Supplemental Digital Content is available in the text.
Abstract
Background:
Coliphages have been proposed as indicators of fecal contamination in recreational waters because they better mimic the persistence of pathogenic viruses in the environment and wastewater treatment than fecal indicator bacteria. We estimated the association between coliphages and gastrointestinal illness and compared it with the association with culturable enterococci.
Methods:
We pooled data from six prospective cohort studies that enrolled coastal beachgoers in California, Alabama, and Rhode Island. Water samples were collected and gastrointestinal illness within 10 days of the beach visit was recorded. Samples were tested for enterococci and male-specific and somatic coliphages. We estimated cumulative incidence ratios (CIR) for the association between swimming in water with detectable coliphage and gastrointestinal illness when human fecal pollution was likely present, not likely present, and under all conditions combined. The reference group was unexposed swimmers. We defined continuous and threshold-based exposures (coliphage present/absent, enterococci >35 vs. ≤35 CFU/100 ml).
Results:
Under all conditions combined, there was no association between gastrointestinal illness and swimming in water with detectable coliphage or enterococci. When human fecal pollution was likely present, coliphage and enterococci were associated with increased gastrointestinal illness, and there was an association between male-specific coliphage level and illness that was somewhat stronger than the association between enterococci and illness. There were no substantial differences between male-specific and somatic coliphage.
Conclusions:
Somatic coliphage and enterococci had similar associations with gastrointestinal illness; there was some evidence that male-specific coliphage had a stronger association with illness than enterococci in marine waters with human fecal contamination.
Fecal indicator bacteria, such as enterococci, are the basis for beach monitoring programs around the world.1 Their concentrations have been linked to increased risk of gastrointestinal illness in swimmers.2 Because they are easier to measure and are abundant in the human intestinal tract, fecal indicator bacteria can serve as a proxy for the numerous pathogens present in human waste. They are measured in preference to pathogens because measuring pathogens directly is expensive and presents substantial technical challenges. However, it has been suggested that viruses are responsible for most illness resulting from recreational water exposure.3,4 Consistent with this hypothesis, some studies have found that the greatest excess illness among swimmers occurs in the 2 days following ocean exposure, which aligns with the incubation periods for common waterborne viruses, such as norovirus or adenovirus.5–8
Fecal indicator bacteria have shortcomings as indicators of viral contamination,1 including their persistance9 and in many cases growth10–13 in the environment. Moreover, modern wastewater treatment facilities are designed to reduce fecal indicator bacteria levels to meet water quality standards, but many human enteric pathogens are less susceptible than bacteria to disinfection regimes and may remain infectious in discharged wastewater effluent.14,15 Due to their smaller size and disparate physiology, viruses can persist in the environment and move through sand, sediment, and groundwater, while bacteria are typically filtered by soil.13,16,17 Consistent with these shortcomings, several studies have found weak associations between enteric viruses and fecal indicator bacteria in marine waters.18–21
An alternative to measuring fecal indicator bacteria is to measure coliphages, viruses that infect Escherichia coli whose environmental degradation characteristics more closely mimic that of viruses.22–27 Coliphages meet many of the criteria for an ideal indicator of fecal contamination of water28,29: (1) They are present when enteric viruses are present in marine and other waters29–32; (2) The density of coliphages is typically much greater than that of human viruses,33–35 making them easier to detect; (3) They are specific to fecal contamination of water36–38; (4) Their resistance and response to disinfection is similar to that of pathogens of interest31,32,39; and (5) They are nonpathogenic37 and can easily be detected and enumerated with inexpensive methods.29,40–45
There are two types of coliphages: male-specific coliphages, which infect E. coli via the F sex pilus, and somatic coliphages, which attach to the exterior of E. coli cells. Associations with illness may vary by coliphage type. Male-specific coliphage morphology resembles that of many enteric viruses; fewer somatic coliphages closely resemble enteric viruses.46 In some studies, male-specific coliphages had a stronger association with pathogens than somatic coliphage.21 Associations with illness might also vary by the assay used to detect coliphage. Two commonly used culture-based methods are Environmental Protection Agency (EPA) Method 1601 and 1602. EPA 1601 includes an enrichment step, which may affect the diversity of coliphage strains detected.47 Coliphage counts from EPA 1601 may be more variable because the method utilizes the most probable number technique for enumeration, whereas EPA 1602 relies on direct counts.
Nine studies have examined whether coliphages are associated with increased gastrointestinal illness,48–56 and six studies found a positive association.48,50–52,55,56 However, the type of coliphage, coliphage enumeration method, and strength of associations varied, and the sample size was limited in several studies. We combined coliphage data from six prospective cohort studies at beaches from the Pacific, Atlantic, and Gulf of Mexico coasts that used a common sampling design. Using this data, we evaluated whether coliphages alone or as a combined indicator with culturable enterococci predicted gastrointestinal illness as well as enterococci, one of the nationally recommended fecal indicator bacteria for protecting public health in ambient waters designated for primary contact recreation.57
METHODS
Study Sites
We pooled data from six prospective cohort studies at coastal beaches in southern California, Alabama, and Rhode Island: Doheny State Beach in Dana Point, Malibu Surfrider State Beach, Mission Bay in San Diego, and Avalon Beach on Catalina Island in southern California, Fairhope Municipal Beach in Alabama, and Goddard Memorial State Park Beach in Rhode Island (eFigure1 and eTable 1; http://links.lww.com/EDE/B203).5,7,8,50,51,55
Enrollment
Studies enrolled beach visitors between May and September from 2003 to 2009. Eligibility criteria included: (1) no previous participation in the study beaches and (2) at least one household member at the beach ≥18 years old (see eTable 2; http://links.lww.com/EDE/B203 for eligibility details). At enrollment, interviewers recorded participants’ beach location, current health status, and recreational water exposure. Interviewers contacted participants 10–14 days later by phone to ascertain illness, demographic information, pre-existing health conditions, and water exposure since enrollment. Studies received approval from the institutional review boards at the University of California, Berkeley, the University of North Carolina at Chapel Hill, and the Centers for Disease Control and Prevention. Participation of human subjects did not occur until after informed consent was obtained.
