Abstract
Background
Standard diagnosis of urinary tract infection (UTI) via urine culture for pathogen identification (ID) and antimicrobial susceptibility testing (AST) takes 2–3 d. This delay results in empiric treatment and contributes to the misuse of antibiotics and the rise of resistant pathogens. A rapid diagnostic test for UTI may improve patient care and antibiotic stewardship.
Objective
To develop and validate an integrated biosensor assay for UTI diagnosis, including pathogen ID and AST, with determination of the minimum inhibitory concentration (MIC) for ciprofloxacin.
Design, setting, and participants
Urine samples positive for Enterobacteriaceae (n = 84) or culture-negative (n = 23) were obtained from the Stanford Clinical Microbiology Laboratory between November 2013 and September 2014. Each sample was diluted and cultured for 5 h with and without ciprofloxacin, followed by quantitative detection of bacterial 16S rRNA using a single electrochemical biosensor array functionalized with a panel of complementary DNA probes. Pathogen ID was determined using universal bacterial, Enterobacteriaceae (EB), and pathogen-specific probes. Phenotypic AST with ciprofloxacin MIC was determined using an EB probe to measure 16S rRNA levels as a function of bacterial growth.
Measurements
Electrochemical signals for pathogen ID at 6 SD over background were considered positive. An MIC signal of 0.4 log units lower than the no-antibiotic control indicated sensitivity. Results were compared to clinical microbiology reports.
Results and limitations
For pathogen ID, the assay had 98.5% sensitivity, 96.6% specificity, 93.0% positive predictive value, and 99.3% negative predictive value. For ciprofloxacin MIC the categorical and essential agreement was 97.6%. Further automation, testing of additional pathogens and antibiotics, and a full prospective study will be necessary for translation to clinical use.
Conclusions
The integrated biosensor platform achieved microbiological results including MIC comparable to standard culture in a significantly shorter assay time. Further assay automation will allow clinical translation for rapid molecular diagnosis of UTI.
Patient summary
We have developed and validated a biosensor test for rapid diagnosis of urinary tract infections. Clinical translation of this device has the potential to significantly expedite and improve treatment of urinary tract infections.
Keywords: Urinary tract infection, Molecular diagnostics, Biosensing techniques, Microbial sensitivity tests, Point-of-care system
1. Introduction
Urinary tract infections (UTIs) are among the most common bacterial infections. Enterobacteriaceae species account for >80% of UTIs, with Escherichia coli accounting for approximately 75% and 65% of UTIs in ambulatory and hospitalized settings, respectively [1]. Diagnosis via urine culture requires a centralized clinical microbiology laboratory for pathogen identification (ID) and associated antimicrobial susceptibility testing (AST), expressed as the minimum inhibitory concentration (MIC) for an individual antibiotic. AST, based on antibiotic disk diffusion or microdilution, provides an interpretation of MIC results as sensitive, intermediate, or resistant for clinician guidance. The entire process for pathogen ID and AST typically requires 2–3 d. This gap between clinical presentation and the microbiology report leads to empiric prescription of antibiotics.
Evidence-based antibiotic selection for UTI may help to stem increases in antibiotic resistance [2]. Emblematic of poor antibiotic stewardship is trimethoprim/sulfamethoxazole (TMP/SMX). This oral antibiotic was previously the first-line treatment for UTI. However, since TMP/SMX resistance exceeds 20% [3] in North America, ciprofloxacin is recommended in current guidelines for complicated UTI as the resistance rate is lower for this antibiotic [4].
