Abstract
The long noncoding RNAs (lncRNAs), which constitute a large portion of the transcriptome, have gained intense research interest because of their roles in regulating physiological and pathophysiological functions in the cell. We identified from RNA-Seq profiling a set of lncRNAs in cultured human umbilical vein endothelial cells (HUVECs) that are differentially regulated by atheroprotective vs. atheroprone shear flows. Among the comprehensively annotated lncRNAs, including both known and novel transcripts, LINC00341 is one of the most abundant lncRNAs in endothelial cells. Moreover, its expression level is enhanced by atheroprotective pulsatile shear flow and atorvastatin. Overexpression of LINC00341 suppresses the expression of vascular cell adhesion molecule 1 (VCAM1) and the adhesion of monocytes induced by atheroprone flow and tumor necrosis factor-alpha. Underlying this anti-inflammatory role, LINC00341 guides enhancer of zest homolog 2, a core histone methyltransferase of polycomb repressive complex 2, to the promoter region of the VCAM1 gene to suppress VCAM1. Network analysis reveals that the key signaling pathways (e.g., Rho and PI3K/AKT) are co-regulated with LINC00341 in endothelial cells in response to pulsatile shear. Together, these findings suggest that LINC00341, as an example of lncRNAs, plays important roles in modulating endothelial function in health and disease.
Keywords: shear stress, long noncoding RNA, LINC00341, EZH2, VCAM1
with the emergence of RNA-Seq, the transcriptomics approach has become commonly used to decipher the roles of RNAs in a variety of physiological and pathophysiological conditions such as stem cell development (4, 5, 11, 15), cancer progression (13, 20), and cardiovascular diseases (1, 19, 22). Unlike microRNAs that act mostly on messenger RNA through seed region complementation, long noncoding RNAs (lncRNAs) have both sequence-dependent and -independent actions. Their actions may involve a variety of RNA-binding proteins, thus making their cellular functions extremely diverse and challenging to predict and identify.
Several lncRNAs have been found to be associated with vascular endothelial cell (EC) functions. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is an abundant nuclear lncRNA in ECs that regulates EC functions. Inhibition of MALAT1 suppresses HUVEC proliferation and promotes their migration, and reduces vascular growth in mice (17). Ablation of MALAT1 mitigates retinopathy by attenuating retina endothelial inflammation in diabetic mice (12). These studies suggest that lncRNAs play critical roles in regulating vascular homeostasis. However, the mechanisms by which lncRNA regulates relevant gene expression remain unclear. Several trans-acting mechanisms for lncRNAs have been proposed. In cancer cells, lncRNA HOTAIR has been reported to be incorporated into and guides polycomb repressive complex 2 (PRC2) to the promoter region of its targets such as Jam2, Pcdh10, and Pcdhb5, where the PRC2 core methyltransferase EZH2 methylates the Lysine 27 of histone H3 to repress these genes (7). However, there is a paucity of information on the roles of lncRNAs in the shear-flow and pharmacological regulations of ECs in health and disease. The limited documentation of lncRNA-protein interactions in public databases greatly hinders the analysis to identify the molecular mechanisms of physiological regulations. A feasible approach is through bioinformatics to explore the co-expressions of lncRNA and protein-coding genes, thus identifying the potential pathways in which lncRNA are involved.
In this study, following the identification of a set of lncRNAs in HUVECs that are differentially regulated by distinct flow patterns, we investigated the functional role of LINC00341 as an example of lncRNAs induced by atheroprotective flow and atorvastatin. We have demonstrated that LINC00341 guides the repressive complex PRC2 to the VCAM1 promoter to decrease VCAM1 expression and inhibit monocyte adhesion. Thus, LINC00341 serves as an anti-inflammatory mediator in ECs.
MATERIALS AND METHODS
Cell culture and shear stress experiment.
Human umbilical vein ECs (HUVECs) were maintained in medium M199 supplemented with 10% Endothelial Cell Growth Medium (Cell Application), 10% FBS, 1% sodium pyruvate, 1% L-glutamine, and 1% penicillin-streptomycin, and used within passages 5–8. To apply mechanical shear forces on HUVECs, an in vitro circulating flow chamber system (24) was used to impose fluid shear stress to the cultured ECs. The HUVECs cultured on collagen I-coated slides were exposed to pulsatile shear (PS; 12 ± 4 dyn/cm2, 1 Hz) or oscillatory shear (OS; 0.5 ± 4 dyn/cm2, 1 Hz), or kept as static control.
Monocyte adhesion assay.
