Abstract
ClusPro-DC (https://cluspro.bu.edu/) implements a straightforward approach to the discrimination between crystallographic and biological dimers by docking the two subunits to exhaustively sample the interaction energy landscape. If a substantial number of low energy docked poses cluster in a narrow vicinity of the native structure of the dimer, then one can assume that there is a well-defined free energy well around the native state, which makes the interaction stable. In contrast, if the interaction sites in the docked poses do not form a large enough cluster around the native structure, then it is unlikely that the subunits form a stable biological dimer. The number of near-native structures is used to estimate the probability of a dimer being biological. Currently the server examines only the stability of a given interface rather than generating all putative quaternary structures as accomplished by PISA or EPPIC, but complements the information provided by these methods.
Keywords: Crystallographic dimer, biological dimer, solution structure, energy landscape, interface discrimination
Introduction
Many proteins function as assemblies of several polypeptide chains where homologous chains exhibit a high degree of symmetry. Over 80% of such structures have been determined by X-ray crystallography, and the arrangement of the subunits in an oligomeric protein often may not be reliably inferred from crystallographic studies. In fact, determining the quaternary structure and biological relevance of subunit interactions based on the X-ray structure alone is not straightforward [1, 2]. The contents of the asymmetric unit (ASU), the fraction of the crystallographic unit cell that has no crystallographic symmetry and is deposited in the PDB, can describe one or several copies of a macromolecule without indicating the oligomeric state (e.g., monomer, dimer) that is most relevant in solution. Although crystallographic interfaces are generally smaller (<1000 Å2) than biologically relevant ones, this is frequently not the case, and there is a substantial overlap between the distributions of these two types of interactions. In addition, oligomerization may be affected by truncation or mutation, and depends on conditions such as concentration and pH. Thus, experiments such as native gel electrophoresis, gel permeation chromatography, ultracentrifugation, or electrospray ionization time-of-flight mass spectrometry are need to reliably establish the multimeric state of a protein [3].
Since experimental validation is often not available for a specific protein of interest, distinguishing biologically relevant interfaces from lattice contacts in protein crystals under native conditions has become a well-recognized problem in structural bioinformatics, and a number of computational tools have been developed. The methods belong to two broad classes. The first class is based on estimating the stability of interaction based on the properties of the two proteins, using mostly, but not exclusively, descriptors of the interface. Among the first methods published in this class was PQS (Protein Quaternary Structure), which used an empirical scoring function based on several contributions such as interface contact area, number of interfacial buried residues, salt bridges, disulfide bonds, and the solvation energy of quaternary structure formation [4]. PQS has been developed into PISA, which uses approximations of the enthalpic and entropic contributions to the binding free energy to predict the biological relevance of a macromolecular assembly [5]. The method considers buried surface area, hydrogen bonds, salt bridges and disulfide bonds in order to estimate changes in enthalpy. For the entropic part, the translational, rotational, vibrational and surface entropy components are estimated using subunit mass, surface area, symmetry number, and inertia moments. PISA has been implemented as a server (http://www.ebi.ac.uk/pdbe/pisa/) that, in addition to determining the strength of the interactions, generates quaternary structure considering the symmetry mates. The server is very useful, and PISA has become the essential reference method, as it is currently used to predict quaternary structures of every entry in the Protein Data Bank (PDB) [6]. A number of similar methods have been developed based on various linear and nonlinear combinations of geometric and energetic descriptors of the protein-protein interface, in some cases involving machine learning and other statistical tools [7–11]. However, due to its importance we still consider PISA as providing the “golden” standard for quaternary structure prediction.
The second class of methods is distinguished by relying mainly on evolutionary information, although descriptors of the interface may also be included in the decision process [2, 12–15]. The most frequently used method in this class is EPPIC (Evolutionary Protein–Protein Interface Classifier) by Duarte et al [14]. EPPIC uses a collection of classifiers based on evolutionary features and a simple geometric measure [15]. The evolutionary conservation of residues is assessed by constructing multiple sequence alignment of all sequence homologs to the target protein structure under study. For the geometric analysis, the interface core residues, defined as fully buried residues, provide fundamental determinants of biological interfaces: their number is in itself a powerful discriminator of interface character and helps the evolutionary measures to distinguish biological contacts from crystal ones. The evolutionary and geometric scores are combined to form a consensus call through a simple-majority voting scheme. EPPIC is also available as a server at http://www.eppic-web.org/ewui/, which provides detailed information on all interfaces present in protein crystal structures in order to determine whether they are biologically relevant or not. Because the method used by EPPIC is substantially differ from the method in PISA, and because the availability of the server, we also consider EPPIC as a very important contribution to quaternary structure prediction.