Water Quality Sampling and Analysis
Each day, studies collected 125 ml to 1 L water samples in sterile containers at shin (0.3–0.5 m) or waist (1 m) depth. The total number of water samples collected and analyzed for coliphage per day ranged from one composite sample (a combination of individual samples collected at different locations within a given beach) at Mission Bay to 18 at Fairhope and Goddard beaches. See eTable 3 (http://links.lww.com/EDE/B203) for further details. Studies detected male-specific and somatic coliphage in water samples using culture-based methods (EPA 1601 and 1602).40,41 EPA 1601 was modified for use as a most probable number (MPN) procedure. Assays conducted to detect indicators varied by beach; somatic (EPA 1601) was analyzed at Avalon, Doheny, and Mission Bay; somatic (EPA 1602) was analyzed at Avalon and Doheny; male-specific (EPA 1601) was analyzed at all six beaches; male-specific (EPA 1602) was analyzed at Avalon and Doheny (eTable 4; http://links.lww.com/EDE/B203). Studies measured the level of enterococci in water samples at all six beaches using culture-based EPA Method 1600 on mEI agar except at Mission Bay, which used Enterolert (IDEXX, Westbrook, ME). We imputed values below the detection limit with 0.1 colony forming units (CFU) per 100 ml for enterococci and 0.1 plaque-forming units (PFU) per 100 ml for coliphages. Further details about water sample analysis are in eTable 3 (http://links.lww.com/EDE/B203).
Exposure Definitions
We defined “beachgoers” as individuals who recreated at the beach, regardless of whether they entered the water, “swimmers” as beachgoers who had water contact above the waist, and “nonswimmers” as those who had no water contact. We matched the daily geometric mean of coliphage and enterococci levels at Avalon, Doheny, and Malibu beaches to participants based on their swim location because there was greater heterogeneity in water quality at different sites at these beaches; for Mission Bay, we matched the level of coliphage in the single composite sample at each beach to swimmers; for the other beaches, we averaged over all samples present on the beach visit day. At Fairhope and Goddard beaches, where there was less heterogeneity in water quality across sample locations, we matched participants to the daily geometric mean at each beach, consistent with how the original authors classified exposure.58 The original studies indicated no substantial differences in the associations between the daily averages of fecal indicators and averages specific to a swimmer’s time and location. We assigned indicator levels below the detectable limits a value of 0.1 MPN/100 ml for coliphage and 0.1 CFU/100 ml for enterococci.
Outcome Definition
The primary outcome for this study was incident gastrointestinal illness within 10 days of exposure. Gastrointestinal illness was defined as: (i) diarrhea or (ii) vomiting or (iii) nausea and stomachache, or (iv) nausea or stomach ache and missed regular activities as a result of illness.5,7,8,55
Beach Conditions Classification
We classified study days by whether human fecal contamination was likely to be present (“human-impacted conditions”). At Fairhope and Goddard beaches, we considered all study days to be human-impacted because of the presence of nearby wastewater treatment facilities and discharges.55 At Doheny Beach, during the spring and summer, a sand berm forms that blocks the flow of San Juan Creek into the surf zone. We classified days when the berm was open as human-impacted.50 At Avalon Beach, wastewater from a faulty sanitary sewer system discharges in submarine groundwater through the sand and is moderated by tidal conditions.8 We classified days when groundwater flow was above the median as human-impacted and those when it was below median flow as not human-impacted. We classified all days at Malibu and Mission Bay beaches as not human-impacted because there were no known sources of fecal discharge at those sites. See eAppendix 1 (http://links.lww.com/EDE/B203) for more additional information on our beach conditions classification.
Statistical Analysis
Primary Analysis
We performed two types of statistical analyses. The first was a threshold analysis using an indicator for coliphage presence/absence; the reference group was swimmers recreating in water without detectable coliphage (eAppendix 2; http://links.lww.com/EDE/B203). Coliphage was considered present if any samples on the beach visit day contained detectable coliphage and absent if none did. For enterococci, the threshold was a geometric mean >35 CFU/100 ml, corresponding to the present water quality standard.57 We also created a joint indicator for coliphage and enterococci classified as 1 if coliphage was detected and the enterococci level was >35 CFU/100 ml and 0 if coliphage was not detected and the enterococci level was ≤35 CFU/100 ml. For both the single and joint indicators, we estimated cumulative incidence ratios (CIRs) that pooled across coliphage detection method (EPA 1601 or 1602). The second approach used continuous log10 levels of enterococci (CFU/100 ml) and coliphage (PFU/100 ml) as the exposure. We estimated associations with a 1-log10 increase in coliphage or enterococci levels with reference levels of −1 CFU/100 ml (log10(0.1)) for enterococci and −1 PFU/100 ml for coliphage corresponding to nondetects. We stratified coliphage analyses by EPA Method because levels from EPA 1601 and 1602 are not directly comparable. Because the number of beaches contributing to each coliphage analysis varied by coliphage type and detection method, we repeated enterococci analyses for the subset of beaches included in each coliphage analysis to ensure comparability.