THP-1 cells were maintained in RPMI-1640 medium containing 10% FBS. To assess the binding of THP-1 cells to ECs, the THP-1 cells were labeled with CellTracker Green CMFDA Dye (Thermo Fisher) and then incubated with the ECs for 15 min. The unattached THP-1 cells were washed away with endothelial growth medium and the number of attached cells was counted under fluorescence microscopy.
RNA-Seq analysis.
Total RNA was extracted with TRIzol (Thermo Fisher) and profiled on the Illumina Genome Analyzer IIx and HiSeq platform. The resulting fastq reads were checked for sequencing quality, and high-quality reads were mapped to human transcriptome (Gencode v19) with Tophat2 aligner. Novel lncRNAs were identified with Cufflinks suites and merged with known annotations in Gencode v19 to form a comprehensive set of lncRNA annotations. Determination of gene expression levels, normalization, and statistical differential expression analysis were conducted with HTseq (2) and DESeq2 (14). Raw fastq reads in this study are available at the Gene Expression Omnibus database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92506).
For LINC00341 coexpression gene network analysis, the Pearson’s correlation of the RNA expression levels of each gene (normalized by DESeq2) to LINC00341 was calculated. Genes with significantly positively correlation (Pearson’s correlation coefficient > 0.75, adjusted P value < 0.001) with LINC00341 were selected and subjected to functional enrichment analysis with Cytoscape ReactomeFIViz plugin (26).
Crosslink RNA-IP.
The HUVECs were treated with 0.3% formaldehyde to preserve protein-RNA and protein-chromatin interactions, and the nuclei were isolated with Nuclei isolation buffer (1.28 M sucrose, 40 mM Tris·HCl pH 7.5, 20 mM MgCl2, 4% Triton X-100). The nuclei were sheared with Bioruptor and mixed with EZH2 antibody (Abcam ab3749) to recognize PRC2 protein complex. The immunocomplex was isolated with pierce protein A/G magnetic beads and washed six times with RIP buffer (150 mM NaCl, 25 mM Tris pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, fresh 1× proteinase, and RNase inhibitors). The immunoprecipitates were decross-linked for 4 h at 55°C followed by proteinase K treatment. The RNA and chromatin were collected with Trizol and alcohol precipitation, respectively. The collected RNA was reverse transcribed to cDNA with M-MLV Reverse Transcriptase (Thermo Fisher). The resulting cDNA and chromatin DNA were subjected to quantitative (q)PCR for the detection of LINC00341 and VCAM1 promoter.
cDNA expression plasmids, siRNAs, real-time qPCR, and antibodies.
LINC00341 expression plasmid was constructed by restriction cloning of full-length LINC00341 sequence into pcDNA3.1 plasmid. LNA siRNA against LINC00341 and Negative Control A were purchased from Exiqon. qPCR were performed on the CFX Connect Real-Time PCR Detection System (Bio-Rad) with iTaq Universal SYBR Green Supermix (Bio-Rad). Primer sequences used in this study are listed in Supplementary Table S1. (The online version of this article contains supplemental material.) Antibodies against GAPDH (#2118) and VCAM1 (#12367) were purchased from Cell Signaling and used in Western blot analysis.
Promoter activity assay.
Putative promoter regions (−2,500 to 1 bp of the transcription start site) of the VCAM1 gene were cloned into the multiple cloning sites of pGL4.20 plasmid (Promega). The reporter plasmids and LNA oligos were transfected into HEK293T cells with Lipofectamine RNAiMAX (Thermo Fisher). The luciferase activities were measured with a Dual-Luciferase Reporter Assay System (Promega), following the manufacturer’s instruction.
RESULTS
Flow regulation of lncRNAs in HUVECs.
To identify shear stress-regulated lncRNAs, we first performed RNA-Seq with HUVECs subjected to different shear flow conditions, including atheroprotective PS or atheroprone OS for four time periods (1, 4, 12, and 24 h). The high-throughput sequencing data were analyzed following the pipelines illustrated in Supplementary Fig. S1.