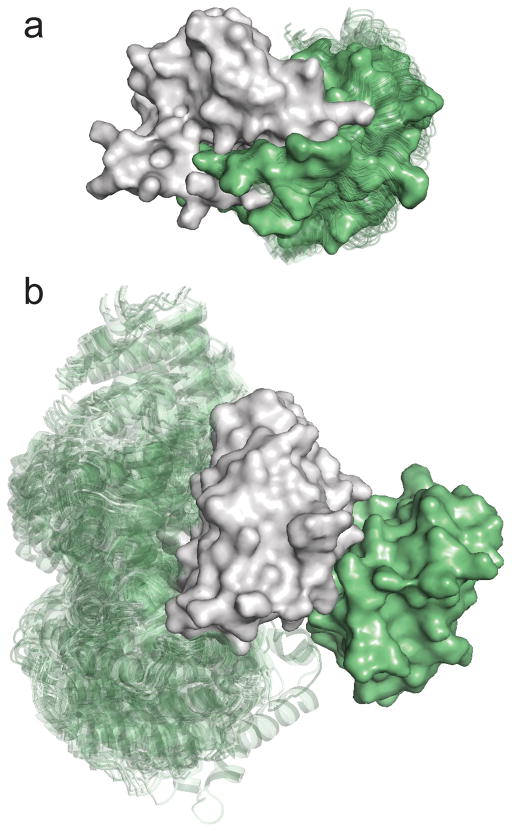
In this paper we introduce a straightforward method that, similarly to PISA, estimates the stability of the interaction between two protein subunits, but is based on exhaustive sampling of the interaction energy landscape using a docking method, rather than approximating the enthalpic and entropic contributions. The basic idea is extremely simple: we separate the two units of the dimer, consider one of the units and dock it to itself without any a priori assumption or restraint, evaluating the energy for billions of docked structures in the process. If a substantial number of low energy docked poses cluster in a narrow vicinity of the native structure, then we can assume that there is a well-defined free energy well around the native complex, which makes the interaction stable. In contrast, if the interaction sites in the docked structures do not form any cluster around the native state, then it is unlikely that the subunits form a stable biological dimer. As illustration of this discrimination strategy, Fig. 1a shows the docking of Escherichia coli met repressor (PDB ID 1cmb, solid surface in grey) to itself. The 100 lowest energy poses (transparent cartoons in green) closely match the actual position of the second subunit, shown as surface in green. Accordingly, the biological assembly as a homodimer was assigned by the authors [16] and supported by PISA. As the other extreme, Fig. 1b shows docking results for soybean leghemoglobin A (PDB ID 1bin, grey surface), demonstrating a case where no low energy docked pose overlaps with the X-ray structure of the second subunit in the dimer (green surface). Such result would be very unlikely for a protein that forms a dimer, and hence we conclude that the C2 symmetry between the two subunits occurs only in the crystal. This prediction is correct, since soybean leghemoglobin A is indeed a monomer in solution [17].
Figure 1.
Docking results for biological and crystallographic dimers. (a) Docking of E. coli met repressor (PDB ID 1cmb, solid surface in grey) to itself. The 100 lowest energy poses (transparent cartoons in green) closely match the actual position of the second subunit, shown as surface in green. Met repressor is a homodimer in solution. (b) Docking of soybean leghemoglobin A (PDB ID 1bin, grey surface) to itself. No low energy docked pose overlaps with the X-ray structure of the second subunit in the dimer (green surface), indicating that there is no stable dimer in solution. Accordingly, soybean leghemoglobin A is a monomer.
The advantage of the classifier presented here is that it is based on the well established docking method PIPER [18] and its energy function as implemented in the ClusPro 2.0 server [19]. ClusPro has been very successful in all rounds of the Critical Assessment of Predicted Interactions (CAPRI) protein-protein docking challenge [20], and has thousands of regular users. Adding dimer discrimination to ClusPro required only two adjustable parameters, the radius of the near-native region, defined in terms of the root mean square deviation (RMSD) from the x-ray structure of the dimer, and the number of docked structures that are expected to cluster in the near-native region in order to classify the dimer biological rather than crystallographic. As will be shown, the need for only two parameters provides remarkable robustness to the method. Furthermore, we also estimate the probability of a dimer being biological, a continuous measure rather than only a yes or no decision. The classifier is freely available for academic and governmental use at https://cluspro.bu.edu/ as part of the ClusPro server. We emphasize that at this point the ClusPro-CD server is able to examine only the stability of an interface specified by the user, rather than generating all putative quaternary structures as accomplished by both PISA or EPPIC. While in this paper we focus on methodology and describe a new prediction tool, our analysis also reveals that data on the quaternary structure of proteins are highly uncertain, and hence comparing the performance of different methods using the available data has limited validity.
