Abstract
A unique pH/redox dual-sensitive cationic unimolecular nanoparticle (NP) enabling excellent endosomal/lysosomal escape and efficient siRNA decomplexation inside the target cells was developed for tumor-targeted delivery of siRNA. siRNA was complexed into the cationic core of the unimolecular NP through electrostatic interactions. The cationic core used for complexing siRNA contained reducible disulfide bonds that underwent intracellular reduction owing to the presence of high concentrations of reduced glutathione (GSH) inside the cells, thereby facilitating the decomplexation of siRNA from the unimolecular NPs. The cationic polymers were conjugated onto the hyperbranched core (H40) via a pH-sensitive bond, which further facilitated the decomplexation of siRNA from the NPs. In vitro studies on the siRNA release behaviors showed that dual stimuli (pH = 5.3, 10 mM GSH) induced the quickest release of siRNA from the NPs. In addition, the imidazole groups attached to the cationic polymer segments enhanced the endosomal/lysosomal escape of NPs via the proton sponge effect. Intracellular tracking studies revealed that siRNA delivered by unimolecular NPs was efficiently released to the cytosol. Moreover, the GE11 peptide, an anti-EGFR peptide, enhanced the cellular uptake of NPs in MDA-MB-468, an EFGR-overexpressing triple negative breast cancer (TNBC) cell line. The GE11-conjugated, GFP-siRNA-complexed NPs exhibited excellent GFP gene silencing efficiency in GFP-MDA-MB-468 TNBC cells without any significant cytotoxicity. Therefore, these studies suggest that this smart unimolecular NP could be a promising nanoplatform for targeted siRNA delivery to EFGR-overexpressing cancer cells.
Keywords: Unimolecular NP, siRNA delivery, Tumor-targeted, Endosomal/lysosomal escape, pH/redox dual-sensitive
1. Introduction
Specific silencing of target genes using short interfering RNA (siRNA) is of significant interest to the treatment of cancers and other diseases [1,2]. siRNA molecules are double-stranded short chain oligonucleotides that post-transcriptionally regulate protein synthesis by sequence-specific matching with mRNA molecules and thus cause specific gene silencing [3,4]. Currently, several dozen potential siRNA therapies are undergoing clinical trials (https://clinicaltrials.gov/ct2/results?term=sirna&no_unk=Y&pg=1). However, due to their highly negatively charged nature, insufficient chemical stability, short plasma half-time, and off-target effects, naked siRNAs show poor therapeutic efficacy [5–7]. Various viral and non-viral delivery systems have been developed to improve the efficacy of siRNA therapy [8–10]. Although viral vectors can provide high transfection efficiency, concerns associated with insertional mutagenesis, immunogenicity, and high toxicity have limited their use [11,12]. Non-viral delivery systems offer a safer alternative to viral vectors [13,14].
Nanoparticles (NPs) have been actively investigated as non-viral siRNA delivery systems. NPs can greatly enhance the stability and cellular uptake of the siRNA molecules [15,16]. Furthermore, NPs are attractive for targeted cancer therapy due to their passive (attributed to the enhanced permeability and retention (EPR) effect) and active (via specific ligand conjugation) tumor-targeting abilities [17,18]. However, in order to achieve high gene silencing efficiency, once the NPs are taken up by the target cancer cells, efficient siRNA escape from the acidic endosome/lysosome compartments and efficient siRNA decomplexation from its carrier in the cytosol are essential [6,19–22]. siRNA-complexed NPs are taken up by the cells via endocytosis typically involving the endolysosomal pathways. Without efficient endosomal/lysosomal escape of the siRNA-complexed NPs, degradation of siRNA can occur in these compartments and thus hinder the siRNA from functioning in the cytosol [20–22]. Besides endosomal/lysosomal escape, decomplexation of siRNA from the NPs inside of the cytosol is yet another determining factor [6,19]. siRNA molecules are commonly complexed onto the NPs via electrostatic interactions. Strong electrostatic interactions can enhance the stability of siRNA-complexed NPs, but they can also result in poor siRNA release, thus leading to limited gene silencing efficacy [19,23,24]. Hence, efficient decomplexation of siRNA from the cationic NPs is also of vital importance to enhance gene silencing efficiency.