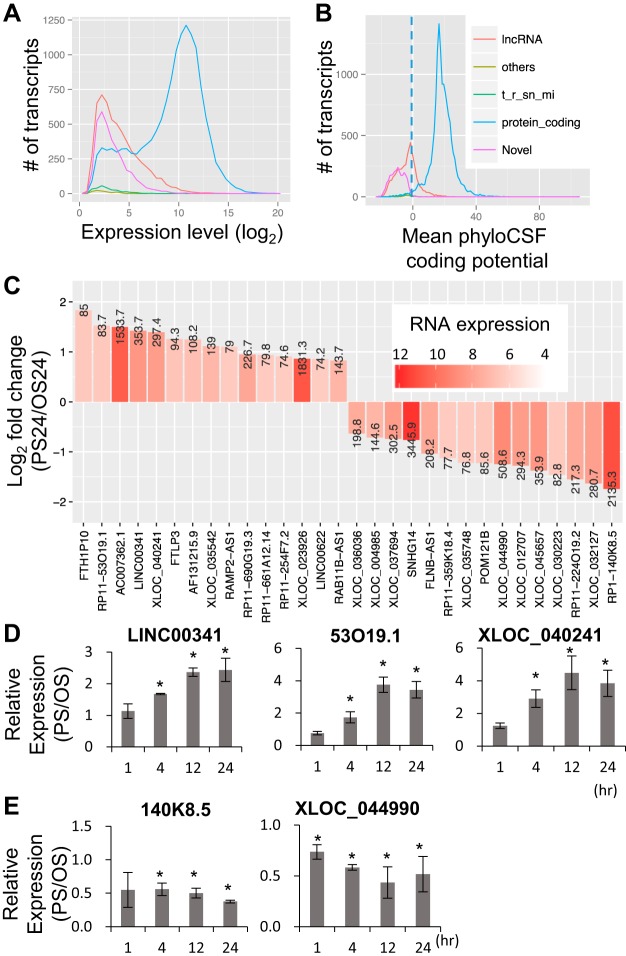
We interrogated the RNA-Seq data for novel RNA transcripts that have not yet been documented in public database (Ensembl GRCh37.p13). To reduce the potential false-positive results, the poorly expressed loci (DESeq2 normalized mean RNA expression level < 50) were excluded; this resulted in 3,368 novel transcripts. In particular, the top 100 novel transcripts expressed are listed in Supplementary Table S2. It is notable that the majority of these newly identified transcripts exhibited the characteristics of lncRNAs, such as relatively low abundance (Fig. 1A) and negative coding potential (Fig. 1B). Reads of the novel transcripts XLOC_040241 and XLOC_044990 were verified in an existing data set GSE71164 (16) (Supplementary Fig. S2), demonstrating the confidence of the novel transcript discovery from our RNA-Seq experiments.
Fig. 1.
Differential expression of long noncoding (lnc)RNAs in pulsatile shear (PS)-treated endothelial cells (ECs). The abundance level (A) and protein-coding potentials (B) of the expressed transcripts. Number of expressed transcripts in different categories are summarized based on their overall expression levels and coding potentials. C: top 15 differentially regulated lncRNAs in ECs exposed to PS or oscillatory shear (OS) for 24 h. The bars represent the log2 fold changes, while the color gradients represent the RNA expression level. RT-quantitative (q)PCR analysis of the lncRNA level in ECs confirming that these lncRNA are increased (D) or repressed (E) by PS, when compared with those in OS groups. Bars represent means ± SE. *P < 0.05, PS compared with OS, Student’s t-test, n = 3.
It has been established that PS maintains EC homeostasis, whereas OS impairs EC function (8). To investigate the functional roles of shear stress-regulated lncRNAs in ECs, we focused on the lncRNAs that increased progressively over the duration of shearing. The top 15 lncRNAs differentially regulated at PS 24 h are listed in Fig. 1C. Among these, five of them were confirmed by RT-qPCR (Fig. 1, D and E). Among these verified lncRNAs, LINC00341 was abundant and significantly upregulated by PS, with a suitable size (2.5 kb) for cloning. Hence, we proceeded to study its functional role in ECs.
LINC00341 suppresses VCAM1 expression and inhibits monocyte adhesion.
