization assays.
(B) Neutralization of wild type and 4-glycan-deleted viruses by week 18 sera of guinea pigs immunized with fully glycosylated and glycan-deleted immunogens. Immunogenicity is expressed as log10(ID50) with respect to autologous and heterologous strains for each immunogen. The standard error for individual animals is displayed following the “±” sign. WT, wild type virus; ΔGly4, 4-glycan-deleted virus.
(C) Targeted glycan-removal increases sensitivity of 4-glycan-deleted viruses to antibodies targeting the CD4 supersite.
To gain insight into recognition of the CD4-binding site by elicited sera, we created 4-glycan-deleted Env-pseudoviruses for 23 viruses including each of the parent strains of the vaccine immunogens. Notably, 17 of 23 viruses were not sensitive to antibodies 17b, 447-52d, or F105, which recognize open tier-1 viruses. By contrast, tier-2 viruses are defined by their moderate sensitivity to sera from HIV-1-infected donors (Seaman et al., 2010) and are not sensitive to CD4-induced, V3, or CD4-binding site antibodies that recognize open conformations of Env. By these criteria, the 17 4-glycan-deleted viruses have tier-2-like neutralization character (Table S3A). However, although these viruses did not show increased sensitivity to most HIV-1-neutralizing antibodies, including those against the V1V2-apex or glycan-V3, they did show substantially greater sensitivity to CD4-binding site-directed antibodies (Figure 3C). We then assessed neutralization titers at week 18 using both wild-type and 4-glycan-deleted viruses with tier-2 neutralization characteristics.
Guinea pigs immunized with wild-type BG505 DS-SOSIP showed neutralization against the wild-type BG505 Env-pseudovirus (titers ranging from 575-19,903) (Figure 3B). Against 4-glycan-deleted BG505 Env-pseudovirus, titers increased by less than 3-fold in most of the animals, though in one animal, titers increased by ~50-fold. An absence of heterologous neutralization was observed on the other 5 viruses, with the exception a single animal showing titers against the 4-glycan-deleted CH505 virus (Figure 3B).
Guinea pigs immunized with soluble 4-glycan-deleted DS-SOSIP.ΔGly4 Env trimers showed sporadic low-level autologous neutralization of two wild-type viruses, BG505 and 16055 (Figure 3B, Table S4). When assessed on 4-glycan-deleted viruses, potent neutralization was observed in five of the seven groups (Figure 3B). Notably, autologous virus titers averaged well over 100,000 in these five groups, with select animals exceeding 1,000,000 in three groups. Heterologous titers were also very high in select cases; animals immunized with the clade C 16055 and CH505 DS-SOSIP.ΔGly4 trimers showed neutralization of the clade A BG505 and cross-neutralized each other, with titers averaging over 50,000 (Figure 3B). With animals immunized with the 3-glycan-deleted 45_01dg5 DS-SOSIP.ΔGly3, sera showed moderate titers against tier-1 isolates (Table S3B), but no neutralization against tier-2 strains, and no neutralization of even 4-glycan-deleted viruses, except for only a single animal with the homologous 4-glycan deleted 45_01dg5 (Figure 3B).