In this study, in order to overcome the limiting factors associated with endosomal/lysosomal escape and decomplexation of siRNA inside of the target cell, a unique pH/redox dual-sensitive cationic unimolecular NP containing imidazole residues was developed for siRNA delivery (Fig. 1). The unimolecular NP was formed by a multi-arm star H40-poly[(N4-(2-((2-aminoethyl)disulfanyl)ethyl)aspartamide)-r-(N4-(2-((2-(1H-imidazole-5-carboxamido)ethyl) disulfanyl)ethyl)aspartamide)]-b-poly(ethylene glycol) (also referred to as H40-poly(aspartic acid-(2-aminoethyl disulfide)-(4-imidazolecarboxylic acid))-poly(ethylene glycol, and abbreviated as H40-P(Asp-AED-ICA)-PEG) block copolymer in an aqueous solution. Because of its covalent nature, the unimolecular NP has excellent stability in vitro and in vivo [25–30]. The cationic core formed by P(Asp-AED-ICA) blocks was used for siRNA complexation through electrostatic interactions, while the PEG shell was used to provide good water solubility and reduced opsonization of NPs during blood circulation. NPs are taken up by cells through endocytosis [31]. The imidazole groups in the cationic segment have a pKa of ∼6.0 and can thus absorb protons in the acidic endocytic compartments (endosomes/lysosomes), leading to osmotic swelling and endosome/lysosome-membrane disruption (i.e., the proton sponge effect), thereby facilitating the endosomal/lysosomal escape of the siRNA-complexed NPs [21,32,33]. Moreover, siRNA molecules were complexed within the NPs by electrostatic interactions with the cationic P(Asp-AED-ICA) segments containing cleavable disulfide bonds. Furthermore, the cationic segments were conjugated onto the hyperbranched polymer (H40) via a pH-sensitive aromatic imine bond, which can be hydrolyzed in the endosome/lysosome, but stays relatively stable at physiological conditions (pH 7.4) [34,35]. Once inside the cells, it was expected that the pendant mercaptoethylamine group (SH—CH2—CH2—NH2) would be cleaved from the P(Asp-AED-ICA) block by highly concentrated GSH (2–10 mM) in the cytosol [36–39]. The GSH concentration in the cytosol is 100–1000 times higher than that in bodily fluids, including blood and extracellular milieu (2–20 μM GSH) where the disulfide bonds are stable [38]. It should be noted that the enzyme, gamma-interferon-inducible lysosomal thiol reductase (GILT in the endosomes/lysosomes), in combination with cysteines, can also potentially trigger the cleavage of disulfide bonds [38,40,41]. Hence, we expect this pH/redox dual-sensitive characteristic of the NPs to facilitate the release of siRNA from the NPs. The NPs were also functionalized with GE11 peptide, which can efficiently bind to the epidermal growth factor receptor (EGFR) to achieve active tumor targeting [42–44]. EGFR is one of the most common receptors overexpressed in many types of cancer cells, including triple negative breast cancers (TNBCs), ovarian cancers, and pancreatic cancers [45–48]. Our studies showed that these pH/redox dual-sensitive unimolecular NPs, with excellent endosomal/lysosomal escape and siRNA decomplexation abilities, can be promising nanocarriers for the targeted delivery of siRNA.
Fig. 1.
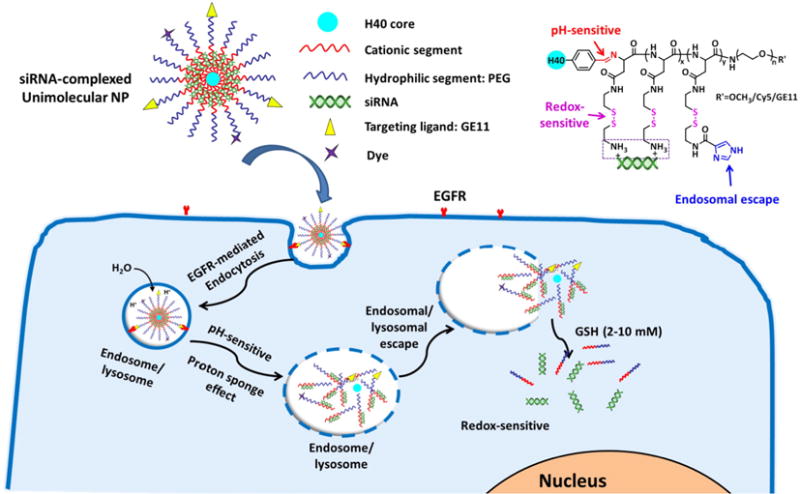
pH/redox dual-sensitive unimolecular NPs with excellent endosomal/lysosomal escape capabilities for efficient targeted delivery of siRNA. A schematic diagram of the cellular uptake of siRNA-complexed NPs and the subcellular release of siRNA into the cytosol.
2. Methods
2.1. Materials
Boltron® H40 (a hyperbranched polyester with 64 hydroxyl terminal groups; Mn: 2833 Da) was kindly provided by Perstorp Polyols Inc., USA, and purified by fractional precipitation in acetone and tetrahydrofuran (THF). β-Benzyl L-aspartate N-carboxyanhydride (BLA-NCA) was prepared as previously reported by our lab [49]. The heterobifunctional poly(ethylene glycol) (PEG) derivatives, methoxy–PEG–NH2 (CH3O-PEG–NH2, Mn = 5 kDa) and maleimide–PEG–NH2 (Mal–PEG–NH2, Mn = 5 kDa), were purchased from JenKem Technology (Allen, TX, USA). Cy5 dye was obtained from Lumiprobe Corporation (Hallandale Beach, FL, USA). GE11 peptide (YHWYGYTPQNVIGGGGC) was synthesized by Tufts University Core Facility (Boston, MA, USA). GFP-siRNA-Cy5.5, GFP-siRNA, dimethyl sulfoxide (DMSO), 2-carboxybenzylaldehyde, 2-aminoethyl disulfide, 4-imidazolecarboxylic acid, and stannous (II) octoate (Sn(Oct)2) were purchased from Sigma–Aldrich (St. Louis, MO, USA). 4-Dimethylamino pyridine (DMAP) and 1,3-dicyclohexylcarbodiimide (DCC) were purchased from ACROS and used without further purification. Other reagents, including RNAiMAX, were purchased from Thermo Fisher Scientific (Fitchburg, WI, USA) and used as received unless otherwise stated.