Point-of-care (POC) diagnosis of UTI has potential to improve patient care by rapidly identifying the causative pathogen and the best treatment choice. Electrochemical biosensors are an ideal basis for POC diagnostics as they provide rapid results and high sensitivity, and are small in size and inexpensive. We previously described a 1-h electrochemical biosensor assay for molecular identification of uropathogens [5,6]. Using a biosensor array functionalized with a panel of DNA probes targeting conserved and unique bacterial 16S rRNA sequences, urine samples containing single or multiple bacterial species (polymicrobial infections) can be identified directly [6] in a urine sample. Since the biosensor assay provides quantitative detection, we further adapted the assay for phenotypic AST by detecting differential 16S rRNA levels after brief culture of a sample in the presence and absence of antibiotic [7]. Using the biosensor-based approach, phenotypic AST of common uropathogens against standard bactericidal and bacteriostatic antimicrobials, including ampicillin, TMP/SMX, ciprofloxacin, gentamicin, and ceftriaxone, was demonstrated in prospectively collected urine samples [7].
For the biosensor assay, cells are lysed to release the rRNA that binds to capture and detector oligonucleotide probes at the sensor surface. Probe-rRNA hybridization is facilitated by electrokinetic (EK) hybridization. EK is a microfluidic sample preparation technique that generates electrothermal flow via Joule heating, thereby reducing the assay time and improving the signal-to-noise ratio [8]. Probe-rRNA complex formation is detected using an antibody-horseradish peroxidase HRP conjugate for generation of an electrochemical signal. With EK-facilitated hybridization, a limit of detection of 103 colony-forming units (cfu)/ml has been demonstrated [9].
In our previous report, ID and AST were conducted on separate biosensor arrays with different starting samples, urine for ID and cultured urine for AST. Here we report the development and validation of ID and AST assays integrated in a single biosensor assay focusing on detection of Enterobacteriaceae and AST for ciprofloxacin. We also demonstrate that growth quantitation by the integrated biosensor can provide ciprofloxacin MIC results comparable to those achieved by clinical microbiology. The combination of pathogen ID and AST in a single assay provides an effective strategy toward integration into a fully automated POC diagnostic for UTI.
2. Materials and methods
2.1. Clinical samples
Between November 2013 and September 2014, de-identified clinical samples containing ≥105 cfu/ml of a single Enterobacteriaceae species (n = 84) or without bacteria (n = 23) were obtained from the Stanford Clinical Microbiology Laboratory (Table 1). Samples had been preserved with boric acid and stored at 4 °C before testing. CHROMagar Orientation (BD Diagnostic Systems, Hunt Valley, MD, USA) was used for identification of Escherichia coli and MALDI Biotyper (Bruker, Billerica, MA, USA) for identification of other species. AST was performed via broth microdilution using VITEK2 (bioMerieux, Marcy-l’Étoile, France) for a panel of antibiotics.
Table 1.
Pathogen species and number of urine samples tested using the biosensor array
| Uropathogen | Samples tested | Ciprofloxacin suscepibility | ||
|---|---|---|---|---|
| (n) | S | I | R | |
| Escherichia coli | 24 | 12 | 12 | |
| Klebsiella pneumoniae | 24 | 24 | ||
| Proteus mirabilis | 11 | 9 | 1 | 1 |
| Enterobacter cloacae | 6 | 5 | 1 | |
| Citrobacter koseri | 4 | 3 | 1 | |
| Citrobacter freundii | 4 | 4 | ||
| Enterobacter aerogenes | 4 | 4 | ||
| Klebsiella oxytoca | 3 | 3 | ||
| Morganella morganii | 2 | 1 | 1 | |
| Raoultella ornithinolytica | 1 | 1 | ||
| Serratia marcescens | 1 | 1 | ||
S = sensitive; I = intermediate; R = resistant.
2.2. Urine culture for biosensor assay
Urine samples were diluted 100× in Muller-Hinton broth and inoculated into wells of ciprofloxacin MIC strips (Merlin Diagnostics, Berlin, Germany). The MIC strips comprised one well without antibiotic and five wells containing lyophilized ciprofloxacin for rehydration with 100 μl of sample to final ciprofloxacin concentrations of 0.25, 0.5, 1, 2, and 4 μg/ml. MIC strips were incubated for 5 h at 37 °C and then immediately tested by biosensor assay or frozen at −80 °C for later assay. Previous studies found no difference in biosensor signal between fresh and frozen cultures [7,10].