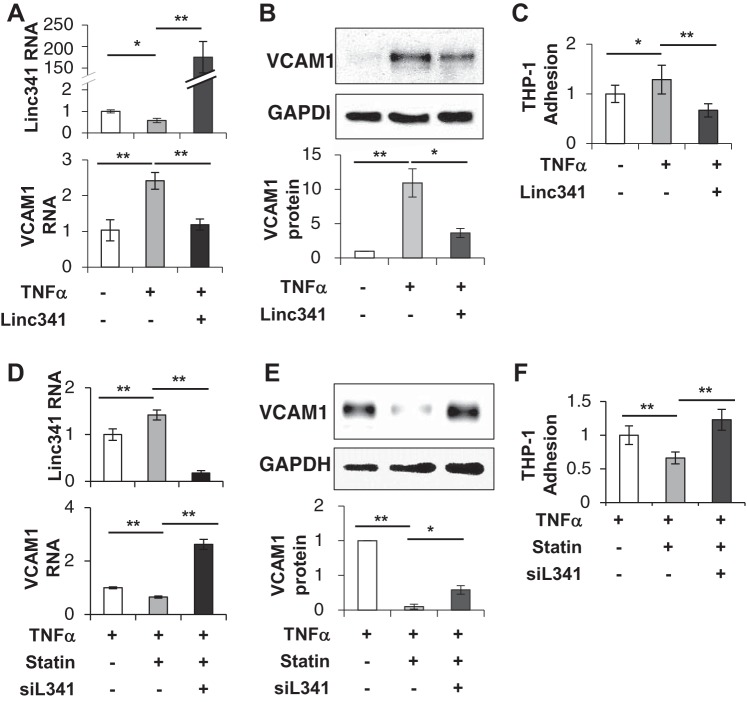
Because atheroprotective PS is known to maintain EC homeostasis, in part through its anti-inflammatory effect, we assessed the role of LINC00341 in flow-mediated endothelial inflammation. HUVECs were transfected with pcDNA3.1-LINC00341 overexpression plasmid or siLINC00341 LNA and then evaluated for monocyte adhesion and VCAM1 expression. As shown in Fig. 2, elevated LINC00341 (Fig. 2A) in HUVECs suppressed the OS-induced THP-1 monocyte adhesion (Fig. 2B). The RNA and protein levels of endothelial inflammation marker gene VCAM1 (Fig. 2, C and D) show concordant expression pattern to the THP-1 adhesion capability of HUVECs. However, knocking down LINC00341 in PS-treated HUVECs by LNA gapmer did not elevate the expression level of VCAM1, nor did altering the THP-1 adhesion (Supplementary Fig. S3). These results indicate that LINC00341 has an anti-inflammatory function in ECs, but the inhibition of LINC00341 per se is insufficient to activate ECs in the quiescent state.
Fig. 2.
Overexpression of LINC00341 represses monocyte adhesion to human umbilical vein endothelial cells (HUVECs) under OS. HUVECs were transfected with LINC00341 or vector control, and the cells were then subjected to PS or OS for 24 h. A: HUVECs were transfected with LINC00341 plasmid (10 μg/106 cells) or vector control. The overexpression efficiency, as indicated by LINC00341 expression level, was tested by RT-qPCR. Numbers above the bars indicate fold over PS-Vec. B: the quantitative analyses of THP-1 adhesion to HUVECs under OS with/without LINC00341 overexpression. The OS-induced VCAM1 mRNA (C) and protein (D) expression levels in HUVECs with/without LINC00341 overexpression. Vec, vector control. *P < 0.05, **P < 0.01, 1-way ANOVA and Tukey post hoc test, n = 3.
LINC00341 is anti-inflammatory in HUVECs.
Because LINC00341 acts as an anti-inflammatory regulator in HUVECs by suppressing VCAM1 expression and THP-1 adhesion (Fig. 2), we hypothesized that LINC00341 also modulates the endothelial inflammation in response to proinflammatory cytokines such as TNF-α. As seen in Fig. 3, A–C, stimulating HUVECs with TNF-α reduced the level of LINC00341, while increasing VCAM1 mRNA/protein expression and THP-1 adhesion (Fig. 3, A–C, column 1 vs. 2). On the other hand, LINC00341 overexpression suppressed the TNF-α-induced VCAM1 mRNA/protein expression and THP-1 adhesion (Fig. 3, A–C, column 2 vs. 3). Statins exert their beneficial effect in atherosclerosis treatment via reducing EC inflammation (3). Thus, we compared the gene expression in TNF-α-stimulated HUVECs with or without the treatment of atorvastatin. The cells treated with atorvastatin showed an increase of LINC00341 expression, decrease of VCAM1 expression, and suppression of THP-1 adhesion (Fig. 3, D–F, column 1 vs. 2). Furthermore, loss-of-function experiments with LINC00341 knock-down abolished the atorvastatin-induced reductions of VCAM1 expression and THP-1 adhesion (Fig. 3, D–F, column 2 vs. 3), when compared with siRNA negative control groups. These results indicate that LINC00341 is important for endothelial inflammation in the anti-inflammatory context.
Fig. 3.
LINC00341 is involved in the endothelial inflammation modulated by TNF-α and atorvastatin. A–C: HUVECs were transfected with LINC00341 or vector control followed by the treatment of TNF-α (2 ng/m) for 12 h, and the subsequent gene expression and functional consequence were then analyzed. A: RT-qPCR analysis for LINC00341 and VCAM1 RNA expression. B: protein expression level of VCAM1. C: monocyte adhesion assays. D–F: HUVECs were transfected with siLINC00341 (siL341), followed by treatment with TNF-α (2 ng/ml) and atorvastatin (2–8 μM) for 24 h, and the expression of LINC00341 and VCAM1 levels, as well as monocyte adhesion, were then examined. D: RT-qPCR analysis for LINC00341 and VCAM1 expression. E: representative images of Western blot and quantitative analysis of VCAM1 protein levels. F: monocyte adhesion assays under the indicated treatments. Bars represent means ± SE; n = 3. *P < 0.05, **P < 0.01, 1-way ANOVA and Tukey post hoc test.