Results and discussion
Theoretical basis
PIPER is a docking program that performs an exhaustive evaluation of simplified energy functions in discretized 6D space of mutual orientations of the protein partners [18]. The center of mass of the receptor is fixed at the origin of the coordinate system, and the possible orientational and translational positions of the ligand are evaluated at the given level of discretization. The rotational space is sampled at 70,000 rotations, which corresponds to about 5 degrees step size in terms of the Euler angles. The translational space is represented as a grid of 1Å displacements. It is easy to see that for an average size protein this amounts to sampling 109–1010 conformations. In view of this global and systematic sampling on a dense grid we can calculate an approximation of the overall partition function by Q = Σj exp(-Ej/RT), where Ej is the energy of the jth pose, and we sum over all poses. Similarly we can approximate the partition function in a near-native region of the native complex by Qnn = Σj exp(-Ej/RT), where we sum over only the near-native poses [19]. Based on these partition functions, the probability of the near-native state, Pnn, is Pnn = Qnn/Q. However, at this point ClusPro routinely retains only the 1000 lowest energy docked structures. Fortunately the dominant part of the partition function is provided by these 1000 structures, and hence the probability of the near-native state is approximated by Pnn ≈ Q′nn/Q′, where Q′ is the approximation of the partition function using the lowest energy 1000 structures. Similarly, Q′nn is the approximation of Qnn in a near-native region of the native complex, but using only the near-native structures among the 1000 low energy ones retained. Furthermore, since the low energy structures are from an energy range that is very narrow relative to the overall energy variation, and the energy values are calculated with considerable error that is comparable to the energy range considered, it is reasonable to assume that these energies do not differ, i.e., Ej = E for all j. Although neglecting the energy differences among the low energy structures seems to be arbitrary, we employ this approximation in in our docking server ClusPro with success. Thus, the approximation seems to be adequate for proteins that are amenable to rigid body docking, i.e., are subject to only moderate conformational changes upon binding [19]. This implies that Q = exp(-E/RT)×N and Qnn = exp(-E/RT)×Nnn, where N=1000 and Nnn is the number of the low energy structures in the near-native region. Therefore, the probability of the near-native state is approximated by Pnn ≈ Nnn/1000, and thus the probability of the ligand protein finding stable near-native binding position on the receptor protein is proportional to the number Nnn of the near-native structures among the 1000 retained. Accordingly, we will use Nnn for predicting the probability of forming a stable dimer that is independent of the crystal lattice and hence also occurs in solution. To obtain this predictor we need to select only the radius that defines the appropriate neighborhood of the native state in terms of the RMSD from the latter. To have a biological versus crystallographic classifier, comparable to PISA or EPPIC, we also select a threshold T on the number of structures in the near-native region such that Nnn ≤ T implies crystallographic whereas Nnn > T means biological dimer. As will be discussed, we also derive an interaction between Nnn and the probability P of a dimer considered biological, and show that the selected threshold value T occurs at P=0.5, which is thus used as the actual threshold.
Training set selection and results
For developing the method we used a set of biological dimers [21] and a set of large interface crystal dimers [22], both manually selected from the Protein Data Bank (PDB). The dimerization state of each protein in solution was checked with the biochemical literature [21, 22]. It was also verified that the sequence of the crystallized fragment was the one used for multimeric studies. Indeed, experimental results show that the full length protein forming a stable dimer cannot guaranty that a fragment will also form a stable dimer [8]. Any dimer was rejected if more than 5% of the interface area was contributed by ligands, prosthetic groups, or other nonprotein elements [21]. The original set of homodimers contained 122 entries [21], but we have removed alpha-chymotrypsin (PDB ID 4cha) because it is not a homodimer [23], as well as glutathione reductase (PDB ID 3grs) because the PDB file lacked the symmetry information needed to generate a dimeric structure for docking. The PDB IDs of the remaining 120 structures are listed in Table S1. We note that this set includes most of the homodimers from the Ponstingl dataset [24], frequently used for training and testing dimer discrimination methods. Some of the structures from the Ponstingl set were updated by Bahadur et al. [21] to consider higher resolution structures. In addition, we replaced the structure of aldehyde oxidoreductase from desulfovibrio gigas (PDB ID 1alo) by a newer one (PDB ID 1vlb). As for the set of crystal dimers, we considered the 103 structures with 2-fold symmetry that were selected by Bahadur et al. [22] to have an interface area greater than 800 Å2. The PDB structure 1hfv of the G-protein ARF6 was superseded by PDB structure 2j5x. Some of the proteins in the Bahadur set [22] had several interfaces that satisfied this condition, but we have retained only the largest interface per PDB entry, reducing the set to 89 entries also listed in Table S1. As in the case of the homodimers, many of these proteins were also in the Ponstingl dataset [24]. However, the latter included structures with packing interfaces that buried less than 800 Å2, and hence were not considered in our training set.