2.3. Biosensor functionalization
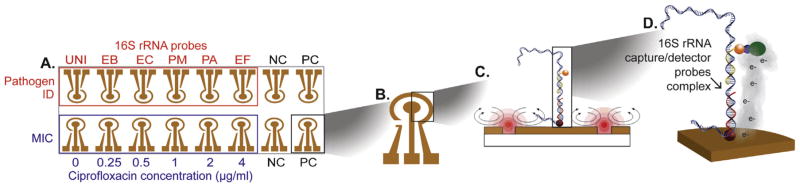
The biosensor chip (GeneFluidics, Irwindale, CA, USA) is composed of 16 individually addressable sensors. Sensors were functionalized with a ternary monolayer interface consisting of thiolated oligonucleotide capture probes, hexanedithiol, and 6-mercapto-1 hexanol [11]. Sensors 1–8 were used for pathogen ID and sensors 9–16 for AST (Fig. 1). Probes for pathogen ID included a universal (UNI) probe targeting a conserved region of bacterial 16S rRNA for detection of all eubacteria; an EB probe targeting a 16S rRNA sequence conserved in Enterobacteriaceae; and specific probes for E. coli (EC), Proteus mirabilis (PM), Pseodomonas aeruginosa (PA), and Enterococcus faecalis (EF) [12]. The EB probe was used to quantify 16S rRNA to measure bacterial growth for AST. Detector probes were dried on the auxiliary sensor surface to allow use of matched probe sets.
Fig. 1.
Schematic showing the integrated biosensor assay for pathogen identification (ID) and antimicrobial susceptibility testing (AST). (A) The biosensor array consists of 16 sensors functionalized with DNA probes for pathogen ID (top row). Sensors were functionalized with a universal bacterial probe (UNI), an Enterobacteriaceae (EB) probe, and probes for Escherichia coli (EC), Proteus mirabilis (PM), Pseudomonas aeruginosa (PA), and Enterococcus faecalis (EF). To determine the phenotypic AST (ciprofloxacin minimum inhibitory concentration [MIC]), the bottom row sensors were functionalized with EB probe to measure 16S rRNA levels after culture in the presence of increasing ciprofloxacin concentrations. The negative control (NC) and positive control (PC) represent electrochemical detection of matched or mismatched synthetic target oligonucleotides. (B) Each sensor is composed of a central working electrode and peripheral reference and auxiliary electrodes. (C) Sandwich hybridization between capture and detector probes with target rRNA binding is facilitated by electrokinetic (EK) heating and mixing to improve hybridization stringency. (D) Electrochemical signal output. Binding of the electrochemical signal transducer enzyme horseradish peroxidase (HRP) to the hybridization complex. Oxidation of the HRP substrate H2O2 and electron mediator under a fixed voltage generates an electroreduction current.
2.4. Biosensor assay
Cultured urine samples were lysed as previously described [7,12]. For pathogen ID, a no-antibiotic control culture was used and lysate was neutralized with 1 M Tris, pH 7.2, containing 2.5% bovine serum albumin (BSA). For AST, urine cultures were lysed and neutralized with 1 M Tris, pH 7.2, containing 2.5% BSA and 0.25 μM EB detector probe. Neutralized lysate was applied to the sensors and subjected to EK mixing and heating at 200 kHz and 7 V peak-to-peak for hybridization. EK was applied using a function generator (Hewlett Packard, Palo Alto, CA, USA) as follows: 10 min of EK, 5 min of rest, and 5 min of EK [12]. After hybridization, the assay was completed and the current output was read as previously described [6,10]. The biosensor assay required less than 1 h to complete.