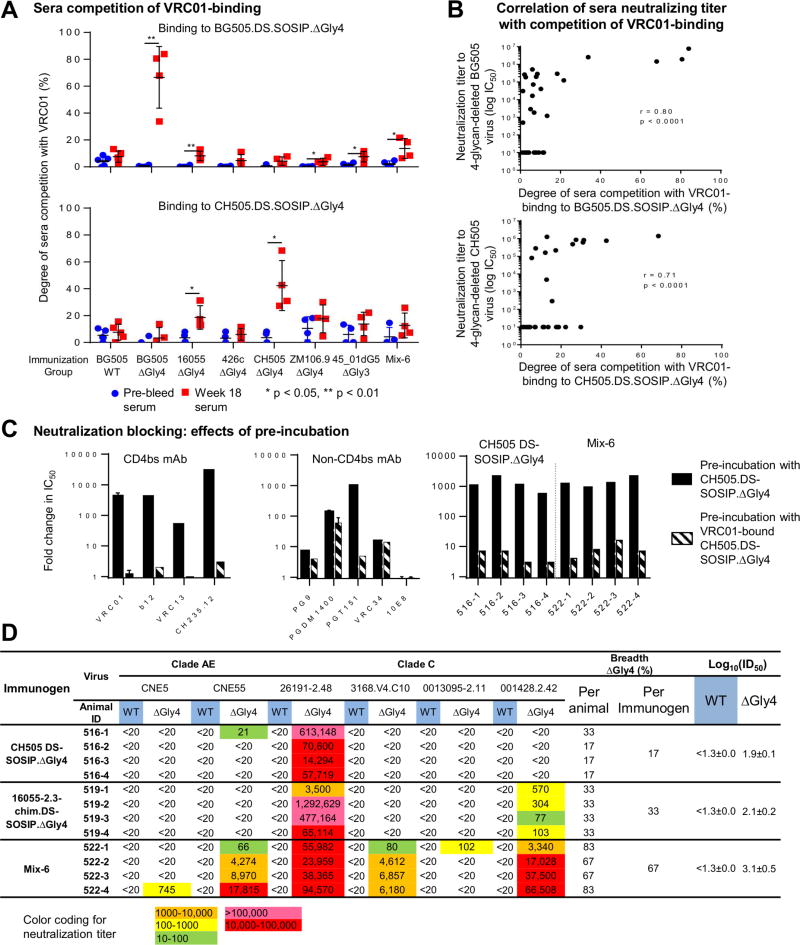
We mapped the guinea pig-serum responses via competition for binding and neutralization. Binding of antibody VRC01 to 4-glycan-deleted BG505 or CH505 DS-SOSIP.ΔGly4 trimers was strongly competed by the trimer-immune sera as compared to naive sera (Figure 4A, Figure S5). The degree of sera competition with VRC01 on binding to 4-glycan-deleted BG505 or CH505 trimers correlated with week 18 sera neutralization titers to the same 4-glycan-deleted virus (Figure 4B). Additionally, neutralization of 4-glycan-deleted CH505 virus was reduced by more than 100-fold by pre-incubating sera with the 4-glycan-deleted CH505.DS-SOSIP.ΔGly4 trimer versus a trimer with VRC01-attached covalently (Figure 4C, Figure S6). Altogether, these data revealed high neutralization titers against glycan-deleted viruses could be elicited by 4-glycan-deleted Env-trimer immunogens and demonstrated this neutralization to be directed almost exclusively at the CD4 supersite.
Figure 4. Mapping of guinea pig immune responses.
(A) Sera competition of VRC01 binding to 4-glycan-deleted BG505.DS-SOSIP and CH505.DS-SOSIP.
(B) Correlation between sera neutralizing titers against 4-glycan-deleted virus and the degrees of sera competition with VRC01 on binding to the same 4-glycan-deleted HIV-1 DS-SOSIP trimer.
(C) Effects of sera pre-incubation with 4-glycan-deleted or VRC01-bound 4-glycan-deleted CH505.DS-SOSIP trimers on neutralization indicated immune responses in sera are directed almost exclusively at the CD4 supersite.
(D) Neutralization of heterologous wild type and 4-glycan-deleted viruses. Immunogenicity is expressed as log10(ID50) with respect to autologous and heterologous strains for each immunogen. The standard error over individual animals is displayed after the “±” sign. The breadth per immunogen is calculated with heterologous neutralization threshold being 50% of animals having an IC50 greater than 20 for each virus. The breadth per immunogen is calculated with heterologous neutralization threshold being 50% of animals having an IC50 greater than 20 for each virus. Note that all of the 4-glycan-deleted viruses assessed here have tier-2-like neutralization character (Table S3A).
See also Figure S5, S6 and Table S3, S4.