LINC00341 attenuates VCAM1 expression via PRC2.
The subcellular localization of lncRNAs may provide evidence for their actions on downstream targets. The ubiquitous nucleus and cytosolic distribution of LINC00341 (Supplementary Fig. S4A) suggest that it may act either in cis or in trans to regulate target genes. Since SYNE3 (a LINC00341 neighboring gene) was not co-regulated with LINC00341 under PS, nor did the overexpression of LINC00341 alter the expression of SYNE3 (Supplementary Fig. S4B), it is inferred that the LINC00341 does not act in cis to regulate its downstream targets. Therefore, we hypothesized that LINC00341 acts in trans to repress VCAM1. In terms of the in trans mechanism regulated by lncRNAs, HOTAIR has been shown to bind to EZH2 to epigenetically regulate the expression of the target genes (7, 27). Hence, we tested whether LINC00341 can bind and then guide EZH2 to the VCAM1 promoter region. Using cross-linking RNA-IP assay, LINC00341 was coimmunoprecipitated with EZH2 from HUVEC nuclear extracts (Fig. 4A), indicating that LINC00341 is associated with EZH2 in HUVECs. Furthermore, atorvastatin treatment of HUVECs enriched the amounts of LINC00341 (Fig. 4A) and the VCAM1 promoter sequence (Fig. 4B) in the PRC2 complex. Complementarily, knocking down LINC00341 reduced the enrichment of VCAM1 promoter sequence in the PRC2 complex (Fig. 4B). These results indicate that LINC00341 can guide EZH2 to the VCAM1 promoter with the consequential repression of VCAM1 expression. We further used a luciferase reporter driven by the VCAM1 promoter to demonstrate the LINC00341 suppression of VCAM1 at the transcriptional level. As shown in Fig. 4C, the VCAM1 promoter activity in TNF-α-treated cells was decreased by atorvastatin but resumed following LINC00341 knock-down. These data demonstrate that LINC00341 functions as a PRC2-guiding lncRNA, which is required for transcriptional suppression of VCAM1.
Fig. 4.
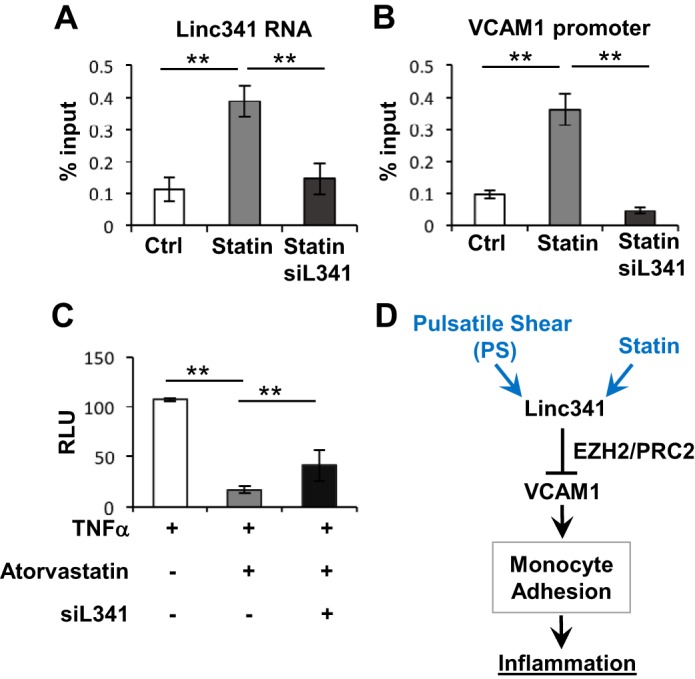
LINC00341 guides polycomb repressive complex 2 (PRC2) to the VCAM1 promoter. A, B: HUVECs were transfected with siLINC00341 (siL341) or control siRNA (Ctrl) and treated with atorvastatin for 24 h. The EZH2 and its associated complex were immunoprecipitated from nuclear lysates with anti-EZH2 antibody. The amount of LINC00341 RNA (A) and VCAM1 promoter DNA (B) in PRC2 immunoprecipitates was determined by RT-qPCR. C: HUVECs were transfected with the VCAM1 promoter construct in the presence or absence of siL341 and treated with TNF-α and atorvastatin as indicated. Promoter activity was then assessed by luciferase assays. RLU, relative luminescence units. D: schematic illustration of the mechanism by which LINC00341 regulates the endothelial inflammatory status.
Analysis of the LINC00341 co-regulated networks.