2.5. Data analysis
Clinical microbiology ID and MIC were used as the comparison standard. For analysis, raw biosensor signals were log10-transformed for comparison to controls, and cutoffs were based on best concordance with standard, as indicated on an empirical receiver operating characteristic curve as previously described [6]. Pathogen ID signals that exceeded six time the standard deviation over the negative control were considered positive, and MIC signals 0.4 log10 units lower than the no-antibiotic growth control indicated sensitivity to ciprofloxacin. Analysis of biosensor data was categorized as the presence or absence of bacteria based on the biosensor signal from UNI and EB probes. The probe configuration on the biosensor allowed for speciation of E. coli, P. mirabilis, P. aeruginosa, and E. faecalis; other pathogens could be identified as Enterobacteriaceae or not. Sensitivity and specificity were calculated using data from all probes. AST results were analyzed only for samples positive for bacteria in the biosensor ID assay. AST results were evaluated on the basis of categorical agreement, with classification of sensitive, resistant, or intermediate in agreement with clinical microbiology; and essential agreement, with MIC the same or within a twofold dilution of the clinical microbiology result.
3. Results
We combined electrochemical biosensor assays for pathogen ID and AST into a single biosensor array. For integrated ID and AST assays, patient urine was diluted and cultured without and with ciprofloxacin. The culture without ciprofloxacin was used for pathogen ID and as the benchmark for growth for phenotypic AST. Growth inhibition with antibiotic, as measured by a lower biosensor signal, was indicative of an inhibitory concentration of ciprofloxacin. To further simplify the assay, we found that detector probes dried on the sensor surface were efficiently reconstituted on EK mixing. Whereas individual samples were prepared for each probe pair in our previous work, the dried probes allowed application of one lysate to each sensor for pathogen ID [12].
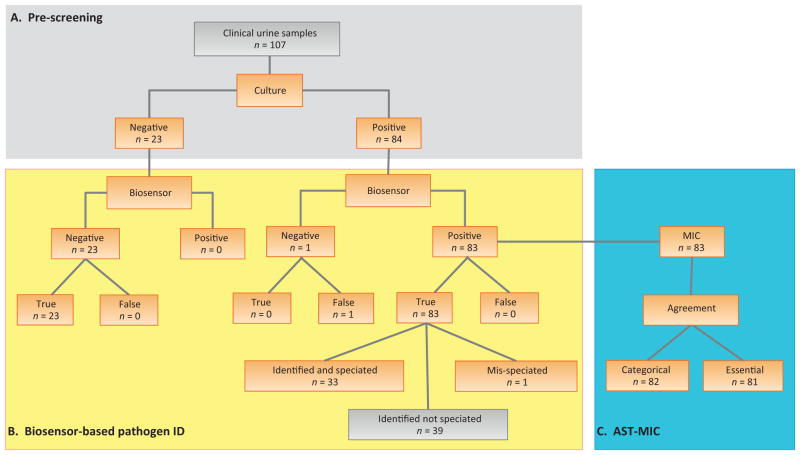
To validate the integrated ID-AST assay, we tested 107 patient-derived urine samples: 84 culture-positive samples and 23 culture-negative samples. Prescreening by clinical microbiology ensured that the validation set contained a variety of Enterobacteriaceae and susceptibility levels (Table 1). Negative samples were included to assess specificity. Analysis of 107 samples on the six sensors for pathogen ID yielded sensitivity and specificity results for a total of 642 probes pairs. Another six sensors were used to quantitate growth for AST, with four sensors for assay controls. While the ID and AST assays were integrated in a single biosensor chip, the data were analyzed independently. Pathogen ID was analyzed for all samples, but AST data were analyzed only for biosensor-positive samples.