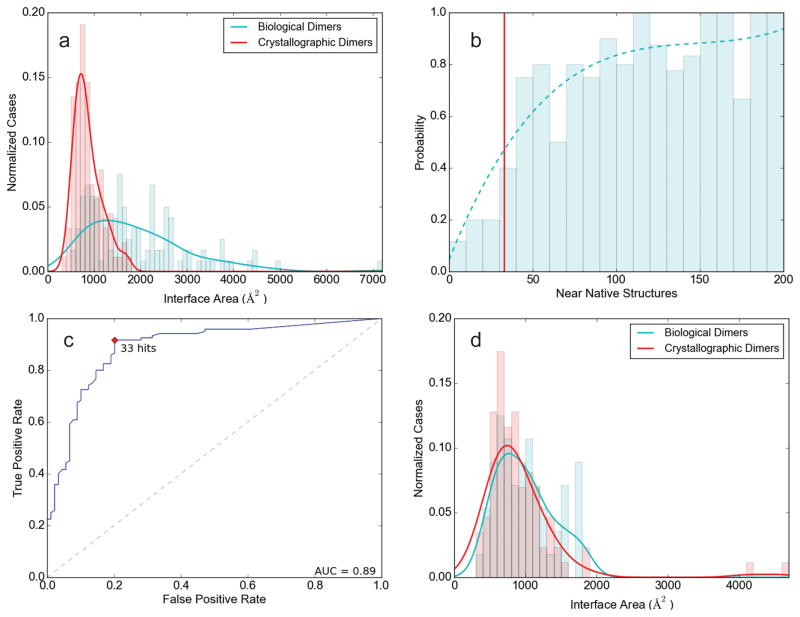
We have used the PISA server to select the interface with the largest area. Symmetry mates were generated using PyMOL when the PDB file didn’t already have the largest interface. In spite of considering crystal dimers with large interfaces, the average interface area is still substantially smaller than for the biological dimers (863.7 Å2 versus 1923.7 Å2, see Table S2). Although the standard deviations are large, based on the t-test the difference is significant (p<0.0001). However, the two distributions significantly overlap as many biological dimers have interface area below 1000 Å2 (see Fig. 2a), and hence discrimination on the basis of interface area alone is only moderately successful. We have used the ClusPro server with the standard PIPER energy function to dock the proteins to their own copies in both biological and crystallographic dimer sets and retained the 1000 lowest energy docked structures as usual in ClusPro (see Methods). Near-native structures were defined as having less than 7 Å2 Cα interface root mean square deviation (IRMSD) from the X-ray structure of the complex (see Methods). As expected, biological dimers were found to have more near-native docked poses than crystal dimers within the top 1000 structures. Fig. 2b shows the fraction of biological dimers as a function of Nnn, the number of near-native structures, with details for individual proteins listed in Table S2. At low values of Nnn (<30), this fraction is relatively small, but biological dimers become dominant for Nnn > 40 or so. Indeed, the average values of Nnn are 25.33 and 129.40, respectively, for crystallographic and biological dimers (Table S2). Although the data are noisy, smoothing the relationship provides a curve that, for any given Nnn, can be used to predict the probability of a dimer being biological. As mentioned, a classifier between crystallographic and biological dimers can be introduced by selecting a threshold value T such that dimers with Nnn < T are predicted to be crystallographic, whereas with Nnn ≥ T are predicted to be biological. Fig. 2c shows the receiver operating characteristic (ROC) curve for the above binary classifier as the value of Nnn is varied. Based on this curve, T = 33 appears to be a reasonable choice for the threshold between crystallographic and biological dimers. In good agreement with this selection, at Nnn = 33 the probability P of being a biological dimer is between 0.48 and 0.52, depending on the level of smoothing of the probability curve, and we select P = 0.5 as the probability threshold between crystallographic and biological dimers. We note that the Matthew correlation coefficient (MCC) also reaches its maximum at Nnn = 33 (see Table S3). Table 1 compares the results obtained by the docking based approach using this threshold and the results of the two most established methods of dimer classification, PISA and EPPIC from their server implementations. Tables S4 and S5, respectively, show detailed results for individual proteins in the multimer and monomer sets. For biological multimers, all three methods work equally well, with over 90% success rate. As shown in Table S4, PISA and EPPIC disagree for 16 structures in the multimer set, and ClusPro provides correct classification in 15 of these, which shows the motivation for using ClusPro as an additional method in case of uncertainty. Results for the proteins with large crystallographic interfaces that are considered monomeric by Bahadur et al. [22] are shown in Table S5. While both EPPIC and ClusPro predict close to 80% of these dimers as merely crystallographic, according to PISA more than 50% of these interactions are biological and stable (Table 1). We originally believed that this is because PISA introduces the class of uncertain structures, in addition to biological multimers and dimers. According to the PISA server, the quaternary structure falls into a grey region of the complex formation criteria and may or may not be stable in solution for 14 proteins. Two of these uncertain predicted structures are dimers, and the other 12 are putative monomers. However, even adding all uncertain structures as correctly predicted monomers, PISA would still predict fewer structures to be monomers than EPPIC or ClusPro (55 vs. 68 and 71). According to Bahadur et al. [22], the monomeric state of each protein in this set was first assessed from the BIOLOGICAL_UNIT record if present in the PDB entry, then checked against the Protein Quaternary Structure server [4] and against the literature, and only entries for which the monomeric state could be confirmed by