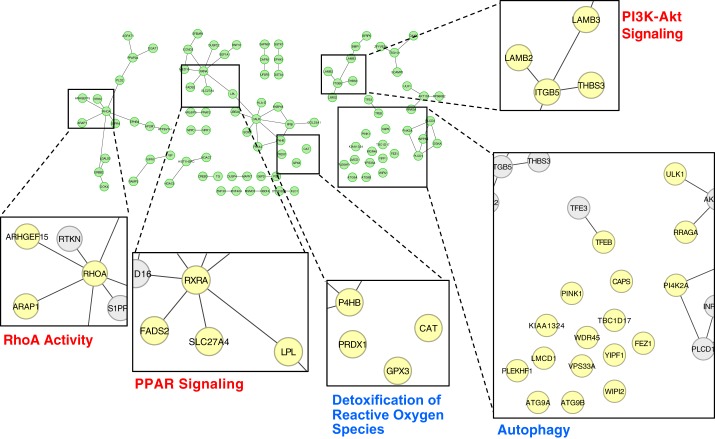
To delineate the detailed LINC00341-regulated networks, we performed in silico coexpression network analysis. The top genes positively correlated with LINC00341 (Pearson’s correlation coefficient > 0.75, Supplementary Table S3) were further subjected to functional enrichment analysis. Such analysis revealed that the coexpressed genes are enriched in the bioprocesses of autophagy and detoxification of reactive oxygen species; this is in line with the functional role of LINC00341 in HUVECs (Fig. 5 and Supplementary Fig. S5). Flow regulations of Ras homolog family member A (RhoA) and PI3 kinase/AKT serine threonine kinase 1 (PI3K/Akt) pathways (28) were also among the enriched bioprocesses in our gene list (Fig. 5), suggesting that LINC00341 may share common regulatory mechanisms with these pathways in regulating the endothelial cell inflammatory status.
Fig. 5.
Functional network analysis of LINC00341 coexpressed genes. The top LINC00341 coexpressed genes were subject to functional analysis with Reactome FIViz. Refer to Supplementary Fig. S5 for the complete networks.
DISCUSSION
We performed RNA-Seq experiments to delineate the shear stress regulation of the HUVEC transcriptome. With the RNA-Seq data demonstrating the significant regulation of lncRNAs, we focused on studying the role of lncRNAs in regulating EC functions. LINC00341, with no previously known biological functions, is one of the most abundant lncRNAs in ECs significantly induced by PS. Functional analyses demonstrated that overexpression of LINC00341 is sufficient to reduce the HUVEC inflammation triggered by OS and TNF-α, as evidenced by the significant suppressions of VCAM1 expression and THP-1 adhesion. In contrast, LINC00341 knock-down reversed the anti-inflammatory effects of atorvastatin in TNF-α-treated HUVECs, indicating that the statin-induced beneficial effects on endothelium are mediated through LINC00341. However, the knock-down of LINC00341 in HUVECs did not abrogate the PS inhibition of VCAM1 expression and monocyte adhesion. It is possible that although PS and statin are both atheroprotective stimuli that share many similarities, there are intricate differences between them that may reflect the nature that chemical stimuli such as statins induce relatively specific responses, while the actions of mechanical stimuli such as PS are more complex and affect multiple sensors and pathways. Hence the reduction of HUVEC inflammation by PS, which involves a large spectrum of gene/pathway modulations, cannot be sufficiently abrogated by disabling only one molecule, i.e., LINC00341.
Genes with significant functions are often conserved across different species. We identified murine and bovine orthologs of LINC00341 from the comparative genomics track of UCSC genome browser (Supplementary Fig. S6A) and confirmed their expression in the ECs of these species by RT-qPCR (Supplementary Fig. S6B). These results suggest that LINC00341 can be a key regulator in ECs across species.
Histone modification is an important epigenetic mechanism involved in the regulation of endothelial gene expression (6, 9). Thus, one of the mechanisms by which inhibitory lncRNAs exert their effect is to shuttle the repressive protein complex to alter histone methylation status at the promoter regions of target genes. Related to the current study, Maleszewska et al. (16) demonstrated that EZH2 level is reduced in ECs by high laminar shear stress, as a consequence of the activation of MAP2K5-MAPK7 signaling pathway, and consequently regulates cell cycle of ECs. In our RNA-Seq data, we also found that PS decreased the expressions of EZH2 and EC inflammation-related genes (i.e., VCAM1) and that the cell cycle-related genes were highly affected as well. The flow modulations of EZH2 in ECs suggest that the changes of histone methylation status are important in EC response to shear stress. Regarding LINC00341, we found that it can bind to the EZH2/PRC2 repressive complex to guide the LINC00341/EZH2/PRC2 to the VCAM1 promoter. To reconcile the increased LINC00341 but decreased EZH2 expression, it is likely that LINC00341 modulates the specificity of EZH2 and PRC2, although the EZH2 expression is generally suppressed in PS. The recruitment of EZH2/PRC2 to the promoter leads to the statin-repressed VCAM1 in ECs.