We estimated CIRs among swimmers using log-linear, modified Poisson models with robust standard errors to account for clustering within households.59 To estimate 95% confidence bands for the probability of illness across levels of coliphage and enterococci, we used a nonparametric bootstrap with 1,000 replicates. We adjusted statistical models for the following potential confounders, consistent with previous studies5,7,8,50,51,55: age; sex; race (white vs. not white); chronic gastrointestinal illness; contact with a person with gastrointestinal illness at enrollment; contact with any animals; and consumption of undercooked or raw eggs, meat, or fish in the 3 days before enrollment. We did not adjust for sand contact because it could be a mediator of the effect of coliphage on illness.60 Models included fixed effects for each beach.61 With the exception of age, which was coded as a categorical variable, we coded all potential confounders as binary (yes/no). We assessed effect modification by whether conditions were human-impacted. Our analysis excluded individuals with missing outcomes and assumed they were missing at random conditional on covariates in our model. We excluded individuals who had gastrointestinal illness in the 3 days before enrollment to ensure the analysis included incident episodes. We conducted a log-linear trend test to assess whether gastrointestinal illness risk increased linearly from (1) not swimming, (2) swimming with no coliphage exposure, (3) swimming with exposure to coliphage, to (4) swimming with exposure to coliphage and enterococci >35 CFU/100 ml.62
Secondary Analyses
We conducted secondary analyses with alternative exposure and outcome and reference group definitions to assess the robustness of our findings. First, we estimated CIRs using nonswimmers as the reference group. Second, to assess whether greater water exposure had stronger associations with illness, we estimated CIRs among swimmers who immersed their head and who swallowed water. Third, we estimated CIRs for diarrhea instead of gastrointestinal illness. Finally, to detect residual confounding and/or differential outcome reporting bias, we conducted a negative control analysis among nonswimmers,63,64 expecting that coliphage presence assigned to nonswimmers would not be associated with increased illness among nonswimmers if no such confounding occurred. Finally, because the number of samples collected per day varied between beaches, our categorization of coliphage presence/absence may have diluted CIRs by ignoring the frequency of coliphage detection each day. We estimated the association between illness and swimming on days when >25% of samples contained detectable coliphage compared with days when no coliphage was detected. We repeated this analysis for days when >50% and >75% of samples had detectable coliphage.
RESULTS
Study Population
The studies enrolled 7,317 beachgoers at Avalon, 11,719 at Doheny, 7,254 at Malibu, 12,469 at Mission Bay, 2,977 at Goddard, and 2,022 at Fairhope Beach. Forty-four percent of beachgoers entered the water to waist depth or deeper and were classified as swimmers (eTable 5; http://links.lww.com/EDE/B203). The percentage of swimmers who swallowed water at each beach ranged from 7% to 14%. The self-reported average time in the water among people with any water contact at each beach ranged from 46 to 118 minutes.
Water Quality
A total of 1,818 water samples were analyzed for coliphage across the six beaches. Somatic coliphage (EPA 1601) was detected in 42%–81% of samples across beaches; somatic coliphage (EPA 1602) was detected in 27%–54% of samples; male-specific coliphage (EPA 1601) was detected in 11%–64% of samples, and male-specific coliphage (EPA 1602) was detected in 2%–3% of samples (Table 1). At Avalon and Doheny beaches, the only beaches where assays were run for both somatic and male-specific coliphage, the geometric mean of somatic coliphage levels was higher than for male-specific coliphage. Combining data from Avalon and Doheny, the geometric mean for somatic coliphage was 1.3 MPN/100 ml (SD = 99) for EPA 1601 and 1.3 MPN/100 ml (SD = 22) for EPA 1602; the geometric mean for male-specific coliphage was 0.6 MPN/100 ml (SD = 4) for EPA 1601 and 1.0 MPN/100 ml (SD = 2) for EPA 1602. The geometric mean of each type of coliphage was similar whether or not conditions were human-impacted except for male-specific coliphage (EPA 1601), for which the geometric mean was 1.34 MPN/100 ml when conditions were human-impacted and 0.88 MPN/100 ml otherwise (eTable 6; http://links.lww.com/EDE/B203).
TABLE 1.
Coliphage Concentrations at Each Beach Where They Were Measured (PFU/100 ml)
When enterococci levels were >35 CFU/100 ml, somatic coliphage (EPA 1601 or 1602) was detected in 78% of samples (N = 2 beaches) and male-specific coliphage (EPA 1601 or 1602) in 73% of samples (N = 5 beaches). When enterococci levels were ≤35 CFU/100 ml, studies detected somatic coliphage (EPA 1601 and 1602) in 72% of samples (N = 2 beaches) and male-specific coliphage (EPA 1601 and 1602) in 79% of samples (N = 5 beaches). The Spearman rank correlation for log10 somatic coliphage and log10 enterococci levels was 0.12 (EPA 1601) and 0.22 (EPA 1602); for male-specific coliphage, the coefficient was 0.08 (EPA 1601) and 0.13 (EPA 1602).