Immunogenicity of 4-glycan-deleted Env trimers in rhesus macaques
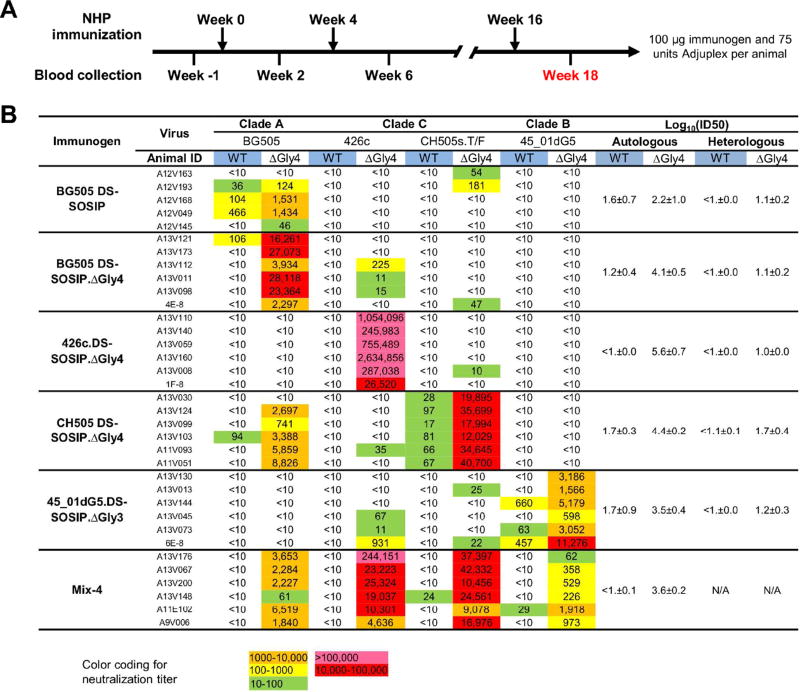
We next assessed Env-trimer immunogenicity in rhesus macaques. We used wild-type glycosylated BG505 DS-SOSIP trimer to compare adjuvants, Adjuplex, Alum, and Matrix M, and observed Adjuplex-elicited titers to be superior (Figure S4). We then used an immunization regimen with Adjuplex comprising 100 µg/injection at weeks 0, 4, and 16 (Figure 5A) with five additional experimental groups: three groups comprising 4-glycan-deleted DS-SOSIP.ΔGly4 Env trimers from strains BG505, 426c, and CH505; one group comprising 3-glycan-deleted DS-SOSIP.ΔGly3 Env trimer from strain 45_01dG5; and a “Mix-4” group comprising all four of these glycan-deleted trimers, and assessed neutralization titers at week 18, against both wild-type and 4-glycan-deleted viruses. Animals immunized with wild-type BG505 SOSIP showed autologous neutralization of BG505 in 3/5 cases, with increased titers in 4/5 cases against the 4-glycan-deleted BG505; heterologous neutralization was not observed except for low level titers against 4-glycan-deleted CH505. Animals immunized with 4-glycan-deleted trimers showed a related pattern of neutralization. On autologous wild-type viruses, glycan-deleted BG505, 45_01dG5 and CH505-trimer vaccinated groups showed low-level or sporadic neutralization (Figure 5B and Table S5). On 4-glycan-deleted viruses, potent neutralization was observed. All groups showed high titer autologous neutralization (two of the 426c–immunized animals showed titers that exceeded 1:1,000,000); substantial heterologous neutralization was observed for the CH505-immunized group and sporadic neutralization in the other groups (Figure 5B and Table S5).
Figure 5. High titers of autologous and heterologous neutralization achieved in rhesus macaque.
(A) Immunization scheme for rhesus macaque. Week 18 sera were used for neutralization assays.
(B) Neutralization of wild type and 4-glycan-deleted viruses by week 18 sera of rhesus macaque immunized with fully glycosylated and glycan-deleted immunogens. Immunogenicity is expressed as log10(ID50) with respect to autologous and heterologous strains for each immunogen. The standard error over individual animals is displayed after the “±” sign.