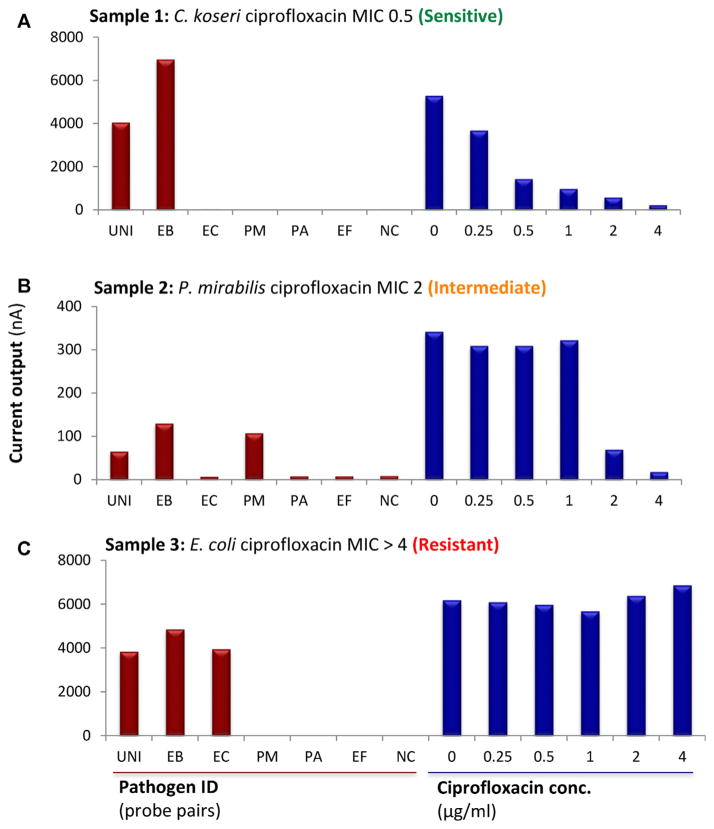
Figure 2 shows biosensor assay results for three representative urine samples. Sample 1 (Fig. 2A) was identified as Citrobacter koseri (an Enterobacteriacea) sensitive to ≤0.5 μg/ml ciprofloxacin by clinical microbiology. As there was no species-specific probe for C. koseri on the biosensor, only EB and UNI probes were appropriately positive. For AST, a significantly lower signal on addition of ciprofloxacin indicated sensitivity (MIC 0.5 μg/ml). For samples 2 and 3 (Fig. 2B,C) species-specific probes were available and appropriately positive, along with EB and UNI probes. The biosensor assays agreed with clinical microbiology in that the E. coli in sample 3 showed intermediate susceptibility to ciprofloxacin (MIC 2 μg/ml) and P. mirabilis in sample 2 was resistant to ciprofloxacin (MIC ≥4 μg/ml). Consistent with the quantitative nature of the biosensor, positive signals were higher for samples containing >108 cfu/ml of bacteria (Fig. 2A,C) and lower for a sample containing 4 × 107cfu/ml of bacteria (Fig. 2B).
Fig. 2.
Representative results for integrated biosensor pathogen identification and ciprofloxacin minimum inhibitory concentration (MIC) in clinical urine samples. (A) Sample 1 was positive for Citrobacter koseri (8 × 108 cfu/ml). The sensor array correctly detected C. koseri (an Enterobacteriacea) with the universal (UNI) and Enterobacteriaceae (EB) probes, but did not speciate the pathogen because the sensor did not include a specific probe for Citrobacter. Consistent with clinical microbiology results, the biosensor revealed a ciprofloxacin MIC of 0.5 μg/ml, whereby the signal decreased with increasing ciprofloxacin concentration. (B) Sample 2 was positive for Proteus mirabilis (4 × 107 cfu/ml). The sensor array identified and speciated the pathogen, showing expected positive signals for UNI, EB and Proteus mirabilis (PM) probes and intermediate sensitivity to ciprofloxacin, showing a reduced signal only at μg/ml. (C) Sample 3 was positive for Escherichia coli (2 × 108 cfu/ml). The array identified and speciated E. coli using the UNI, EB, and EC probes, and revealed resistance to ciprofloxacin, with no reduced signals on the MIC sensors, consistent with clinical microbiology results.
3.1. Pathogen ID
The ability to speciate pathogens in the biosensor assay depends on the availability of specific probes. The current biosensor configuration can discern two Enterobacteriaceae species: E. coli and P. mirabilis. In this set we correctly identified all E. coli (n = 24) and 10 of the 11 P. mirabilis samples. In one sample the predominant species was correctly identified as P. mirabilis; however, the biosensor assay also indicated a low level of E. coli that was not reported by clinical microbiology. One sample identified as P. mirabilis by clinical microbiology was misidentified as E. coli in the biosensor assay. However, plating on CHROMagar revealed that this sample appeared to contain a mixture of E. coli and P. mirabilis.