Gastrointestinal Illness Onset During Follow-up
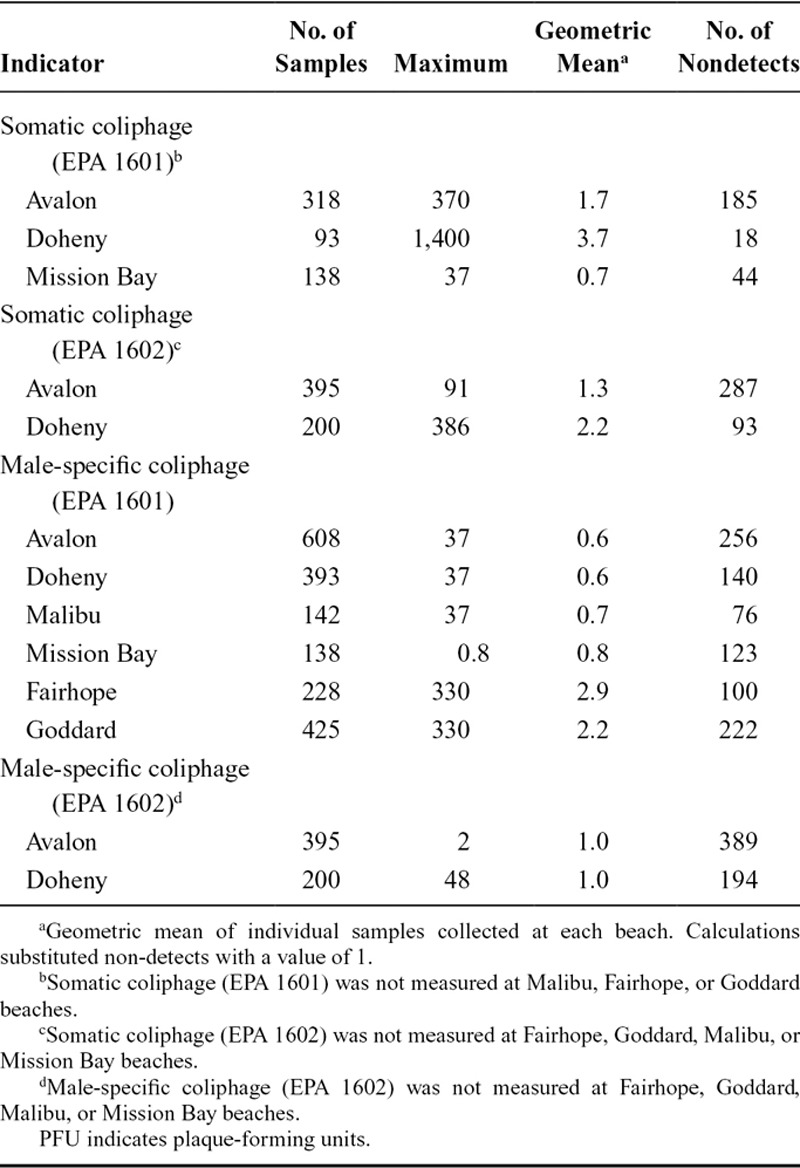
The cumulative incidence of gastrointestinal illness was 6.5% among all beachgoers (N = 33,261), 5.8% among nonswimmers (N = 12,633), and 7.2% among swimmers (N = 15,276). Among swimmers, the incidence was 7.3% at Avalon, 6.4% at Doheny, 9.0% at Fairhope, 6.2% at Goddard, 7.4% at Malibu, and 8.0% at Mission Bay beach. The incidence was lowest among nonswimmers and highest among swimmers in waters with detectable coliphage and/or enterococci (Figure 1). We found evidence of a log-linear trend in illness for both types of coliphage when comparing nonswimmers to swimmers in waters with and without coliphage and enterococci; the P values for the tests of trend were 0.017 for somatic coliphage and 0.013 for male-specific coliphage.
FIGURE 1.
Probability of gastrointestinal illness among beachgoers in waters with and without detectable coliphage. These results combine results from EPA 1601 and 1602 assays. The probability estimates are unadjusted, and confidence intervals were constructed with robust standard errors adjusted for clustering at the household level.
Association Between Coliphage and Gastrointestinal Illness
Threshold Analysis
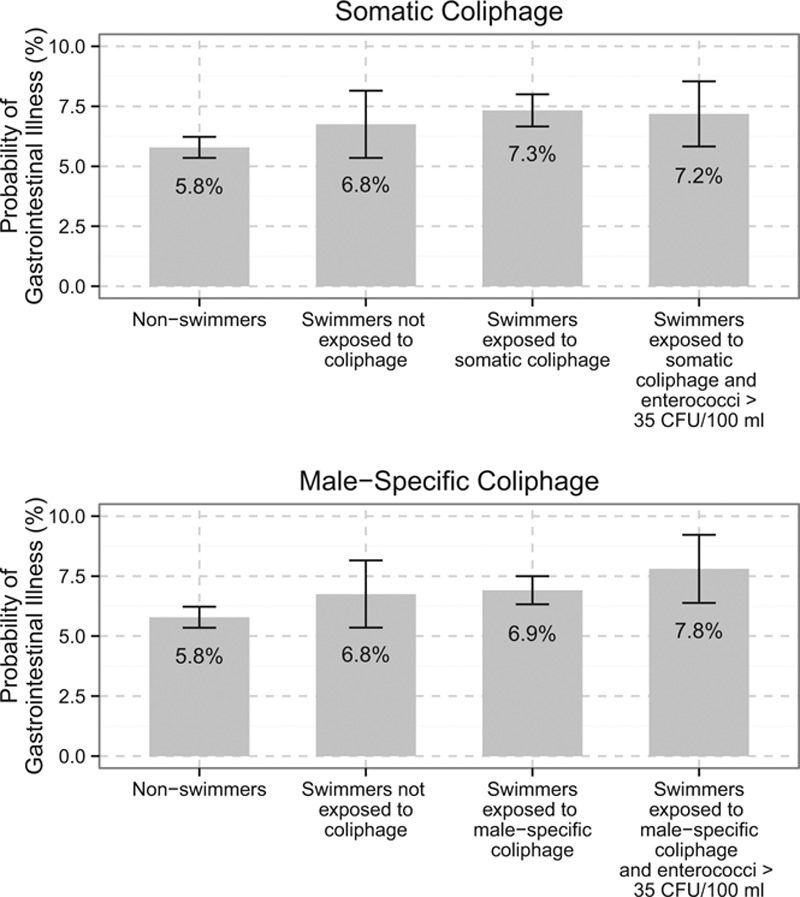
Approximately 75% (N = 10,678) and 65% (N = 14,422) of body immersion swimmers swam in waters where somatic and male-specific coliphage, respectively, were present. Under human-impacted conditions, coliphage presence was associated with increased gastrointestinal illness (Figure 2, eTable 7; http://links.lww.com/EDE/B203); the CIR was 1.39 (95% confidence interval [CI] 0.95, 2.03) for somatic coliphage and 1.28 (95% CI 0.83, 1.97) for male-specific coliphage. Results were similar when we stratified by EPA 1601 and 1602 (eFigure 2; http://links.lww.com/EDE/B203). This was similar to the pattern for the enterococci threshold analysis (>35 CFU/100 ml), where we observed no association with illness under not-human-impacted conditions, but an association with illness was present under human-impacted conditions. We found no evidence of increased gastrointestinal illness associated with the joint indicator for the coliphage presence and enterococci levels >35 CFU/100 ml under not human-impacted conditions or across all conditions (Figure 2, eTable 7; http://links.lww.com/EDE/B203). However, there was an association under human-impacted conditions: the CIR for somatic coliphage presence and enterococci >35 CFU/100 ml was 1.83 (95% CI 1.19, 2.82), and the CIR for male-specific coliphage presence and enterococci >35 CFU/100 was 1.48 (95% 1.04, 2.11) relative to days when coliphage was absent and enterococci was <35 CFU/100 ml.