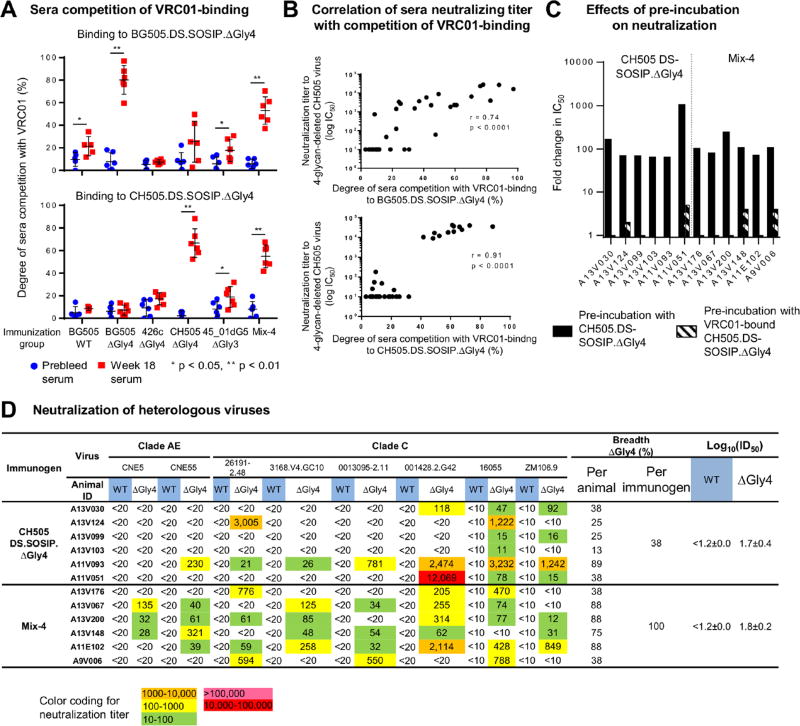
Overall, these results were reflected in ID50 immunogenicity values, with the 4-glycan-deleted CH505 DS-SOSIP.ΔGly4 immunogen yielding the highest heterologous immunogenicity, as assessed by neutralization of both WT and 4-glycan-deleted viruses (Figure 5B, right column). We also mapped these rhesus macaque responses using the same assays as for guinea pig sera. Notably, all data were consistent with the neutralization being directed at the CD4 supersite (Figure 6A, 6B, 6C and Figure S6 and S7).
Figure 6. Mapping of rhesus macaque responses.
(A) Sera competition of VRC01 binding to 4-glycan-deleted BG505.DS-SOSIP and CH505.DS-SOSIP.
(B) Correlation between sera neutralizing titers against 4-glycan-deleted virus and the degrees of sera competition with VRC01 on binding to the same 4-glycan-deleted HIV-1 DS-SOSIP trimer.
(C) Effects of sera pre-incubation with 4-glycan-deleted or VRC01-bound 4-glycan-deleted CH505.DS-SOSIP trimers on neutralization indicated immune responses in sera are directed almost exclusively at the CD4 supersite. Controls are shown in Figure 4B.
(D) Neutralization of heterologous wild type and 4-glycan-deleted viruses. Immunogenicity is expressed as log10(ID50) with respect to autologous and heterologous strains for each immunogen. The standard error over individual animals is displayed after the “±” sign. The breadth per immunogen is calculated with heterologous neutralization threshold being 50% of animals having an IC50 greater than 20 for each virus. Note that all of the 4-glycan-deleted viruses assessed here display tier-2 neutralization character (Table S3A).
See also Figure S6, S7 and Table S3, S5.
Broad heterologous neutralization of 4-glycan-deleted viruses
The immunizations that elicited the greatest breadth against heterologous 4-glycan-deleted viruses involved the CH505 and mixed-trimer immunogen in both guinea pigs and rhesus, as well as the 16055 immunogen in guinea pigs. We therefore tested the sera from these groups on a panel of six additional heterologous isolates that were not included in any of the immunogens and which retained tier-2-like neutralization character, even with 4-glycan-deleted from the CD4 supersite (Table S3a); with the rhesus macaque sera, we also tested 16055 and ZM106 viruses.
With the guinea pig immune sera, no neutralization of wild-type viruses was observed. However, substantial neutralization was observed against select 4-glycan-deleted viruses (Figure 4D and Table S4). Neutralization was broadest for the “Mix-6” group, with extensive neutralization observed in 4 of the 6 heterologous viruses, with titers exceeding 10,000 on 3 of the viruses. CH505 and 16055 immunogens also elicited high titers against 1 or 2 of the 6 heterologous viruses, respectively. This neutralization did not correlate with similarity to immunized strain, as the 4-glycan-deleted virus from clade AE strain CNE55 was neutralized well by clade C immunogens, whereas some clade C viruses showed no neutralization.
With the rhesus macaque immune sera, no neutralization of wild-type viruses was observed. However, on 4-glycan-deleted viruses, extensive neutralization was observed for all 8 heterologous viruses with the Mix-4-immune sera (Figure 6D and Table S5). The CH505-immune sera showed sporadic neutralization against 7 of 8 viruses, with sera from animal A11B093 neutralizing 7 of 8 heterologous viruses. Consistent with the data for autologous viruses, titers were generally lower in rhesus macaques than in guinea pigs; however, the breadth of the neutralization by the sera from the Mix-4-immunized rhesus macaques did appear to be substantial on 4-glycan-deleted viruses, with at least half of the animals showing neutralization of all 8 viruses (Figure 6D and Table S5). Thus, while neutralization breadth could not be assessed with wild-type viruses, the enhanced CD4 supersite sensitivity of the four-glycan-deleted viruses enabled the assessment of neutralization breadth as well as the ranking of immunogens, with the Mix-4 and Mix-6 groups yielding the greatest breadth.