biochemical or biophysical data were retained. In spite of these assurances, in five cases all three methods predict the multimers to be biological (Table S5). Among these five, the author determined biological unit is monomeric for 1ehy and 830c, but dimeric for 1c02, 2scp, and 1mss. In addition, both ClusPro and PISA predict 7 more structures as stable multimers (Table S5), and the author’s determination shows similar variation between momomeric and dimeric. Thus, we conclude that in spite of the analysis by Bahadur et al. [22], the reliability of quaternary structure assignment is limited even in the heavily used classic dataset. However, further analysis of this problem is beyond the scope of this paper. Considering the assignments correct, the overall success rate of quaternary structure prediction is 85.6% and 86.6%, respectively, for EPPIC and ClusPro, but only 72.7% for PISA, primarily due to the discussed overprediction of multimers. For the ClusPro based method the area-under-the-curve (AUC) value based on Fig. 2c is 0.89, which is comparable to the performance reported for the other two methods [5, 14].
Figure 2.
Selected results for training and “difficult” test sets. (a) Distributions of interface areas for biological dimers (green curve) and crystallographic dimers (red curve) in the training set. (b) Fraction of biological dimers as a function of Nnn, the number of low energy docked structures within 7Å Cα interface RMSD of the native structure. The direct data are shown as bar graph, with the smoothed probability values as the continuous curve. The red vertical line shows the threshold value T = 33 for Nnn. (c) Receiver operating characteristic (ROC) curve for the binary classifier as the value of Nnn is varied. Based on this curve, T = 33 appears to be a reasonable choice for the threshold Nnn value to discriminate between crystallographic and biological dimers. The area-under-the-curve (AUC) value is 0.89. (d) Distributions of interface areas for biological dimers (green curve) and crystallographic dimers (red curve) in the “difficult” subset of the test set.
Table 1.
Comparing the performance of the three servers.
| Set | Property | PISA | EPPIC | ClusPro |
|---|---|---|---|---|
| Training set | ||||
| Dimers: 120 | Dimer correct | 111 (92.5 %) | 111 (92.5 %) | 110 (91.7%) |
| Monomers: 89 | Monomer correct | 41 (46.1%) | 68 (76.4%) | 71 (79.8%) |
| Total: 209 | Total correct | 152 (72.7%) | 179 (85.6%) | 181 (86.6%) |
| Sensitivity & Specificity | 0.93 & 0.46 | 0.93 & 0.76 | 0.92 & 0.80 | |
| F1 value | 0.80 | 0.88 | 0.89 | |
| DC set | ||||
| DC Bio: 63 | Dimer correct | 42 (66.7%) | 59 (93.7%) | 58 (92.1%) |
| DC Xtal: 78 | Monomer correct | 42 (53.8%) | 51 (65.4%) | 47 (60.3%) |
| Total: 141 | Total correct | 84 (59.6%) | 110 (78.0%) | 105 (74.5%) |
| Sensitivity & Specificity | 0.67& 0.54 | 0.94 & 0.65 | 0.92 & 0.60 | |
| F1 value | 0.60 | 0.79 | 0.76 | |
| Test set | ||||
| Dimers: 293 | Dimer correct | 208 (69.8%) | 223 (74.8%) | 223 (74.8%) |
| Monomers: 490 | Monomer correct | 378 (77.1%) | 385 (78.6%) | 395 (80.6%) |
| Total: 783 | Total correct | 586 (74.8%) | 608 (77.7%) | 618 (78.9%) |
| Sensitivity & Specificity | 0.71 & 0.77 | 0.76 & 0.79 | 0.76 & 0.81 | |
| F1 value | 0.68 | 0.72 | 0.73 | |
| “Difficult” subset | ||||
| Dimers: 56 | Dimer correct | 34 (60.7%) | 15 (26.8%) | 31 (55.4%) |
| Monomers: 86 | Monomer correct | 31 (36.0%) | 39 (45.3%) | 55 (64.0%) |
| Total: 142 | Total correct | 65 (45.8%) | 54 (38.0%) | 86 (60.6%) |
| Sensitivity & Specificity | 0.61 & 0.36 | 0.27 & 0.45 | 0.55 & 0.64 | |
| F1 value | 0.47 | 0.25 | 0.53 | |
Test set selection and results
We tested the methods on three different sets of proteins. Table 1 compares classification results by ClusPro, PISA and EPPIC for all three sets. The first set, collected by Duarte et al. [14] includes the DCxtal set of proteins with large crystal contacts (78 entries validated as monomers) and the DCbio set of proteins with small biological interfaces (63 validated homodimers). For the entries in these sets the oligomeric structure was experimentally verified, the crystal entries were checked to fulfill a series of quality criteria [14], and the focus was on the range of interface areas where it was really difficult to distinguish crystal from biological contacts. Indeed, the interface areas are similar, 1309.0 Å2 and 1212.5 Å2, respectively, for DCbio and DCxtal. Nevertheless, both EPPIC and ClusPro perform fairly well (78.0% and 74.5% overall success rates), whereas PISA is again biased toward multimers, resulting in 59.6% overall success rate (see Tables 1, S6, S7, and S8).