FIGURE 2.
Cumulative gastrointestinal illness incidence ratios for presence of coliphage and enterococci levels >35 CFU/100 ml. aThese results combine results from EPA 1601 and 1602 assays. bBeaches included in adjacent point estimates: A, Avalon beach; D, Doheny beach; Ma, Malibu beach; Mb, Mission Bay beach; F, Fairhope beach; G, Goddard beach. cNot human-impacted conditions: The berm was closed at Doheny beach or the groundwater flow was below the median at Avalon beach. Human fecal pollution was considered to be unlikely on all study days at Mission Bay and Malibu beaches. dHuman-impacted conditions: The berm was open at Doheny beach or the groundwater flow was above median at Avalon beach. Human fecal pollution was considered to be likely on all study days at Fairhope and Goddard beaches. eCumulative incidence ratios were estimated for gastrointestinal illness among swimmers and were adjusted for age, sex, race, presence of chronic gastrointestinal illness, any contact with animals, and consumption of undercooked eggs, meat, or fish.
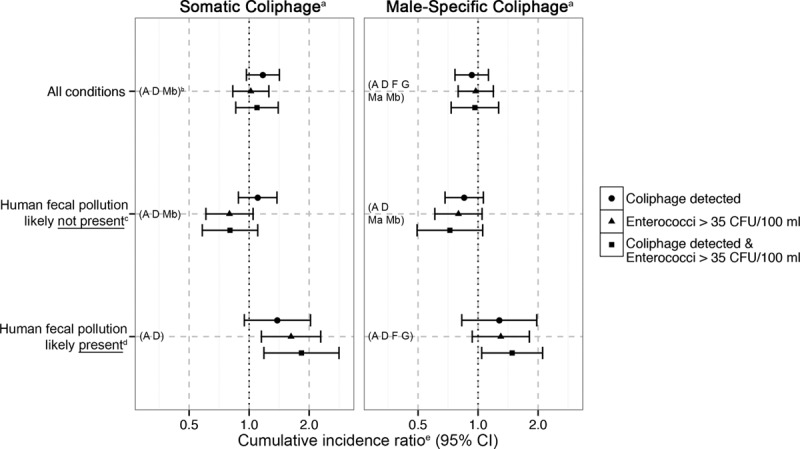
Continuous Analysis
Neither somatic coliphage level (PFU 100/ml) measured by EPA 1601 nor enterococci level (CFU/100 ml) measured at the same subset of beaches was associated with gastrointestinal illness. At the two beaches where somatic coliphage level was measured with EPA 1602, illness risk increased as the level of coliphage or enterococci increased under human-impacted conditions (Table 2, eFigure 3; http://links.lww.com/EDE/B203): for coliphage, the CIR for a 1-log10 increase was 1.27 (95% CI 0.92, 1.76), and for enterococci it was 1.21 (95% CI 1.01, 1.46). Under human-impacted conditions, there was no association with illness for male-specific coliphage (EPA 1601) or enterococci levels (Table 2, eFigure 4; http://links.lww.com/EDE/B203). Under human-impacted conditions, the association with illness was stronger for male-specific coliphage (EPA 1602) levels than for enterococci levels, but the CIs overlapped substantially: the CIR for a 1-log10 increase in male-specific coliphage (EPA 1602) was 2.20 (95% CI 1.30, 3.71); for enterococci levels measured at the same two beaches, the CIR was 1.21 (95% CI 1.01, 1.46).
TABLE 2.
Cumulative Incidence Ratio for Gastrointestinal Illness and Levels of Coliphage and Enterococci
Secondary Analyses
Our secondary analyses produced similar results to our primary analysis and in some cases associations were stronger. Using nonswimmers as the reference group, CIRs were slightly higher than when using swimmers not exposed to coliphage as the reference (eFigure 5; http://links.lww.com/EDE/B203). CIRs for swimmers who immersed their head and swallowed water were similar under not human-impacted conditions; under human-impacted conditions, associations were stronger than for swimmers, and somatic coliphage was associated with a 1.70-fold increase in gastrointestinal illness risk (95% CI 1.07, 2.69) for head immersion swimmers and 3.08-fold increase in risk (95% CI 1.40, 6.78) for swimmers who swallowed water. CIRs for diarrhea were similar overall to those for gastrointestinal illness and slightly higher under human-impacted conditions. Our negative control analysis among nonswimmers found no association with coliphage presence (eFigures 6–8; http://links.lww.com/EDE/B203). Our analysis using indicators for whether >25%, >50%, or >75% of samples per day contained detectable coliphage produced similar results to the threshold analysis (eFigure 9; http://links.lww.com/EDE/B203).
DISCUSSION
For both coliphage and enterococci, associations with gastrointestinal illness were only observed under human-impacted conditions. Under those conditions, we found some evidence that the gastrointestinal illness risk associated with a log10 increase in male-specific coliphage was greater than the risk associated with a log10 increase in culturable enterococci. Somatic coliphage and enterococci results were similar. Associations between coliphage presence and illness were stronger when we examined swimmers who immersed their head or swallowed water. Prior studies have also primarily found associations between illness and coliphage48,49,52,53,56 or enterococci7,8,50,55,58,65 when human fecal pollution was present. A possible explanation for this pattern is that few pathogens were present when there were no known sources of human fecal contamination (not human-impacted conditions). We found a slightly stronger association with gastrointestinal illness for the joint indicator for coliphage presence plus enterococci levels >35 CFU/100ml compared with the associations for single indicators (Figure 2). When enterococci levels were ≤35 CFU/100 ml, coliphage was detected in the majority of samples, indicating that viruses may be present below the water quality monitoring criterion level for enterococci.