Quantification of site-by-site impact of the glycan shield
To obtain greater insight into the impact of individual glycans, we created a panel of 3-glycan-deleted viruses, in which a single glycan site corresponding to N197, N276, N363 or N462 was individually restored. In total, the panel comprised 19 3-glycan-deleted viruses. We used this panel to assess the impact of adding a single N-glycan on the neutralization from the guinea pig immunized sera (Figure 7A and Table S3 and S6) and rhesus macaques immunized sera (Table S7). Results varied considerably by immunogen and by immunized animal. For example, with the four guinea pigs immunized with 4-glycan-deleted BG505 DS-SOSIP.ΔGly4 trimers, we observed only moderate effects by adding glycans N197 or N363. However, in three of the animals, substantial reduction in neutralization was observed with N462 added, and in the fourth animal a substantial reduction in neutralization was observed with N276 but not with N462. These results indicate a dominant response in each of the animals, which could be substantially reduced by the addition of a single glycan.
Figure 7. Mapping of guinea pig immune responses with 3-glycan-deleted viruses.
(A) Mapping immune responses by neutralization of wild type, 4-glycan-deleted and 3-glycan-deleted viruses that restored each of the individual glycans from the 4-glycan-deleted viruses (marked with “+” and the glycan position).
(B) Visualization of elicited antibody responses against BG505 wild type, 4-glycan-deleted and 3-glycan-deleted viruses for the guinea pig group immunized with 4-glycan-deleted BG505.DS-SOSIP. Antibody accessible - protein surface areas at different glycan-deleted states are shown along with antigenicity to VRC01-like antibodies and immunogenicity are colored as indicated.
(C) Pearson correlation of mean antibody accessible-protein surface area extracted from Man5 glycosylated BG505 SOSIP molecular dynamics simulation (calculated using a 10 Å probe by Naccess) with BG505-DS SOSIP ΔGly4 immunogenicity (log(ID50)), for WT, ΔGly4, and ΔGly3 BG505 viruses.
(D) Pearson correlations between antibody accessible-protein surface area and immunogenicity (log(ID50) for WT, ΔGly4, and ΔGly3 viruses, left to right in the order of 16055-2.3-chim.DS-SOSIP ΔGly4, 426c.DS-SOSIP ΔGly4, CH505 DS-SOSIP ΔGly4, and ZM106.9-chim.DS-SOSIP ΔGly4. Accessible surface areas were calculated based on Man5-glycosylated homology models with a 10Å probe using Naccess.
We quantified the impact of the single glycan additions on antigenicity, on immunogenicity, and on the calculated protein surface-area accessible to a probe with 10 Å radius, the estimated radius of a protruding antibody loop (Figure 7B). Notably, the amount of protein accessible-surface area correlated with autologous immunogenicity. In specific, glycan 276 which had a large effect on neutralization is situated not only next to the CD4 supersite, but also proximal to a glycan-free surface: its addition (or absence) thus alters access to not only the CD4 supersite, but also to this glycan-free surface. Glycan 462, which is proximal to glycan 276, appeared to share in the impact of glycan 276. Glycan 197 is peripheral to the accessible protein surface, and glycan 363 even more peripheral. Notably for BG505 the mean antibody accessible-protein surface area from molecular dynamics simulation (calculated with a 10 Å probe radius and modeled with Man5 glycans) correlated with both guinea pig (r=0.986, p=0.0003) (Figure 7C) and rhesus macaque immunogenicity (r=0.925, p=0.0082) (Figure S7C). Similar trends were also observed for other strains with antibody accessible-protein surface area calculated based on homology models (Figure 7D and Figure S7D). Altogether, these results quantify the site-by-site impact of N-linked glycosylation on CD4 supersite immunogenicity, and revealed direct correlation between immunogenicity and antibody accessible-protein surface.