2.2. Synthesis of H40-poly[(N4-(2-((2-aminoethyl)disulfanyl)ethyl)aspartamide)-r-(N4-(2-((2-(1H-imidazole-5-carboxamido)ethyl)disulfanyl)ethyl)aspartamide)]-b-poly(ethylene glycol)-methoxy/Cy5/GE11 (i.e., H40-poly(aspartic acid-(2-aminoethyl di-sulfide)-(4-imidazolecarboxylic acid))-poly(ethylene glycol)-OCH3/Cy5/GE11, abbreviated as H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5/GE11)
2.2.1. Synthesis of poly(β-benzyl L-aspartate)-poly(ethylene glycol)-Mal (i.e., PBLA-PEG-Mal)
PBLA-PEG-Mal was prepared by ring-opening polymerization of BLA-NCA using NH2-PEG-Mal as the macro-initiator. Briefly, BLA-NCA (53 mg), and NH2-PEG-Mal (25 mg) were dissolved in DMF (5 mL). The reaction was carried out at 55 °C under argon for 48 h. The resulting mixture was then added dropwise into a 10-fold volume of cold diethyl ether. The precipitate was collected by filtration using a Büchner funnel, washed with diethyl ether, and dried under vacuum. The poly(β-benzyl L-aspartate)-poly(ethylene glycol)-methoxy (PBLA-PEG-OCH3) was synthesized following a similar method using OCH3-PEG-NH2 instead. The chemical structure was confirmed by 1H NMR (400 MHz, DMSO-d6). PBLA-PEG-OCH3: 8.23 (20H, s, C(O)NHCH2), 7.28–7.40 (100H, m, Ar-H), 5.15 (40H, s, CH2-Ar), 4.55–4.68 (20H, m, COCHCH2), 3.35–3.53 (450H, m, CH2CH2O from PEG), and 2.60–2.80 (41H, m, COCHCH2) ppm. PBLA-PEG-Mal: 8.23 (20H, s, C(O)NHCH2), 7.28–7.40 (100H, m, Ar-H), 6.95 (2H, s, Mal), 5.15 (40H, s, CH2-Ar), 4.55–4.68 (20H, m, COCHCH2), 3.35–3.53 (450H, m, CH2CH2O from PEG), and 2.60–2.80 (41H, m, COCHCH2) ppm.
2.2.2. Synthesis of H40-carboxybenzaldehyde (i.e., H40-CHO)
H40-OH (10 mg), 2-carboxybenzylaldehyde (82 mg), DCC (135 mg), and DMAP (8.3 mg) were dissolved in anhydrous DMF (3 mL). The solution was stirred at room temperature under argon for 48 h. Thereafter, the dicyclohexylurea was removed by filtration using a Büchner funnel. The solution was collected and poured into a 10-fold volume of cold diethyl ether. The precipitate was collected by filtration using a Büchner funnel, washed with diethyl ether, and dried under vacuum. The chemical structure was confirmed by 1H NMR (400 MHz, DMSO-d6). H40-CHO: 8.23 (20H, s, C(O)NHCH2), 7.60–7.80 (100H, m, Ar-H), 3.8–4.1(88H, m, OCH2C(CH3)(CH2)CO(O)), and 0.97–1.2 (66H, m, OCH2C(CH3) (CH2)CO(O)) ppm.
2.2.3. Synthesis of H40-poly(β-benzyl L-aspartate)-poly(ethylene glycol)-methoxy/Mal (i.e., H40-PBLA-PEG-OCH3/Mal)
H40-CHO (0.5 mg), PBLA-PEG-OCH3 (25 mg), and PBLA-PEG-Mal (8 mg) were dissolved in DMSO. The reaction was conducted at room temperature for 24 h. Thereafter, the resulting solution was dialyzed (molecular weight cut-off: 15 kDa) against DMSO for the first 24 h and DI water for another 24 h. The product was obtained after lyophilization. The H40-PBLA-PEG-OCH3 was synthesized following a similar method. The chemical structure was confirmed by 1H NMR (400 MHz, DMSO-d6). H40-PBLA-PEG-OCH3/Mal: 8.23 (20H, s, C(O)NHCH2), 7.28–7.40 (106H, m, Ar-H), 6.95 (2H, s, Mal), 5.15 (40H, s, CH2-Ar), 4.55–4.68 (20H, m, COCHCH2), 3.8–4.1(21H, m, OCH2C(CH3)(CH2)CO(O)), 3.35–3.53 (450H, m, CH2CH2O from PEG), 2.60–2.80 (41H, m, COCHCH2), and 0.97–1.2 (23H, m, OCH2C(CH3) (CH2)CO(O)) ppm.