We found no difference in the association with gastrointestinal illness for somatic and male-specific coliphage, which is inconsistent with several previous studies. Two studies that compared illness associations for both types of coliphage, datasets from which were included in this analysis, found stronger associations with illness for male-specific than somatic coliphage.50,51 There are biologic reasons why male-specific coliphage might have a stronger association with illness. Both types of coliphage have morphologic features similar to different types of enteric pathogens found in recreational water, although male-specific coliphages are morphologically similar to a larger number of enteric viruses than somatic coliphages.46 Some studies have found that male-specific coliphages have a stronger association with pathogens21; for adenovirus there was a stronger association with male-specific coliphage than somatic coliphage. Enterococci were also not associated with any viral pathogen. In this analysis, data from additional beaches beyond those analyzed in past studies50,51 were available for male-specific but not somatic coliphage. It is possible that if somatic coliphage data were available from additional beaches that we would have seen a difference in illness associations between the two types of coliphage.
We found that coliphage detected using EPA 1602 had a slightly stronger association with illness than coliphage detected using EPA 1601. EPA 1601 includes an enrichment step that may mask certain strains of coliphage, and as a result this method may fail to capture the diversity of coliphage strains in a sample.47 Thus, it is possible that EPA 1602, which does not include an enrichment step, is better able to capture the range of coliphages associated with enteric viruses than EPA 1601, which may have led to stronger associations with illness.
Our study includes several limitations typical of a prospective cohort design in which swimmers are not randomly assigned to enter the water with different levels of coliphage or enterococci, creating the potential for unmeasured confounding. However, our negative control analysis among nonswimmers found no association with coliphage presence, indicating that any residual confounding or differential outcome reporting bias was unlikely to explain the associations estimated in this study.63,64 Observational studies are also subject to misclassification due to self-reporting of exposures and outcomes; we would expect outcome misclassification to be independent of coliphage levels (i.e., nondifferential), which would have biased results toward a null finding.66,67
Our finding that the association between coliphage and gastrointestinal illness was not much stronger under all conditions than the association for culturable enterococci could have resulted from two study design attributes that could have caused associations to be underestimated. First, the data we analyzed detected coliphage in 100 ml volumes that are typically used for quantifying enterococci. This volume might be appropriate for routine beach monitoring because clogging of filters can be problematic using higher volumes. However, that volume may be suboptimal for assessing the association between coliphage and illness because coliphage occurs at lower densities than enterococci in the human intestine. Using larger volume methods such as dead-end hollow-tube fiber cartridges that improve detection of coliphage68 might have resulted in fewer nondetects, a more accurate exposure classification, and possibly a stronger association between coliphage and illness.
CONCLUSIONS
This pooled analysis is the largest evaluation to date of the association between coliphage in recreational water and gastrointestinal illness. We found an increased cumulative incidence of gastrointestinal illness among swimmers in waters with detectable coliphage when human fecal contamination was likely present, but not otherwise. Compared with associations with enterococci, associations were similar for somatic coliphage, and there was some evidence for a stronger association with male-specific coliphage. This study highlights the potential utility of coliphage as a predictor of gastrointestinal illness when human fecal contamination is likely present. Given the paucity of data on different coliphage types, coliphages should be included in future ambient recreational water epidemiologic analyses.
ACKNOWLEDGMENTS
We thank the following for their contributions to the processing of water samples for this study: Mark Sobsey, Dave Love, Roberto Rodriguez, Jill Stewart, Jerold W. Dickerson Jr., Laura F. Webster, and the National Oceanic and Atmospheric Administration Oceans and Human Health Initiative for their support of coliphage analyses.
Supplementary Material
Footnotes
This study was funded by the National Institutes of Health (R03-HD076066). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors report no conflicts of interest.
The views expressed in this manuscript are those of the authors and do not necessarily reflect the views or policies of the US Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Data and replication scripts are available from the authors on request.
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
REFERENCES
- 1.Boehm AB, Ashbolt NJ, Colford JM, et al. A sea change ahead for recreational water quality criteria. J Water Health. 2009;7:9–20. [DOI] [PubMed] [Google Scholar]
- 2.Wade TJ, Pai N, Eisenberg JN, Colford JM., JrDo U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ Health Perspect. 2003;111:1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang SC, Chu W, He JW.Seasonal detection of human viruses and coliphage in Newport Bay, California. Appl Environ Microbiol. 2007;73:6468–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinclair RG, Jones EL, Gerba CP.Viruses in recreational water-borne disease outbreaks: a review. J Appl Microbiol. 2009;107:1769–1780. [DOI] [PubMed] [Google Scholar]
- 5.Arnold BF, Schiff KC, Griffith JF, et al. Swimmer illness associated with marine water exposure and water quality indicators: impact of widely used assumptions. Epidemiology. 2013;24:845–853. [DOI] [PubMed] [Google Scholar]