DISCUSSION
The HIV-1 glycan shield covers the entire accessible surface of the prefusion-closed HIV-1 Env trimer, leaving little protein-surface accessible for antibody recognition (Gristick et al., 2016; Lee et al., 2016; Stewart-Jones et al., 2016). With HIV-1 strain BG505, the naturally missing glycans at N241 and N289 form a double-glycan hole, which has recently been reported to comprise the dominant site of autologous neutralizing antibodies elicited by vaccination (McCoy et al., 2016). With HIV-1 strain JR-FL, the naturally missing glycan at N197 forms a glycan hole targeted by vaccine-elicited antibodies capable of neutralizing Tier 2 strains with N197 deleted (Crooks et al., 2015). Analysis of antibodies from donor CAP257 indicate recognition by antibody CAP257-RH1, which recognizes a rare V5-glycan hole adjacent to the CD4-binding site (Wibmer et al., 2016). And immunization with a SHIV naturally missing glycan N160 elicits antibodies against the V1V2-apex, which neutralize tier 2 strains with N160 deleted (Robinson et al., 2010; Wu et al., 2011). Thus, HIV-1 Env-glycan shielding differs from other type I fusion machines on viruses such as influenza and RSV, where glycan shielding on the prefusion trimers are incomplete leaving substantial areas of protein-surface exposed (McLellan et al., 2013; Wilson et al., 1981). Indeed, broadly neutralizing antibodies against influenza hemagglutinin or RSV fusion glycoprotein generally recognize only protein, whereas known broadly neutralizing antibodies against HIV-1 must accommodate glycan (Pancera et al., 2014; Stewart-Jones et al., 2016).
Despite its central importance, the quantitative impact of the glycan shield on antibody elicitation had not previously been quantified. Here we measure the impact of the glycan shield on immunogenicity (Figures 3–6), and observe an exponential relationship between immunogenicity (as measured by ID50) and protein-surface accessible to a penetrating antibody loop (as measured in Å2) (Figure 7). This observed exponential relationship likely extends over only a limited interval, ranging from a fully shielded Env to an exposed Env-protein sufficient to allow unimpeded antibody access. Above this interval, the relationship between immunogenicity and exposed protein surface is likely to lose its exponential character.
Enhancement of Env immunogenicity through glycan removal has been attempted previously. For example, Wyatt and colleagues removed glycans proximal to the CD4 binding site (Koch et al., 2003); however, this experiment was performed in the context of monomeric gp120, where the glycan shield is not assembled. Bradley et al. reported induction of a macaque clonal lineage of CD4 binding site autologous Tier 2 neutralizing antibodies that neutralized only autologous HIV-1 with a glycan hole in V5 at residue 463 (Bradley et al., 2016). Schief, Stamatatos and colleagues observed increased germline priming with a B cell ontogeny-based approach through selective glycan removal (Jardine et al., 2013; McGuire et al., 2016; McGuire et al., 2013); while deglycosylation improved priming, this effect was observed specifically for VRC01-class antibodies, which are known to penetrate the glycan shield, and the effect of deglycosylation in the context of the entire antibody repertoire was not assessed. Here we demonstrate that creating a hole in the Env-glycan shield large enough to allow unimpeded access of antibody to the Env-protein surface induces very high titers of neutralizing antibody. These high neutralization titers were observed with glycan-deleted viruses, but not with wild-type virus; indeed, the increased sensitivity of the 4-glycan-deleted viruses, which we demonstrate to be 10–100-fold more sensitive to neutralization at the CD4 supersite, may provide a tool for detection of responses that would otherwise be too weak to observe. Overall, our results indicate deglycosylation to improve priming in a lineage-independent (polyclonal) manner, suggesting that glycan-deleted immunogens could be used for priming to increase the frequency of desired immune precursors. Such site-specific priming may be especially suited for glycan-free sites of vulnerability, such as the CD4-binding site.
Can this CD4 supersite-directed neutralization be extended to fully glycosylated viruses? Studies of B cell ontogeny show the ability of antibody lineages to evolve tolerance of glycan – by avoidance through shifts in antibody orientation (e.g., antibody PGT121 with glycan N137) (Garces et al., 2015) or by engagement through direct interaction (e.g., antibody VRC01 with glycan N276) (McGuire et al., 2013; Stewart-Jones et al., 2016). One epitope-based strategy to achieve neutralization of wild type viruses would involve first identifying sites of protein vulnerability (as we have done here with the CD4 binding site), inducing high titers of antibody against this site through immunization with selectively deglycosylated Env immunogens, and boosting with Env immunogens with removed glycans restored. The site-by-site assessment of the glycan shield reported here quantifies the link between immunogenicity and exposed protein surface, a case-specific correlation, which nevertheless identifies a physical property that can be manipulated to enhance immunogenicity. More generally, targeted deglycosylation of prefusion-closed Env trimers appears to provide an approach to selectively enhance immunogenicity. Such increased immunogenicity can be utilized in epitope-based strategies as well as B cell ontogeny-based strategies and highlights the use of site-specific deglycosylation as a tool for HIV-1-vaccine design.