For the second test set we collected newly published structures from the Protein Data Bank (PDB) using the following criteria: (1) PDB release date between January 2014 and August 2015; (2) no ligands in structure; (3) only a single type of protein in the structure, i.e., no heterodimers; and (4) the PDB file describes author determined biological units to assess the biological assembly as suggested by the authors. The resulting set, listed in Table S9 and called the test set, contains 783 entries total, with 293 biological multimers and 490 monomers. The interface areas substantially differ: 1635.0 Å2 for the biological and only 793.6 Å2 for the crystallographic multimers. However, the advantage of this set that the proteins were not used to train PISA, EPPIC, or ClusPro. Table 1 compares classification results by the three methods to the assignment of biological assembly provided by the authors in the PDB file, with the detailed assignments shown in Tables S10 and S11. We are aware that the biological assembly assigned by the authors is not necessarily correct, and that in some cases relevant publications may provide more valid classification. However, selecting publications for evaluating the three methods, even when some information is available, would introduce substantial level of subjectivity, and hence we retained the author’s assignment as the “true” state of quaternary assembly. According to Table 1, the three methods perform almost equally well, with PISA only slightly worse than the other two.
For the third test set we selected the “difficult” subset of the test set by adding a fifth selection criterion: (5) results from EPPIC and PISA are conflicting, thus one method considers the dimer biological and the other crystallographic, or the classification by PISA is uncertain. The “difficult” subset contained 142 entries total, with 56 biological multimers and 86 monomers. As shown in Fig. 2d and Table S12, for these two sets the interface areas are small and their distributions are almost identical. Although the average interface area of the biological multimers, 994.3 Å2, is slightly higher than for the crystallographic ones, which is 934.1 Å2, a two-sided t-test shows that the difference is not significant (p>0.1). Thus, this test set is different from the ones used earlier. As shown in Table 1, on the “difficult” set all three methods perform much worse than on the training set and on the other two test sets, but now ClusPro is better than the other two. As on the other sets, PISA works relatively well for multimers (Table S13), but classifies 42 of the 86 monomers as stable multimers, in addition to predicting 14 structures as uncertain multimers (Table S14), resulting in the success rate of only 36.0% (Table 1). In contrast to its good performance on the training and DC sets, EPPIC recognizes only 15 of the 56 multimers as biological (26.8% correct), primarily because many of the more recently crystallized proteins have only a few homologs or no homolog at all, and hence the evolutionary criteria could not be used. Consequently, both PISA and EPPIC have relatively low overall success rates, 45.8% and 38.0%, respectively. In contrast, the overall success rate for ClusPro is 60.6%. However, we note that selecting the “difficult” cases for which PISA and EPPIC contradict to each other makes our analysis on this “difficult” subset biased against these two methods. Nevertheless, the application to this set of proteins is useful for demonstrating that ClusPro can be a valuable tool for improving the reliability of quaternary structure prediction when results obtained by the standard methods are uncertain.
Extending the above analysis, we applied the three methods to subsets of the test set from several interface-area ranges. Predictions were separately analyzed for interface areas below 600, 800, and 1000 Å2 (Table S15). Results show that the identification of very small interface area biological dimers is difficult. For proteins with less than 600 Å2 interface the success rate was only 23.5% (4 out of the 17 cases) for all three methods. However, since the overall percentage of biological dimers with such small interface is low (17 out of 783, thus 2.17%), the overall success rate was over 90%, in spite of the inability to correctly identify most of the dimers. Further results are presented as Supplemental Information. As an additional study we explored how the three methods perform when applied to transient homodimers, with the results also shown in Supplemental Information.