2.2.4. Synthesis of H40-poly[N4-(2-((2-aminoethyl)disulfanyl)ethyl)aspartamide]-b-poly(ethylene glycol)-methoxy/Mal (i.e., H40-poly(aspartic acid-(2-aminoethyl disulfide))-poly(ethylene glycol)-OCH3/Mal, abbreviated as H40-P(Asp-AED)-PEG-OCH3/Mal)
2-Aminoethyl disulfide (13.1 mg) and H40-PBLA-PEG-OCH3/Mal (20 mg) were dissolved in DMSO (10 mL). The reaction was carried out at room temperature for 24 h. Thereafter, the resulting solution was dialyzed (molecular weight cut-off: 15 kDa) against DI water for 48 h. The product was obtained after lyophilization. The H40-P(Asp-AED)-PEG-OCH3 was synthesized following a similar method. The chemical structure was confirmed by 1H NMR (400 MHz, CDCl3). H40-P(Asp-AED)-PEG-OCH3/Mal: 8.23 (20H, s, C(O)NHCH2), 7.28–7.40 (4H, m, Ar-H), 6.70 (2H, s, Mal), 4.55–4.68 (20H, m, COCHCH2), 3.8–4.1(21H, m, OCH2C(CH3)(CH2)CO(O)), 3.35–3.53 (450H, m, CH2CH2O from PEG), 3.15–3.22 (63H, m, NH2CH2CH2SSCH2CH2NHCO), 2.60–2.80 (79H, m, COCHCH2), and 0.97–1.2 (23H, m, OCH2C(CH3) (CH2)CO(O)) ppm.
2.2.5. Synthesis of H40-poly[(N4-(2-((2-aminoethyl)disulfanyl)ethyl)aspartamide)-r-(N4-(2-((2-(1H-imidazole-5-carboxamido)ethyl)disulfanyl)ethyl)aspartamide)]-b-poly(ethylene glycol)-methoxy/Mal (i.e., H40-poly(aspartic acid-(2-aminoethyl disulfide)-(4-imidazolecarboxylic acid))-poly(ethylene glycol)-OCH3/Mal, abbreviated as H40-P(Asp-AED-ICA)-PEG-OCH3/Mal)
4-Imidazolecarboxylic acid (2.2 mg), H40-P(Asp-AED)-PEG-OCH3/Mal (20 mg), DCC (4.4 mg), and N-hydroxysuccinimide (2.9 mg) were dissolved in DMSO (5 mL). The reaction was carried out at room temperature for 24 h. Thereafter, the resulting solution was dialyzed (molecular weight cut-off: 15 kDa) against DI water for 48 h. The product was obtained after lyophilization. The H40-(Asp-AED-ICA)-PEG-OCH3 was synthesized following a similar method. The chemical structure was confirmed by 1H NMR (400 MHz, DMSO-d6). H40-P(Asp-AED-ICA)-PEG-OCH3/Mal: 8.23 (20H, s, C(O)NHCH2), 7.91 (1H, s, C(=)CHNH), 7.28–7.40 (4H, m, Ar-H), 7.23 (1H, s, N(=)CHNH), 6.70 (2H, s, Mal), 4.55–4.68 (20H, m, COCHCH2), 3.8–4.1(22H, m, OCH2C(CH3)(CH2)CO(O)), 3.35–3.53 (450H, m, CH2CH2O from PEG), 3.15–3.22 (61H, m, NH2CH2CH2SSCH2CH2NHCO), 2.60–2.80 (81H, m, COCHCH2), and 0.97–1.2 (22H, m, OCH2C(CH3) (CH2)CO(O)) ppm.
2.2.6. Synthesis of H40-poly[(N4-(2-((2-aminoethyl)disulfanyl)ethyl)aspartamide)-r-(N4-(2-((2-(1H-imidazole-5-carboxamido)ethyl)disulfanyl)ethyl)aspartamide)]-b-poly(ethylene glycol)-methoxy/Cy5/GE11 (i.e., H40-poly(aspartic acid-(2-aminoethyl disulfide)-(4-imidazolecarboxylic acid))-poly(ethylene glycol)-OCH3/Cy5/GE11, abbreviated as H40-(PAsp-AED-ICA)-PEG-OCH3/Cy5/GE11)
Cy5-SH was first prepared by a reaction between Cy5-NH2 and Traut’s reagent. Briefly, Cy5-NH2 (0.3 mg) and Traut’s reagent (0.51 mg) were dissolved in DMSO. The solution was stirred at room temperature in complete darkness for 4 h. H40-P(Asp-AED-ICA)-PEG-OCH3/Mal (20 mg) and GE11 (1.3 mg) were added into the above solution. After 24 h, the reaction solution was dialyzed (molecular weight cut-off: 15 kDa) against DI water for 48 h. The product was obtained after lyophilization. The H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5 and H40-P(Asp-AED-ICA)-PEG-OCH3/GE11 were synthesized following a similar method. Polymers H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5 and H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5/GE11 were only used for the cellular uptake studies. For all other experiments, H40-P(Asp-AED-ICA)-PEG-OCH3 and H40-P(Asp-AED-ICA)-PEG-OCH3/GE11 were used. The chemical structure was confirmed by 1H NMR (400 MHz, DMSO-d6). H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5/GE11: 8.23 (20H, s, C(O)NHCH2), 7.91 (1H, s, C(=)CHNH), 7.20–7.80 (25H, m, GE11 and Cy5), 6.70 (2H, s, Mal), 4.55–4.68 (20H, m, COCHCH2), 3.8–4.1(22H, m, OCH2C(CH3)(CH2)CO(O)), 3.35–3.53 (450H, m, CH2CH2O from PEG), 3.15–3.22 (61H, m, NH2CH2CH2SSCH2CH2NHCO), 2.60–2.80 (81H, m, COCHCH2), and 0.97–1.2 (22H, m, OCH2C(CH3) (CH2)CO(O)) ppm.