- 6.Arnold BF, Wade TJ, Benjamin-Chung J, et al. Acute gastroenteritis and recreational water: highest burden among young US children. Am J Public Health. 2016;106:1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colford JM, Jr, Schiff KC, Griffith JF, et al. Using rapid indicators for Enterococcus to assess the risk of illness after exposure to urban runoff contaminated marine water. Water Res. 2012;46:2176–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yau VM, Schiff KC, Arnold BF, et al. Effect of submarine groundwater discharge on bacterial indicators and swimmer health at Avalon Beach, CA, USA. Water Res. 2014;59:23–36. [DOI] [PubMed] [Google Scholar]
- 9.Anderson KL, Whitlock JE, Harwood VJ.Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol. 2005;71:3041–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ.Enterococci in the environment. Microbiol Mol Biol Rev. 2012;76:685–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson DM, Weisberg SB, Hagedorn C, et al. Enterococcus growth on eelgrass (Zostera marina); implications for water quality. FEMS Microbiol Ecol. 2016;92:fiw047. [DOI] [PubMed] [Google Scholar]
- 12.Fujioka R, Sian-Denton C, Borja M, Castro J, Morphew K.Soil: the environmental source of Escherichia coli and Enterococci in Guam’s streams. J Appl Microbiol. 1998;85(Suppl 1):83S–89S. [DOI] [PubMed] [Google Scholar]
- 13.Yamahara KM, Walters SP, Boehm AB.Growth of enterococci in unaltered, unseeded beach sands subjected to tidal wetting. Appl Environ Microbiol. 2009;75:1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payment P, Plante R, Cejka P.Removal of indicator bacteria, human enteric viruses, Giardia cysts, and Cryptosporidium oocysts at a large wastewater primary treatment facility. Can J Microbiol. 2001;47:188–193. [PubMed] [Google Scholar]
- 15.Sobsey MD.Inactivation of health-related microorganisms in water by disinfection processes. Water Sci Technol. 1989;21:179–195. [Google Scholar]
- 16.Yamahara KM, Sassoubre LM, Goodwin KD, Boehm AB.Occurrence and persistence of bacterial pathogens and indicator organisms in beach sand along the California coast. Appl Environ Microbiol. 2012;78:1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitman RL, Przybyla-Kelly K, Shively DA, Nevers MB, Byappanahalli MN.Hand-mouth transfer and potential for exposure to E. coli and F+ coliphage in beach sand, Chicago, Illinois. J Water Health. 2009;7:623–629. [DOI] [PubMed] [Google Scholar]
- 18.Goyal SM, Adams WN, O’Malley ML, Lear DW.Human pathogenic viruses at sewage sludge disposal sites in the Middle Atlantic region. Appl Environ Microbiol. 1984;48:758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble RT, Fuhrman JA.Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia. 2001;460:175–184. [Google Scholar]
- 20.Vantarakis AC, Papapetropoulou M.Detection of enteroviruses and adenoviruses in coastal waters of SW Greece by nested polymerase chain reaction. Water Res. 1998;32:2365–2372. [Google Scholar]
- 21.Wu J, Long SC, Das D, Dorner SM.Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J Water Health. 2011;9:265–278. [DOI] [PubMed] [Google Scholar]
- 22.Borrego JJ, Moriñigo MA, de Vicente A, Córnax R, Romero P.Coliphages as an indicator of faecal pollution in water. Its relationship with indicator and pathogenic microorganisms. Water Res. 1987;21:1473–1480. [Google Scholar]
- 23.Dhillon TS, Dhillon EK, Chau HC, Li WK, Tsang AH.Studies on bacteriophage distribution: virulent and temperate bacteriophage content of mammalian feces. Appl Environ Microbiol. 1976;32:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Environmental Protection Agency. Review of coliphages as possible indicators of fecal contamination for ambient water quality. Available at: http://water.epa.gov/scitech/swguidance/standards/criteria/health/microbial/upload/coliphages-literature-review-report-2015.pdf. Published 2015. Accessed 2 November 2015.
- 25.Kott Y, Roze N, Sperber S, Betzer N.Bacteriophages as viral pollution indicators. Water Res. 1974;8:165–171. [Google Scholar]
- 26.Silverman AI, Peterson BM, Boehm AB, McNeill K, Nelson KL.Sunlight inactivation of human viruses and bacteriophages in coastal waters containing natural photosensitizers. Environ Sci Technol. 2013;47:1870–1878. [DOI] [PubMed] [Google Scholar]
- 27.Vaughn JM, Metcalf TG.Coliphages as indicators of enteric viruses in shellfish and shellfish raising estuarine waters. Water Res. 1975;9:613–616. [Google Scholar]
- 28.Griffin DW, Lipp EK, McLaughlin MR, Rose JB.Marine recreation and public health microbiology: quest for the ideal indicator. BioScience. 2001;51:817–825. [Google Scholar]
- 29.Jofre J, Lucena F, Blanch AR, Muniesa M.Coliphages as model organisms in the characterization and management of water resources. Water. 2016;8:199. [Google Scholar]
- 30.Mocé-Llivina L, Lucena F, Jofre J.Enteroviruses and bacteriophages in bathing waters. Appl Environ Microbiol. 2005;71:6838–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simková A, Cervenka J.Coliphages as ecological indicators of enteroviruses in various water systems. Bull World Health Organ. 1981;59:611–618. [PMC free article] [PubMed] [Google Scholar]
- 32.Stetler RE.Coliphages as indicators of enteroviruses. Appl Environ Microbiol. 1984;48:668–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabow W.Indicator systems for assessment of the virological safety of treated drinking water. Water Sci Tech. 1986;18:159–165. [Google Scholar]
- 34.Havelaar AH, Pot-Hogeboom WM, Furuse K, Pot R, Hormann MP.F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J Appl Bacteriol. 1990;69:30–37. [DOI] [PubMed] [Google Scholar]
- 35.Leclerc H, Edberg S, Pierzo V, Delattre JM.Bacteriophages as indicators of enteric viruses and public health risk in groundwaters. J Appl Microbiol. 2000;88:5–21. [DOI] [PubMed] [Google Scholar]
- 36.Brion GM, Meschke JS, Sobsey MD.F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 2002;36:2419–2425. [DOI] [PubMed] [Google Scholar]
- 37.Grabow WO, Taylor MB, de Villiers JC.New methods for the detection of viruses: call for review of drinking water quality guidelines. Water Sci Technol. 2001;43:1–8. [PubMed] [Google Scholar]
- 38.Tallon P, Magajna B, Lofranco C, Leung KT.Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Pollut. 2005;166:139–166. [Google Scholar]
- 39.Havelaar AH.Bacteriophages as model organisms in water treatment. Microbiol Sci. 1987;4:362–364. [PubMed] [Google Scholar]
- 40.Environmental Protection Agency. Method 1601: male-specific (F+) and somatic coliphage in water by two-step enrichment procedure. Available at: http://water.epa.gov/scitech/methods/cwa/bioindicators/upload/2008_11_25_methods_method_biological_1601.pdf. Published 2001. Accessed 3 November 2015.