Of the 49 urine samples containing species other than E. coli or P. mirabilis, 48 were correctly identified as containing an Enterobacteriaceae species in the biosensor assay. One sample reported to have Klebsiella pneumoniae appeared negative for all biosensor probes. For 10 samples, Enterobacteriaceae infection was detected by the biosensor but a statistically significant signal was measured for a species-specific probe. In most cases this signal was quite weak compared to the EB and UNI signals and was probably due to cross-reactivity with high concentrations of bacteria. When analyzing the AST portion of the assay for these samples, the biosensor results agreed with clinical microbiology assessment, further suggesting probe cross-reactivity instead of sample contamination.
Overall, the biosensor assay correctly predicted the presence or absence of bacteria for 98% of the samples according to a positive signal for both the UNI and EB probes (Fig. 3). Analysis of all probes in the biosensor assay yielded sensitivity of 98.5%, specificity of 96.6%, positive predictive value of 93.0%, and negative predictive value of 99.3%.
Fig. 3.
Summary of the integrated biosensor assay results for clinical urine samples. Clinical microbiology results were used as the standard. For the minimum inhibitory concentration (MIC), data were analyzed on the basis of categorical and essential agreement. Categorical agreement evaluates the antimicrobial susceptibility profile as sensitive (S), resistant (R), or intermediate (I). Essential agreement evaluates the agreement with the MIC.
3.2. Antimicrobial susceptibility
Among the 83 biosensor-positive samples evaluated for MIC, clinical microbiology results for susceptibility to ciprofloxacin included 66 sensitive (MIC ≤1 μg/ml), 14 resistant (MIC ≥4 μg/ml), and three intermediate (MIC 2 μg/ml) strains. Compared to clinical microbiology, the biosensor MIC profile revealed 97.6% categorical agreement and essential agreement. One minor categorical error was identified as Enterobacter cloaceae with intermediate susceptibility (MIC 2 μg/ml) by clinical microbiology, but as an Enterobacteriaceae species sensitive to ciprofloxacin (MIC 1 μg/ml) in the biosensor assay. One essential agreement error was categorized as sensitive by both tests, but with MIC ≤0.25 μg/ml by clinical microbiology and MIC 1 μg/ml in the biosensor assay. The one P. mirabilis sample that was misidentified as E. coli was also misclassified by both categorical and essential agreement for AST.
4. Discussion
We previously demonstrated robust molecular identification of uropathogens and reliable phenotypic AST in electrochemical biosensor assays [6,7]. However, these assays were run independently, which added complexity to the test. To streamline the assays for integration into a future device with automated reagent delivery and readout, we report here the integration of ID and AST assays on a single biosensor array. Furthermore, we demonstrated additional strategies to facilitate system integration, including drying of the detector probes on the sensor surface to reduce the number and complexity of solutions needed in the assay, and EK-facilitated hybridization to improve probe hybridization and eliminate the need for an incubator. Validation of the integrated ID-AST assay with patient-derived urine samples confirmed that high levels of sensitivity and specificity were maintained, indicating incremental but significant steps toward the development of an integrated device for molecular UTI diagnosis.
Another advance achieved by the current work is the demonstration and validation of biosensor-based MIC measurement. We previously reported biosensor-based AST using a single concentration of antibiotic for classification as resistant or sensitive. For better comparison with clinical microbiology, which reports the MIC, the biosensor assay was further refined. We selected ciprofloxacin for biosensor MIC as it is commonly prescribed for treatment of UTIs. For uncomplicated UTI, TMP/SMX was the first-line treatment, but because of increasing resistance rates in North America and Europe, guidelines suggest prescription of a different antibiotic [4,13] and ciprofloxacin is the first-line treatment in many clinics.