2.3. Preparation of siRNA-complexed unimolecular NPs (i.e., siRNA-complexed NPs) and gel retardation assay
To prepare siRNA-complexed NPs, siRNA and H40-P(Asp-AED-ICA)-PEG were dissolved in PBS and the solution was mixed for 30 min under gentle shaking. The binding ability of siRNA to NPs was studied by agarose gel electrophoresis. The siRNA-complexed NPs were prepared at different N/P ratios (molar ratio of nitrogen in polymers to phosphorus in siRNA: 2, 5, 7, 10, and 15). Electrophoresis was carried out on 1% agarose gel in a TAE (Tris-acetate-EDTA) buffer solution with a current of 100 V for 35 min. The final siRNA concentration was 1 μg per well. The retardation of the complexes was visualized on a UV illuminator (Bio-Rad Baloratories, Inc., Hercules, CA, USA) to show the position of the complexed siRNA band relative to that of naked siRNA.
2.4. Characterization
1H NMR spectra of all intermediate and final polymer products were recorded on a Varian Mercury Plus 300 spectrometer in DMSO–d6 or CDCl3 at 25 °C. Molecular weights (Mn and Mw) and polydispersity indices (PDI) of the polymers were determined by a gel permeation chromatography (GPC) system equipped with a refractive index detector, a viscometer detector, and a light scattering detector (Viscotek, USA). Fourier transform infrared (FT-IR) spectra were recorded on a Bruker Tensor 27 FT-IR spectrometer. The morphologies of the siRNA-complexed NPs were studied by dynamic light scattering (DLS; ZetaSizer Nano ZS90, Malvern Instruments, USA; 0.5 mg/mL) and transmission electron microscopy (TEM, FEI Tecnai G2 F30 TWIN 300 KV, E.A. Fischione Instruments, Inc. USA).
2.5. In vitro siRNA release behavior
The release profiles of siRNA from siRNA-complexed NPs were studied in a glass apparatus at 37 °C in a release medium at four conditions: (1) pH 7.4, (2) pH 5.3, (3) pH 7.4 + 10 mM GSH, and (4) pH 5.3 + 10 mM GSH. siRNA-complexed NP solutions (5 mL; 100 μg/mL) were enclosed in a dialysis bag. The dialysis bag was immersed in 50 mL of the release medium and kept at 37 °C under a horizontal laboratory shaker (Thermo Scientific MaxQ Shaker, USA) at 100 rpm. At specific time points, 3 mL of release media were collected and replaced by the same volume of fresh media. GFP-siRNA-Cy5.5 was used in this experiment. The amount of released siRNA was analyzed based on the UV–vis intensity of Cy5.5 at 649 nm.
2.6. Cellular uptake analyses
The cellular uptake behaviors of the NPs in MDA-MB-468 TNBC cell lines were analyzed using a fluorescence microscope based on the Cy5 dye conjugated on the NPs. Cells were seeded (1 × 105 cells/mL) onto 8-well high-optical-quality plates and grown overnight. Cells were treated with either non-targeted NPs, targeted NPs, or targeted NPs with free GE11 peptide (2 μM; blocking assay) at an NP concentration of 100 μg/mL. After 2 h incubation, cells were washed with PBS twice, fixed with 4% PFA, and stained with DAPI for 4 h. Then the cells were mounted with Prolong Gold anti-fade reagent. The cellular uptake was observed using a fluorescence microscope (Nikon, Melville, NY). Digital monochromatic images were acquired using NIS-Elements BR Software.
2.7. Endosomal/lysosomal escape studies
To study the endosomal/lysosomal escape behaviors of the NPs, MDA-MB-468 cells were incubated with siRNA-complexed NPs for 2 h at 37 °C. Cells treated with pure medium or free siRNA were used as negative controls. The siRNA labeled with Cy5.5 was used for intracellular tracking. The cells were washed three times with PBS, followed by staining with LysoTracker Green DND-26 (100 nM) for endosomes/lysosomes and Hoechst (5 ng/mL) for the nuclei, for 20 min at 37 °C. Cells were then washed three times with PBS. The cellular localization of siRNA was visualized with a fluorescence microscope (Nikon, Japan) or a confocal laser scanning microscope (CLSM, Nikon Eclipse Ti inverted microscope equipped with Nikon A1R confocal diode lasers, Japan).
2.8. In vitro siRNA transfection studies
Cellular transfection in GFP-expressing MDA-MB-468 TNBC cells was investigated using flow cytometry and fluorescence microscopy. For the flow cytometry assay, cells were seeded at a density of 50,000 cells/well on a 24-well plate and incubated overnight. Cells were treated with pure medium (control), siRNA-complexed non-targeted NPs (siRNA-NT), siRNA-complexed targeted NPs (siRNA-T), and siRNA complexed with RNAiMAX (i.e., siRNA-RNAiMAX; positive control). The concentration of siRNA was 40 nM. After 24 h incubation, cells were washed twice with PBS and harvested with 0.25% trypsin. Cells were collected by centrifugation at 200 g for 5 min, washed twice with PBS, fixed with paraformaldehyde (PFA) for 15 min, and resuspended in 500 μL PBS for analysis. The transfection efficiency was examined by quantifying GFP expression levels in the cells using an AccuriTM C6 flow cytometry system (BD Biosciences, USA). A minimum of 10,000 cells was analyzed from each sample.