To comprehend the potential functions of LINC00341 in ECs, we constructed a coexpression network comprising genes with their expression levels positively correlated to that of LINC00341. Several hub genes were identified in this in silico-constructed network, including retinoid X receptor alpha (RXRA), RhoA, and integrin beta 5 subunit (ITGB5). The retinoid receptor family consists of a large number of proteins with diverse biological functions, including cholesterol metabolism and atherosclerosis (23). Among them, RXRA is a pleotropic nuclear receptor that is activated and dimerized upon ligand binding to activate the PPAR signaling pathway. RXRA activity is critical and sufficient to repress TNF-α-induced VCAM1 expression, leukocyte adhesion, and EC growth (21). RhoA was one of the LINC00341 coexpressed genes, which has been reported to be involved in shear-regulated EC functions (18, 25). As shown in Fig. 5, both RhoA and RTKN (a RhoA scaffold protein) were coexpressed with LINC00341, thus further supporting the notion that LINC00341 may be involved in the shear regulation of EC functions in a fine-tuning manner. The ITGB5 and lamin b2 and b3 (LAMB2/3) have been associated with the PI3K/Akt signaling pathway in the KEGG database (10), and we also found that these proteins are coexpressed with LINC00341 (Fig. 5). The PI3K/Akt pathway is activated by shear stress and is essential in mediating the downstream outcomes of atheroprotective shear (28). The coexpression of these pathways further suggests the potentials of LINC00341 network as a key modulation of the atheroprotective effect exerted by PS.
In summary, we have identified, through whole transcriptome RNA-Seq analysis, the shear-responsive lncRNA LINC00341, which can also be modulated by inflammation regulatory reagents such as TNF-α and atorvastatin. We also demonstrated that LINC00341 suppresses endothelial inflammation by recruiting PRC2 to VCAM1 promoter to inhibit VCAM1 expression. The potential LINC00341 gene network was deciphered by in silico coexpression network analysis. These findings provide new insights on the role of noncoding RNA in regulating vascular homeostasis.
GRANTS
This study is supported by the following grants: Taiwan MOST 103-2917-I-564-030 and American Heart Association 17POST33410101 (T.-S. Huang), National Heart, Lung, and Blood Institute K99HL-135416 (K.-C. Wang), HL-106579 and HL-108735 (S. Chien, J. Shyy, S. Subramaniam), and HL-121365 (S. Chien, Yingxiao Wang).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.-S.H., Y.-S.L., and S.C. conceived and designed research; T.-S.H., K.-C.W., S.Q., P.N., and T.-Y.C. performed experiments; T.-S.H. analyzed data; T.-S.H., Z.C., Y.-S.L., and S.C. interpreted results of experiments; T.-S.H. prepared figures; T.-S.H., Y.-S.L., and S.C. drafted manuscript; T.-S.H., T.-Y.C., Z.C., Y.-S.L., J.Y.-J.S., and S.C. edited and revised manuscript; T.-S.H., K.-C.W., S.Q., P.N., Z.C., Y.-S.L., S.S., J.Y.-J.S., and S.C. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
pGL4.20 reporter plasmid is a kind gift from Dr. Yingxiao Wang. We express our sincere appreciation for the valuable advice and encouragement by the late Dr. Hsei-Wei Wang.