To ensure that our validation would test a variety of pathogens and susceptibilities, we used patient urine samples that were first prescreened using clinical microbiology. These samples had been preserved with boric acid and refrigerated according to standard clinical protocol, which necessitated a longer culture period to account for the extended lag phase before bacteria re-enter the growth phase and to achieve an adequate growth differential for AST. On the basis of our previous data, the 5-h culture time reported here can be reduced to 2.5 h for fresh urine samples, even when testing a wide array of antibiotics [7].
Since a no-antibiotic growth control was used for pathogen ID, skin flora in urine samples might be amplified, leading to false positive results. However, use of cultured urine did not increase the false positive rate of the assay. Since we diluted the urine 100-fold before culture, it is likely that any contaminating flora was diluted to below the detection level of our biosensor, even for brief culture. In some culture-positive samples, we detected low-level signals for nonspecific probes. Previous experience suggests this is due to probe cross-reactivity at high concentration rather than amplification of a contaminant. However, in some samples the low secondary signal may indicate a polymicrobial infection. In the eventual clinical use of the biosensor assay, a positive secondary signal may be an indication for standard culture and susceptibility analysis.
One strength of this assay is the flexibility for modification to better suit the needs of the patient population. While this study focused on detection and AST of the most common class of uropathogens, Enterobacteriaceae, we previously developed and validated probes for other common Gram-positive and Gram-negative uropathogens [12]. Each electrode on the biosensor is individually addressable; therefore, the panel of species-specific probes can be specifically designed to meet clinical needs. Antibiotic resistance also varies over time, location, and patient population. Testing of additional antibiotics instead of a single broad-spectrum antibiotic may help to promote more judicious use of ciprofloxacin. Our prior study with a panel of single-concentration antibiotics indicated that this test can be easily modified for MIC of other antibiotics [7].
Prospective testing of a fully automated iteration of the biosensor assay will allow us to standardize data analysis and to assess probes for detection of additional species and culture conditions for other antibiotics. For the automated system, microfluidics necessary for sample manipulation and software for system control are under development. Ideally, the system would allow users to choose a biosensor configuration to best meet clinical needs. Users would then simply insert a biosensor in the manifold, apply a sample, and start a program for sample manipulation, assay, and analysis.
The shortcomings of urine culture and the need for rapid UTI diagnosis have generated wide research interest in developing POC diagnostics for infection. POC diagnostics for infection are unlikely to completely replace clinical microbiology culture and susceptibility analysis in the near future. Patient populations that might significantly benefit from initiation of treatment based on a POC diagnostic for UTI include urology clinic patients who present with complicated UTI and preoperative patients. Many strategies for POC devices are in development and warrant future comparative analysis [14–17]. We have demonstrated that our biosensor assay can provide reliable results for both pathogen ID and ciprofloxacin MIC, thus encouraging future studies for complete system integration and expanded validation with other pathogens and antibiotics with prospectively collected urine samples.
5. Conclusions
This integrated biosensor platform is a promising tool for UTI management, with diagnostic accuracy similar to that of the standard culture-based approach. By significantly reducing the time for pathogen ID and MIC from 2–3 d to < 6 h, this assay has the potential to improve patient care. Future work is needed to automate the integrated assay.
Acknowledgments
Funding/Support and role of the sponsor: Joseph Liao received a National Institutes of Health (NIH) grant (U01 AI082457). Vincent Gau received an NIH grant (R44 AI088756). Emanuela Altobelli received support from the Campus Biomedico Alumni Research Grant. The sponsors played a role in the design and conduct of the study.
Footnotes
Author contributions: Joseph C. Liao had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Liao, Mach.
Acquisition of data: Altobelli, Mohan, Mach, Anikst, Sin.
Analysis and interpretation of data: Altobelli, Mach, Mohan, Sin, Liao.
Drafting of the manuscript: Altobelli, Mach, Liao.
Critical revision of the manuscript for important intellectual content: Altobelli, Mach, Liao, Banaei, Mohan, Buscarini.
Statistical analysis: Mach.
Obtaining funding: Liao.
Administrative, technical, or material support: Wong, Gau, Banaei.
Supervision: Liao.
Other: None.