For fluorescence microscope imaging, cells were seeded (50,000 cells/well) in an 8-well chamber slide system. Cells were treated with the same five groups as described above. After 24 h incubation, cells were washed twice with PBS, fixed with PFA for 15 min, stained with DAPI for 4 h, and mounted with ProLong Gold Antifade Mountant. Images were acquired with a fluorescence microscope (Nikon, Melville, NY) to observe the GFP expression levels in the cells. Digital monochromatic images were acquired using NIS-Elements BR Software.
2.9. Cell viability assays
Cell viability tests were conducted using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. To test the cytotoxicity of the pure (empty) NPs, MDA-MB-468 cells were seeded in quadruplicate on 96-well plates and incubated overnight. Cells were treated with NPs at different concentrations (i.e., 10, 20, 50, 100, and 500 μg/mL). Cells treated with pure medium were used as the control group. After 24 h of incubation, a standard MTT assay was performed by aspirating the treatment media, adding 25 μL of the medium containing 0.5 mg/mL MTT agent, and incubating at 37 °C for 4 h. Thereafter, the medium was aspirated and 75 μL of DMSO was added to each well. The plates were then measured at 570 nm using a spectrophotometer (Quant, Bio-Tek Instruments, Winooski, VT), and the average absorbance and percent of cell viability relative to the control (pure medium) were calculated. The cytotoxicity of siRNA-complexed NP systems was also studied. Similarly, cells were treated with pure medium (control), siRNA-NT, siRNA-T, siRNA-RNAiMAX, and RNAiMAX at the equivalent amount of siRNA (40 nM). After 24 h of incubation, the aforementioned MTT protocol was performed and the cell viabilities relative to the control (pure medium) were calculated.
3. Results and discussion
3.1. Polymer synthesis and siRNA encapsulation
pH/redox dual-sensitive multi-arm star block copolymer H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5/GE11 was synthesized as outlined in Scheme 1. PBLA-PEG-OCH3 and PBLA-PEG-Mal were first synthesized by ring-opening polymerization of BLA-NCA using NH2-PEG-OCH3 and NH2-PEG-Mal as the macro-initiators, respectively. Their chemical structures were confirmed by 1H NMR spectra as shown in Fig. 2 (A) and (B). The peaks at (e) 7.28–7.40 ppm and (d) 5.15 ppm were assigned to the protons in the benzyl and methylene groups in the PBLA side chains, respectively. The signals labeled as (c) at 2.6–2.8 ppm were ascribed to the methylene group of the side chain that connects the main chain in the PBLA segment. The peak located at (a) 3.57 ppm corresponded to the methylene protons of the oxyethylene repeat units in the PEG segment. The Mal group in the PBLA-PEG-Mal at 6.7 ppm was also observed. The number of BLA-NCA repeat units in the polymers was calculated to be 20 based on the relative intensity ratio of the methylene proton (a) of the PEG chain and the methylene proton (d) near the benzyl group of the PBLA chain. The molecular weights of the NH2-PEG-OCH3 and NH2-PEG-Mal polymers as measured by GPC (Table 1) were 9040 and 9105 g/mol, respectively, which was consistent with that determined by the 1H NMR analyses.
Scheme 1.
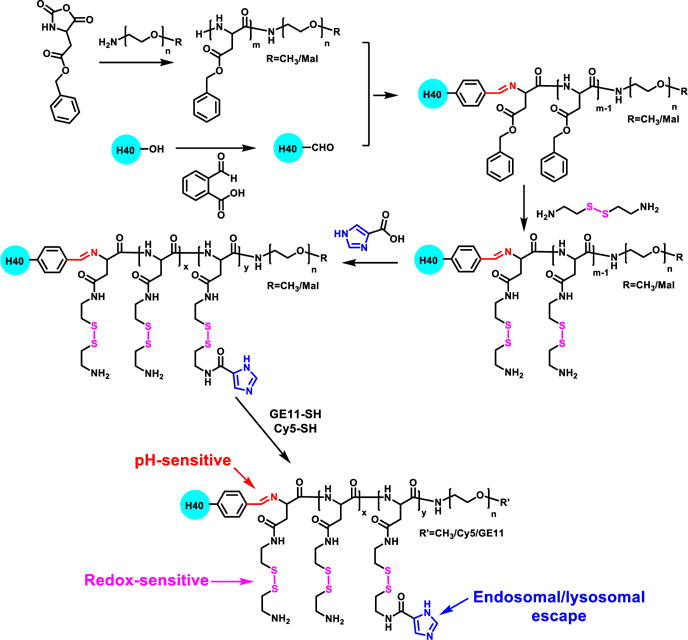
A synthetic scheme of multi-arm star block copolymer H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5/GE11.
Fig. 2.