REFERENCES
- 1.Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J, Mihailovic A, Sauer M, Ji R, Ramarathnam A, Totary-Jain H, Williams Z, Tuschl T, Schulze PC. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA 111: 11151–11156, 2014. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169, 2015. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonopoulos AS, Margaritis M, Lee R, Channon K, Antoniades C. Statins as anti-inflammatory agents in atherogenesis: molecular mechanisms and lessons from the recent clinical trials. Curr Pharm Des 18: 1519–1530, 2012. doi: 10.2174/138161212799504803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang T-Y, Huang T-S, Wang H-W, Chang S-J, Lo H-H, Chiu Y-L, Wang Y-L, Hsiao C-D, Tsai C-H, Chan C-H, You RI, Wu CH, Tsai TN, Cheng SM, Cheng CC. miRNome traits analysis on endothelial lineage cells discloses biomarker potential circulating microRNAs which affect progenitor activities. BMC Genomics 15: 802, 2014. doi: 10.1186/1471-2164-15-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng C-C, Lo H-H, Huang T-S, Cheng Y-C, Chang S-T, Chang S-J, Wang H-W. Genetic module and miRNome trait analyses reflect the distinct biological features of endothelial progenitor cells from different anatomic locations. BMC Genomics 13: 447, 2012. doi: 10.1186/1471-2164-13-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fish JE, Matouk CC, Rachlis A, Lin S, Tai SC, D’Abreo C, Marsden PA. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J Biol Chem 280: 24824–24838, 2005. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076, 2010. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang J, Ing MH, Salazar A, Lassègue B, Griendling K, Navab M, Sevanian A, Hsiai TK. Pulsatile versus oscillatory shear stress regulates NADPH oxidase subunit expression: implication for native LDL oxidation. Circ Res 93: 1225–1232, 2003. doi: 10.1161/01.RES.0000104087.29395.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Illi B, Nanni S, Scopece A, Farsetti A, Biglioli P, Capogrossi MC, Gaetano C. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ Res 93: 155–161, 2003. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- 10.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44, D1: D457–D462, 2016. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, Lachman HM. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One 6: e23356, 2011. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J-Y, Yao J, Li X-M, Song Y-C, Wang X-Q, Li Y-J, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 5: e1506, 2014. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J, Klijn C, Guillory J, Bhatt D, Vartanian S, Walter K, Chan J, Holcomb T, Dijkgraaf P, Johnson S, Koeman J, Minna JD, Gazdar AF, Stern HM, Hoeflich KP, Wu TD, Settleman J, de Sauvage FJ, Gentleman RC, Neve RM, Stokoe D, Modrusan Z, Seshagiri S, Shames DS, Zhang Z. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res 22: 2315–2327, 2012. doi: 10.1101/gr.140988.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukovic D, Moreno-Manzano V, Klabusay M, Stojkovic M, Bhattacharya SS, Erceg S. Non-coding RNAs in pluripotency and neural differentiation of human pluripotent stem cells. Front Genet 5: 132, 2014. doi: 10.3389/fgene.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maleszewska M, Vanchin B, Harmsen MC, Krenning G. The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis 19: 9–24, 2016. doi: 10.1007/s10456-015-9485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114: 1389–1397, 2014. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki T, Honda K, Ohata H. m-Calpain antagonizes RhoA overactivation and endothelial barrier dysfunction under disturbed shear conditions. Cardiovasc Res 85: 530–541, 2010. doi: 10.1093/cvr/cvp311. [DOI] [PubMed] [Google Scholar]
- 19.Qiao C, Meng F, Jang I, Jo H, Chen YE, Zhang J. Deep transcriptomic profiling reveals the similarity between endothelial cells cultured under static and oscillatory shear stress conditions. Physiol Genomics 48: 660–666, 2016. doi: 10.1152/physiolgenomics.00025.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren S, Peng Z, Mao J-H, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, Tian Z, Guan Y, Tang L, Xu C, Wang L, Gao X, Tian W, Wang J, Yang H, Wang J, Sun Y. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res 22: 806–821, 2012. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz M-J, Albertos F, Otero E, Juez M, Morcillo EJ, Piqueras L. Retinoid X receptor agonists impair arterial mononuclear cell recruitment through peroxisome proliferator-activated receptor-γ activation. J Immunol 189: 411–424, 2012. doi: 10.4049/jimmunol.1102942. [DOI] [PubMed] [Google Scholar]
- 22.Song HK, Hong S-E, Kim T, Kim DH. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS One 7: e35552, 2012. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szanto A, Narkar V, Shen Q, Uray IP, Davies PJA, Nagy L. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ 11, Suppl 2: S126–S143, 2004. doi: 10.1038/sj.cdd.4401533. [DOI] [PubMed] [Google Scholar]
- 24.Wang K-C, Yeh Y-T, Nguyen P, Limqueco E, Lopez J, Thorossian S, Guan K-L, Li YJ, Chien S. Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc Natl Acad Sci USA 113: 11525–11530, 2016. doi: 10.1073/pnas.1613121113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C-C, Li Y-S, Haga JH, Kaunas R, Chiu J-J, Su F-C, Usami S, Chien S. Directional shear flow and Rho activation prevent the endothelial cell apoptosis induced by micropatterned anisotropic geometry. Proc Natl Acad Sci USA 104: 1254–1259, 2007. doi: 10.1073/pnas.0609806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Res 3: 146, 2014. doi: 10.12688/f1000research.4431.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry 52: 9519–9527, 2013. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Li Y-S, Chien S. Shear stress-initiated signaling and its regulation of endothelial function. Arterioscler Thromb Vasc Biol 34: 2191–2198, 2014. doi: 10.1161/ATVBAHA.114.303422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.