Financial disclosures: Joseph C. Liao certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Vincent Gau is co-founder and CEO of GeneFluidics. The remaining authors have nothing to disclose.
References
- 1.Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. 2010;7:653–60. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 2.Mach KE, Wong PK, Liao JC. Biosensor diagnosis of urinary tract infections: a path to better treatment? Trends Pharmacol Sci. 2011;32:330–6. doi: 10.1016/j.tips.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhanel GG, Hisanaga TL, Laing NM, et al. Antibiotic resistance in Escherichia coli outpatient urinary isolates: final results from the North American Urinary Tract Infection Collaborative Alliance (NAUTICA) Int J Antimicrob Agents. 2006;27:468–75. doi: 10.1016/j.ijantimicag.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Gupta K, Hooton TM, Naber KG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. 2011;52:e103–20. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 5.Liao JC, Mastali M, Gau V, et al. Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. J Clin Microbiol. 2006;44:561–70. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mach KE, Du CB, Phull H, et al. Multiplex pathogen identification for polymicrobial urinary tract infections using biosensor technology: a prospective clinical study. J Urol. 2009;182:2735–41. doi: 10.1016/j.juro.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mach KE, Mohan R, Baron EJ, et al. A biosensor platform for rapid antimicrobial susceptibility testing directly from clinical samples. J Urol. 2011;185:148–53. doi: 10.1016/j.juro.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sin ML, Liu T, Pyne JD, Gau V, Liao JC, Wong PK. In situ electrokinetic enhancement for self-assembled-monolayer-based electrochemical biosensing. Anal Chem. 2012;84:2702–7. doi: 10.1021/ac203245j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang M, Mohan R, Lu Y, et al. An AC electrokinetics facilitated biosensor cassette for rapid pathogen identification. Analyst. 2013;138:3660–6. doi: 10.1039/c3an00259d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao JC, Mastali M, Li Y, et al. Development of an advanced electrochemical DNA biosensor for bacterial pathogen detection. J Mol Diagn. 2007;9:158–68. doi: 10.2353/jmoldx.2007.060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campuzano S, Kuralay F, Lobo-Castanon MJ, et al. Ternary monolayers as DNA recognition interfaces for direct and sensitive electrochemical detection in untreated clinical samples. Biosens Bioelectron. 2011;26:3577–83. doi: 10.1016/j.bios.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohan R, Mach KE, Bercovici M, et al. Clinical validation of integrated nucleic acid and protein detection on an electrochemical biosensor array for urinary tract infection diagnosis. PLoS One. 2011;6:e26846. doi: 10.1371/journal.pone.0026846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabe M, Bartoletti R, Bjerklund-Johansen TE, et al. Limited update. European Association of Urology; Mar, 2015. Guidelines on urological infections. http://uroweb.org/wp-content/uploads/19-Urological-infections_LR2.pdf. [Google Scholar]
- 14.Bonkat G, Braissant O, Widmer AF, et al. Rapid detection of urinary tract pathogens using microcalorimetry: principle, technique and first results. BJU Int. 2012;110:892–7. doi: 10.1111/j.1464-410X.2011.10902.x. [DOI] [PubMed] [Google Scholar]
- 15.Burillo A, Rodriguez-Sanchez B, Ramiro A, Cercenado E, Rodriguez-Creixems M, Bouza E. Gram-stain plus MALDI-TOF MS (matrix-assisted laser desorption ionization-time of flight mass spectrometry) for a rapid diagnosis of urinary tract infection. PLoS One. 2014;9:e86915. doi: 10.1371/journal.pone.0086915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsson C, Kapoor D, Howard G. A method for the rapid detection of urinary tract infections. Urology. 2012;79:761–5. doi: 10.1016/j.urology.2011.12.040. [DOI] [PubMed] [Google Scholar]
- 17.Wang XH, Zhang G, Fan YY, Yang X, Sui WJ, Lu XX. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J Microbiol Methods. 2013;92:231–5. doi: 10.1016/j.mimet.2012.12.016. [DOI] [PubMed] [Google Scholar]