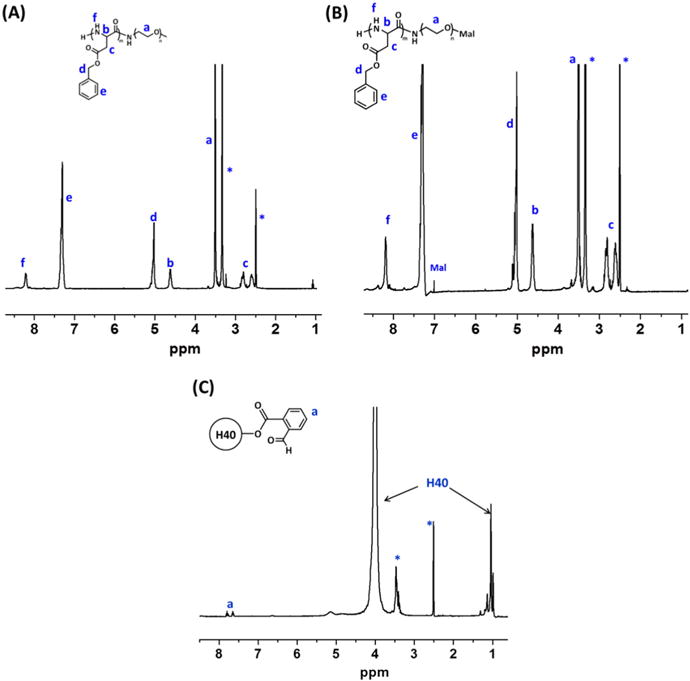
1H NMR spectra of (A) PBLA-PEG-OCH3, (B) PBLA-PEG-Mal, and (C) H40-CHO.
Table 1.
GPC analyses of polymers.
| Polymers | Mn (g/mol) | PDI |
|---|---|---|
| PBLA-PEG-OCH3 | 9040 | 1.4 |
| PBLA-PEG-Mal | 9105 | 1.3 |
| H40-PBLA-PEG-OCH3/Mal | 200,833 | 1.6 |
| H40-P(Asp-AED)-PEG-OCH3/Mal | 186,406 | 1.7 |
| H40-P(Asp-AED-ICA)-PEG-OCH3/Mal | 187,130 | 1.6 |
The benzylaldehyde-functionalized H40 (H40-CHO) was prepared by an esterification reaction. The chemical structure was also confirmed by 1H NMR spectrum (Fig. 2 (C)). The peaks at 0.97–1.2 ppm and 3.8–4.1 ppm were assigned to the protons in H40. The peaks at (a) 7.6–7.8 ppm corresponded to the protons in the phenyl group of H40-CHO as labeled. Thereafter, PBLA-PEG-OCH3 and PBLA-PEG-Mal (molar ratio: 3.1/1) polymers were then conjugated to H40-CHO through imine bonds to form H40-PBLA-PEG-OCH3/Mal. In the 1H NMR spectrum shown in Fig. 3 (A), other than the proton peaks assigned to PBLA-PEG-OCH3 and PBLA-PEG-Mal, proton peaks ascribed to H40 were also observed. The GPC analyses further demonstrated the formation of H40-PBLA-PEG-OCH3/Mal and its molecular weight was measured to be 200,833 Da, which was significantly larger than that of the linear NH2-PEG-OCH3 or NH2-PEG-Mal polymers. The average number of arms in the H40-PBLA-PEG-OCH3/Mal was calculated to be 22 based on the molecular weights of H40-PBLA-PEG-OCH3/Mal, NH2-PEG-OCH3, and NH2-PEG-Mal. Thereafter, the H40-PBLA-PEG-OCH3/Mal polymer underwent aminolysis by using 2-aminoethyl disulfide (AED) to form water-soluble polymer H40-P(Asp-AED)-PEG-OCH3/Mal. As shown in Fig. 3 (B), the absence of proton peaks at 7.28–7.40 ppm and 5.15 ppm, and the presence of proton peaks at 2.71 and 3.17 ppm ascribed to the protons in AED, demonstrated the formation of H40-P(Asp-AED)-PEG-OCH3/Mal.
Fig. 3.
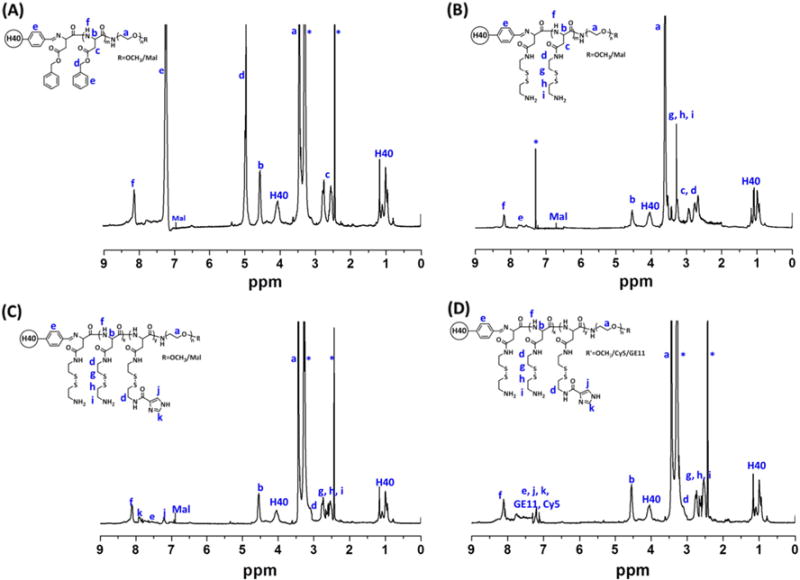
1H NMR spectra of (A) H40-PBLA-PEG-OCH3/Mal, (B) H40-P(Asp-AED)-PEG-OCH3/Mal, (C) H40-P(Asp-AED-ICA)-PEG-OCH3/Mal, and (D) H40-P(Asp-AED-ICA)-PEG-OCH3/Cy5/GE11.