The ClusPro-DC server
Dimer classification has been added as a new option to our protein-protein docking server ClusPro. The server can be used without a user account or with a user account (if having an educational or governmental email address) at https://cluspro.bu.edu/. Users with an account can request an e-mail to be sent when any submitted job is completed. The server opens at the ClusPro home screen and the user can select the option “Dimer Classification” rather than the option ‘Dock”. This opens the dimer classification page (Fig. S2a), where the user can provide a job name for the submission and input the coordinates of a homo-oligomer using PDB format. There are two options for input: importing coordinates from the PDB or uploading a structure. Only atoms of 20 standard amino acid residues are retained. The next step is selecting the two chains of the dimer that define the interface of interest. Multiple chains, separated by whitespace, can be selected in each box. Clicking the “Submit” button will start the calculation. The status of the job can be immediately checked from the “Queue” page. Clicking the job ID opens the status page, which shows the job ID, job name, user name, a status update, and pictorial representations of the uploaded and processed input structures (Fig. S2b). If requested, an email will be sent when the job has completed or if an error occurred. The email will contain a link to the results or error message. One can click the link, or alternatively, locate the results under the Results tab on the server, which shows the number Nnn of near-native docked structures among the lowest energy 1000 structures and the implied probability of the interaction being a biological dimer (Fig. S3a). One can also download a PyMOL session that shows the 100 lowest energy structures as transparent cartoons out of the 1000 retained (Fig S3b).
We demonstrate the application of the server to modulator protein MzrA (PDB ID 4pwu), which was target T70 of the CAPRI protein docking experiment [20]. In Round 30 of CAPRI the challenge was predicting the structure of homo-oligomers based on the sequence of the protein, in advance to the release of the structure to the PDB [25]. Since then the coordinates of most targets, including T70, have been released. According to the author, 4pwu is a dimer, and based on PISA, it is a tetramer. The PDB for 4pwu provides 4 chains (A, B, C, and D), and we first analyzed the stability of A:B, C:D, and A:C interactions. Fig. S2 shows the input and status page for the analysis of the A:B interface, and Fig. S3 shows the result that the probability of the A:B dimer being biological is 97%. The same strong interaction exists for the C:D interface. For the A:C interaction the probability of being stable is only 11% (not shown), but there is strong binding on the other side of the A subunit (Fig. S4). In fact, PyMOL generates a symmetry mate at that position, and it is included in the A2:B2 tetramer constructed by PISA. Therefore we tested the stability of the interaction between two A:B dimers, and found it biological with 75% probability, implying that 4pwu forms a tetramer in agreement with the PISA assessment. Note that although for 4pwu we had four chains in the ASU, direct analysis of these subunits confirmed only a biological dimer, and it was necessary to generate the symmetry mates to determine all biological interfaces. Alternatively one can generate and download the quaternary assemblies using PISA. At this point the ClusPro-CD server is able to examine only the stability of an interface specified by the user, rather than generating all putative quaternary structures as accomplished by both PISA and EPPIC. Thus, we think that the primary application of the server is confirming the results obtained by PISA or EPPIC, particularly if the two contradict to each other.
Methods
Selection of the test set and its “difficult” subset
We selected PDB files with release dates between January 2014 and August 2015 with no ligands and one type of protein only, resulting in 783 structures. To determine the assignment by PISA, for each structure we downloaded the xml for “macromolecular assemblies” using http://www.ebi.ac.uk/pdbe/pisa/cgi-bin/multimers.pisa?pdbcodelist and selected the most probable multimeric state, which was the first assembly listed in the xml. All potentially uncertain assignments were checked by manual submission to the PISA server. To determine the assignment by EPPIC, for each structure downloaded the xml using http://www.eppic-web.org/ewui/ewui/dataDownload?type=xml&id=PDB_code. The multimer was considered biological if any interface was assigned as bio in the consensus column. We have identified 142 structures with conflicting results from EPPIC and PISA, or with uncertain PISA assignment, and these structures were used as the “difficult” subset of the test set.
Dimer classification by ClusPro
ClusPro performs rigid body docking using PIPER [18], a docking program based on the Fast Fourier Transform (FFT) correlation approach. For generating putative dimeric structures we consider the given protein structure as the receptor and a second copy of it as the ligand. The center of mass of the receptor is fixed at the origin of the coordinate system, and the possible orientational and translational positions of the ligand are evaluated on a dense grid, evaluating the energy for billions of poses. ClusPro retains the 1000 lowest energy docked structures. We then determine the number Nnn of such structures with less than 7 Å Cα interface root mean square deviation (IRMSD) from the native state. While other IRMSD values between 5Å and 10Å were also tested, 7Å IRMSD provided the best discrimination between biological and crystallographic dimers in the training set. To calculate the interface RMSD of a docked structure we first select the interface residues in the X-ray structure, defined as the ligand residues that have any atom within 10 Å of any receptor atom. We then superimpose the receptors in the docked and X-ray structures, and calculate the Cα RMSD for the selected interface residues. We have determined the relationship between Nnn and the fraction of biological dimers in the training set (Fig. 2b). After smoothing, the relationship was used to estimate the probability of a specific structure being a biological dimer on the basis of the Nnn value obtained by the docking.