- 41.Environmental Protection Agency. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. Available at: http://water.epa.gov/scitech/methods/cwa/bioindicators/upload/2008_11_25_methods_method_biological_1602.pdf. Published 2001. Accessed 3 November 2015.
- 42.Fong TT, Lipp EK.Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev. 2005;69:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love DC, Sobsey MD.Simple and rapid F+ coliphage culture, latex agglutination, and typing assay to detect and source track fecal contamination. Appl Environ Microbiol. 2007;73:4110–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobsey MD, Schwab KJ, Handzel TR.A simple membrane filter method to concentrate and enumerate male-specific RNA coliphages. J Am Water Works Assoc. 1990;82:52–59. [Google Scholar]
- 45.Wentsel RS, O’Neill PE, Kitchens JF.Evaluation of coliphage detection as a rapid indicator of water quality. Appl Environ Microbiol. 1982;43:430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King A, Adams M, Carstens E, Lefkowitz E.Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. 2011London, UK: Elsevier; [Google Scholar]
- 47.Stewart-Pullaro J, Daugomah JW, Chestnut DE, Graves DA, Sobsey MD, Scott GI.F+ RNA coliphage typing for microbial source tracking in surface waters. J Appl Microbiol. 2006;101:1015–1026. [DOI] [PubMed] [Google Scholar]
- 48.Abdelzaher AM, Wright ME, Ortega C, et al. Daily measures of microbes and human health at a non-point source marine beach. J Water Health. 2011;9:443–457. [DOI] [PubMed] [Google Scholar]
- 49.van Asperen IA, Medema G, Borgdorff MW, Sprenger MJ, Havelaar AH.Risk of gastroenteritis among triathletes in relation to faecal pollution of fresh waters. Int J Epidemiol. 1998;27:309–315. [DOI] [PubMed] [Google Scholar]
- 50.Colford JM, Jr, Wade TJ, Schiff KC, et al. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology. 2007;18:27–35. [DOI] [PubMed] [Google Scholar]
- 51.Griffith JF, Weisberg SB, Arnold BF, Cao Y, Schiff KC, Colford JM., JrEpidemiologic evaluation of multiple alternate microbial water quality monitoring indicators at three California beaches. Water Res. 2016;94:371–381. [DOI] [PubMed] [Google Scholar]
- 52.Lee JV, Dawson SR, Ward S, Surman SB, Neal KR.Bacteriophages are a better indicator of illness rates than bacteria amongst users of a white water course fed by a lowland river. Water Sci Technol. 1997;35:165–170. [Google Scholar]
- 53.Medema GJ, van Asperen IA, Klokman-Houweling JM, Nooitgedagt A, van de Laar MJW, Havelaar AH.The relationship between health effects in triathletes and microbiological quality of freshwater. Water Sci Technol. 1995;31:19–26. [Google Scholar]
- 54.von Schirnding YE, Kfir R, Cabelli V, Franklin L, Joubert G.Morbidity among bathers exposed to polluted seawater. A prospective epidemiological study. S Afr Med J. 1992;81:543–546. [PubMed] [Google Scholar]
- 55.Wade TJ, Sams E, Brenner KP, et al. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ Health. 2010;9:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiedenmann A, Krüger P, Dietz K, López-Pila JM, Szewzyk R, Botzenhart K.A randomized controlled trial assessing infectious disease risks from bathing in fresh recreational waters in relation to the concentration of Escherichia coli, intestinal enterococci, Clostridium perfringens, and somatic coliphages. Environ Health Perspect. 2006;114:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Environmental Protection Agency. 2012 recreational water quality criteria. Available at: http://water.epa.gov/scitech/swguidance/standards/criteria/health/recreation/upload/RWQC2012.pdf. Published 2012. Accessed 2 November 2015.
- 58.Wade TJ, Calderon RL, Sams E, et al. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ Health Perspect. 2006;114:24–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou G.A modified Poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004;159:702–706. [DOI] [PubMed] [Google Scholar]
- 60.Robins JM, Greenland S.Identifiability and exchangeability for direct and indirect effects. Epidemiology. 1992;3:143–155. [DOI] [PubMed] [Google Scholar]
- 61.Hedges LV, Vevea JL.Fixed- and random-effects models in meta-analysis. Psychol Methods. 1998;3:486–504. [Google Scholar]
- 62.Vittinghoff E, Glidden DV, Shiboski SC, McCulloch CE.Regression Methods in Biostatistics: Linear, Logistic, Survival, and Repeated Measures Models. 2011New York, NY: Springer Science & Business Media; [Google Scholar]
- 63.Arnold BF, Ercumen A, Benjamin-Chung J, Colford JM., JrBrief Report: Negative controls to detect selection bias and measurement bias in epidemiologic studies. Epidemiology. 2016;27:637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lipsitch M, Tchetgen Tchetgen E, Cohen T.Negative controls: a tool for detecting confounding and bias in observational studies. Epidemiology. 2010;21:383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wade TJ, Calderon RL, Brenner KP, et al. High sensitivity of children to swimming-associated gastrointestinal illness: results using a rapid assay of recreational water quality. Epidemiology. 2008;19:375–383. [DOI] [PubMed] [Google Scholar]
- 66.Fleisher JM.The effects of measurement error on previously reported mathematical relationships between indicator organism density and swimming-associated illness: a quantitative estimate of the resulting bias. Int J Epidemiol. 1990;19:1100–1106. [DOI] [PubMed] [Google Scholar]
- 67.Hutcheon JA, Chiolero A, Hanley JA.Random measurement error and regression dilution bias. BMJ. 2010;340:c2289. [DOI] [PubMed] [Google Scholar]
- 68.Rhodes ER, Hamilton DW, See MJ, Wymer L.Evaluation of hollow-fiber ultrafiltration primary concentration of pathogens and secondary concentration of viruses from water. J Virol Methods. 2011;176:38–45. [DOI] [PubMed] [Google Scholar]