EXPERIMENTAL PROCEDURES
Molecular Dynamics Simulations and Glycan-Antibody Overlap Analysis
All molecular dynamics simulations with glycan-deleted BG505.SOSIP Env and glycan overlap analysis were performed as described in the Supplemental Experimental Procedures.
Crystallization of Glycan-deleted HIV-1 BG505.SOSIP Env
Expression and purification of glycan-deleted HIV-1 BG505.SOSIP gp140 were performed as described previously (Stewart-Jones et al., 2016). Protein complex of HIV-1 trimer with antibody Fabs of 109L+3H and 35O22 was formed and purified as described in the Supplemental Experimental Procedures.
X-ray Data Collection and Structure Refinement
Structure solution and refinement were carried out with protocols as described in the Supplemental Experimental Procedures.
Construction and Purification of Glycan-deleted DS-SOSIP Immunogens
Glycan-deleted HIV-1 clade B and C DS-SOSIP trimer were made in chimeric format utilizing gp41, N- and C-termini of BG505. DS-SOSIP. Selected glycan sites were removed by mutagenesis. Expression, purification and negative-selection by V3-directed antibodies were performed as described previously and in the Supplemental Experimental Procedures.
Antigenic Characterization of Immunogens
Antigenic characteristics of purified HIV-1 glycan-deleted DS-SOSIP trimers were analyzed with antibodies targeting different HIV-1 sites of vulnerability and conformational states using Meso Scale Discovery technology as described in the Supplemental Experimental Procedures.
Negative-stain Electron Microscopy
Images of glycan-deleted HIV-1 Env collected as described in the Supplemental Experimental Procedures.
Animal Protocols and Immunization
For immunization studies, animals were housed and cared for in accordance with federal and NIH Office of Animal Care and Use (OACU) policies in an AAALACi-accredited facility at the VRC, NIAID, NIH or at a contract facility associated with the VRC (Bioqual Inc, MD). All animal experiments were reviewed and approved by the Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH. Immunization of animals was performed as described in the Supplemental Experimental Procedures.
Serum Competition of VRC01-binding to HIV-1 Env
Antibody specificity of serum from immunized animals were assessed for their ability to compete with biotinylated antibody VRC01 binding to HIV-1 Env trimers as described in the Supplemental Experimental Procedures.
Neutralization Assays
Wild type, 3-glycan-deleted and 4-glycan-deleted pseudoviruses were prepared, tittered, and used to infect TZM-bl target cells as described in the Supplemental Experimental Procedures.
Statistical Analyses
To calculate statistical significance and correlation, t-test and two-tailed Pearson’s correlation were used. Error bars indicate SD. GraphPad Prism was used for statistical analysis.
Supplementary Material
Highlights.
Engineered diverse prefusion-closed selectively deglycosylated HIV-Env trimers
Exponential relationship between exposed protein surface and Env immunogenicity
Broad and potent neutralization by vaccine-elicited sera of glycan-deleted viruses
Site-specific deglycosylated trimers to prime in a lineage-independent manner
Acknowledgments
We thank Bette Korber and Kshitij Wagh for assistance with HIV-1 sequence database, Jonathan Stuckey for assistance with figures, and members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions and comments on the manuscript. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panel. We thank Dennis Burton and Mark Feinberg for PGT121-lineage antibody 3H-109L and Mark Connors for antibody 35O22 used in structural analysis. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, by the International AIDS Vaccine Initiative’s (IAVI’s) Neutralizing Antibody Consortium, by NIH NIAID Grants by P01 AI094419 (to L.Stamatatos) and UM-1 AI100645, Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (to B.F.H), and in part by the National Center for Biomedical Glycomics P41GM103490 (to L.W.). Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Coordinates and structure factors for the 4-glycan-deleterd structure have been deposited with the Protein Data Bank under accession codes: 5V7J.