Supplementary Material
Acknowledgments
This research was supported in part by the National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) under grants R35-GM118078 and R01-GM061867 and by the National Science Foundation (NSF) under grant DBI 1458509.
References
- 1.Janin J. Specific versus non-specific contacts in protein crystals. Nat Struct Biol. 1997;4:973–974. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- 2.Valdar WS, Thornton JM. Conservation helps to identify biologically relevant crystal contacts. J Mol Biol. 2001;313:399–416. doi: 10.1006/jmbi.2001.5034. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald MC, Chernushevich I, Standing KG, Whitman CP, Kent SB. Probing the oligomeric structure of an enzyme by electrospray ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 1996;93:6851–6856. doi: 10.1073/pnas.93.14.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 5.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitra P, Pal D. Combining Bayes classification and point group symmetry under Boolean framework for enhanced protein quaternary structure inference. Structure. 2011;19:304–312. doi: 10.1016/j.str.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Bernauer J, Bahadur RP, Rodier F, Janin J, Poupon A. DiMoVo: a Voronoi tessellation-based method for discriminating crystallographic and biological protein-protein interactions. Bioinformatics. 2008;24:652–658. doi: 10.1093/bioinformatics/btn022. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchiya Y, Nakamura H, Kinoshita K. Discrimination between biological interfaces and crystal-packing contacts. Adv Appl Bioinform Chem. 2008;1:99–113. doi: 10.2147/aabc.s4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo JS, Guo YZ, Fu YY, Wang Y, Li WL, Li ML. Effective discrimination between biologically relevant contacts and crystal packing contacts using new determinants. Proteins. 2014;82:3090–3100. doi: 10.1002/prot.24670. [DOI] [PubMed] [Google Scholar]
- 11.Da Silva F, Desaphy J, Bret G, Rognan D. IChemPIC: A random forest classifier of biological and crystallographic protein-protein interfaces. J Chem Inf Model. 2015;55:2005–2014. doi: 10.1021/acs.jcim.5b00190. [DOI] [PubMed] [Google Scholar]
- 12.Hou QZ, Dutilh BE, Huynen MA, Heringa J, Feenstra KA. Sequence specificity between interacting and non-interacting homologs identifies interface residues - a homodimer and monomer use case. BMC Bioinformatics. 2015;16:325. doi: 10.1186/s12859-015-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scharer MA, Grutter MG, Capitani G. CRK: An evolutionary approach for distinguishing biologically relevant interfaces from crystal contacts. Proteins. 2010;78:2707–2713. doi: 10.1002/prot.22787. [DOI] [PubMed] [Google Scholar]
- 14.Duarte JM, Srebniak A, Scharer MA, Capitani G. Protein interface classification by evolutionary analysis. BMC Bioinformatics. 2012;13:334. doi: 10.1186/1471-2105-13-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capitani G, Duarte JM, Baskaran K, Bliven S, Somody JC. Understanding the fabric of protein crystals: Computational classification of biological interfaces and crystal contacts. Bioinformatics. 2016;32:481–489. doi: 10.1093/bioinformatics/btv622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rafferty JB, Somers WS, Saint-Girons I, Phillips SE. Three-dimensional crystal structures of Escherichia coli Met repressor with and without corepressor. Nature. 1989;341:705–710. doi: 10.1038/341705a0. [DOI] [PubMed] [Google Scholar]
- 17.Hargrove MS, Barry JK, Brucker EA, Berry MB, Phillips GN, Jr, Olson JS, et al. Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J Mol Biol. 1997;266:1032–1042. doi: 10.1006/jmbi.1996.0833. [DOI] [PubMed] [Google Scholar]
- 18.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 19.Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, et al. How good is automated protein docking? Proteins. 2013;81:2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, Vajda S, et al. CAPRI: a Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 21.Bahadur RP, Chakrabarti P, Rodier F, Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- 22.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein - protein interfaces. J Mol Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 23.Tsukada H, Blow DM. Structure of alpha-chymotrypsin refined at 1.68 Å resolution. J Mol Biol. 1985;184:703–711. doi: 10.1016/0022-2836(85)90314-6. [DOI] [PubMed] [Google Scholar]
- 24.Ponstingl H, Kabir T, Thornton JM. Automatic inference of protein quaternary structure from crystals. J Appl Cryst. 2003;36:1116–1122. [Google Scholar]
- 25.Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, et al. Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment, Proteins: Structure, Function, and Bioinformatics. 2016 Apr 28; doi: 10.1002/prot.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.