Supplemental Information includes Extended Experimental Procedures, 7 figures, and 6 tables and can be found with this article online at http://dx.doi.org/xxx.
AUTHOR CONTRIBUTIONS
T.Z. and P.D.K. conceived, designed and coordinated the study; T.Z., N.A.D.-R., C.C, G.B.E.S.-J, C.S., G-Y.C., L.Shapiro, J.R.M. and P.D.K. generated figures and wrote and revised the manuscript; J.R.M. provided intellectual expertise and guidance. Design and purification of glycan-deleted HIV-1 DS-SOSIP immunogens: T.Z., H.G., Y.D.K., M.S., G.B.E.S.-J, K.X., L.C., M.P., B.Z., A.Z. with assistance from I.S.G and M.G.J.; protein expression: A.D.; crystal structure: G.B.E.S.-J.; molecular dynamics: C.S., T.L. and L.Shapiro; negative stain EM: Y.T.; antigenic assays: M.C., S.N., and A.B.M.; glycan analysis: P.Z., M.T. and L.W.; animal protocol: K.F., J.-P.T. and D.S. with C.C. coordinating; ELISA: R.E.C., T.G.W. and C.C.; virus panel and neutralization: K.M., S.O., S.R.S., M.K.L. and T.Z. with N.A.D.-R. coordinating; Bioinformatics and immune correlation: G.-Y.C., K.F., R.R. and C.-H.S.; HIV CH505 and 426c constructs: B.F.H. and L.Stamatatos.
References
- Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, Krumm SA, Struwe WB, Cupo A, Kumar A, Zitzmann N, et al. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley T, Fera D, Bhiman J, Eslamizar L, Lu X, Anasti K, Zhang R, Sutherland LL, Scearce RM, Bowman CM, et al. Structural Constraints of Vaccine-Induced Tier-2 Autologous HIV Neutralizing Antibodies Targeting the Receptor-Binding Site. Cell Rep. 2016;14:43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, Druz A, Narpala S, Doria-Rose NA, McDermott AB, et al. Immunogenicity of a Prefusion HIV-1 Envelope Trimer in Complex with a Quaternary-Structure-Specific Antibody. Journal of virology. 2016;90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, et al. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS pathogens. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Gristick HB, von Boehmer L, West AP, Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nature structural & molecular biology. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313:387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nature structural & molecular biology. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Grant OC, Ito K, Wallace A, Wang S, Zhao P, Wells L, Lu S, Woods RJ, Sharp JS. Structural Analysis of the Glycosylated Intact HIV-1 gp120-b12 Antibody Complex Using Hydroxyl Radical Protein Footprinting. Biochemistry. 2017;56:957–970. doi: 10.1021/acs.biochem.6b00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, et al. Holes in the Glycan Shield of the Native HIV Envelope Are a Target of Trimer-Elicited Neutralizing Antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Gray MD, Dosenovic P, Gitlin AD, Freund NT, Petersen J, Correnti C, Johnsen W, Kegel R, Stuart AB, et al. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat Commun. 2016;7:10618. doi: 10.1038/ncomms10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. The Journal of experimental medicine. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JE, Franco K, Elliott DH, Maher MJ, Reyna A, Montefiori DC, Zolla-Pazner S, Gorny MK, Kraft Z, Stamatatos L. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. Journal of virology. 2010;84:3443–3453. doi: 10.1128/JVI.02617-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. Journal of virology. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for neutralizing antibody assessment. Journal of virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC, et al. IDENTIFICATION AND CHARACTERIZATION OF CONSERVED AND VARIABLE REGIONS IN THE ENVELOPE GENE OF HTLV-III LAV, THE RETROVIRUS OF AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wibmer CK, Gorman J, Anthony CS, Mkhize NN, Druz A, York T, Schmidt SD, Labuschagne P, Louder MK, Bailer RT, et al. Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site. Journal of virology. 2016;90:10220–10235. doi: 10.1128/JVI.01357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Wu X, Changela A, O’Dell S, Schmidt SD, Pancera M, Yang Y, Zhang B, Gorny MK, Phogat S, Robinson JE, et al. Immunotypes of a quaternary site of HIV-1 vulnerability and their recognition by antibodies. Journal of virology. 2011;85:4578–4585. doi: 10.1128/JVI.02585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid J, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.