Imidazole groups were selectively conjugated to H40-P(Asp-AED)-PEG-OCH3/Mal (molar ratio: 5/1) through an amidization reaction for enhanced endosomal/lysosomal escape. The characteristic proton peaks at (j) 7.23 and (k) 7.91 ppm for imidazole groups were observed in Fig. 3 (C). In the last step, GE11 peptide and Cy5 dye were conjugated to H40-P(Asp-AED-ICA)-PEG-OCH3/Mal (molar ratio: 3/2/1) though a Mal-SH reaction. Proton peaks assigned to the GE11 and Cy5 molecules, as labeled in Fig. 3 (D), were also observed.
The cationic polymer H40-P(Asp-AED-ICA)-PEG, which had good solubility in aqueous solutions, was able to form the unimolecular NPs. Because of its covalent nature, the unimolecular NPs had excellent stability in vitro and in vivo. siRNA (GFP-siRNA was used as a model siRNA) was electrostatically complexed with the cationic P(Asp-AED-ICA) polymer to form siRNA-complexed NPs. The complexation was evaluated using agarose gel electrophoresis. As shown in Fig. 4 (A), siRNA-complexed NPs with various N/P ratios were tested and siRNA completely lost mobility in the electric field when the N/P ratio reached 10, which was selected for the following tests. The siRNA loading level, defined by the weight percentage of the siRNA in the siRNA-complexed NP was 16.3 % at N/P ratio of 10, and the loading efficiency is 100%. The average hydrodynamic diameter of the siRNA-complexed NPs was 68.3 nm (PDI = 0.14) as measured by DLS. TEM observation showed that the siRNA-complexed NPs were uniform, with an average size of around 39 nm (Fig. 4 (B)). The stability of the siRNA-complexed NPs was studied in PBS. As shown in Fig. S1, there was no obvious change in terms of the size distribution of the siRNA-complexed NPs over one week, demonstrating excellent stability.
Fig. 4.
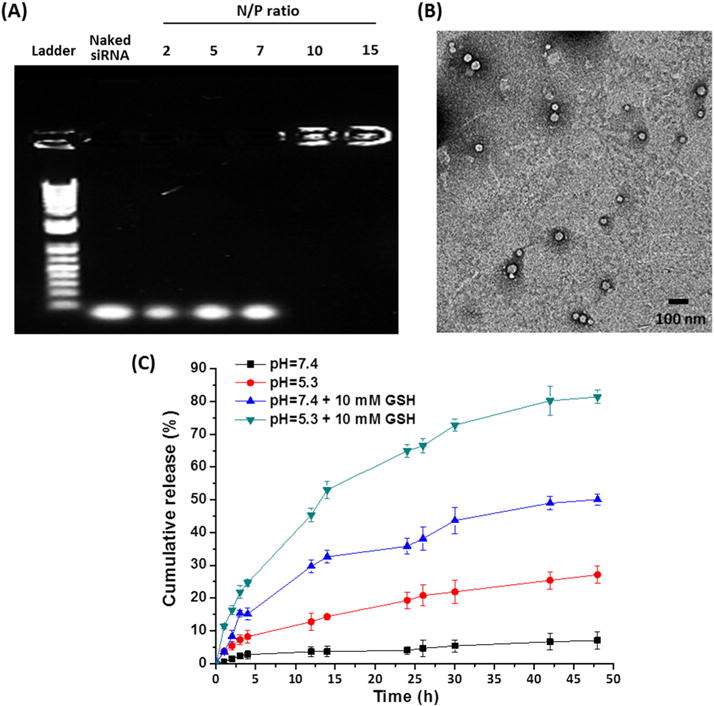
(A) Gel retardation assays of siRNA/NPs with various N/P ratios. (B) TEM images of the siRNA-complexed NPs (N/P = 10). (C) In vitro siRNA release from siRNA-complexed NPs in different buffers at 37 °C. Data represent mean ± SD (n = 3).
3.2. pH/redox dual-sensitive siRNA release
The decomplexation of siRNA from cationic nanocarriers is of vital importance to enhance gene silencing efficiency. Here, NPs with pH/redox dual-sensitive structures were designed to achieve controlled release of siRNA. To verify the pH/redox dual-sensitive release behavior, in vitro release studies were conducted by exploring Cy5.5-labeled siRNA. As shown in Fig. 4 (C), the release rate was very slow at neutral pH (7.4) without adding GSH, with 7.1% of siRNA released after 48 h. In comparison, the addition of GSH (10 mM) to the solution resulted in an increased siRNA release rate (50.1% of siRNA released after 48 h). Meanwhile, 27.2% of siRNA was released at a pH of 5.3 after 48 h, which is much faster compared to that at a neutral pH. Moreover, dual stimuli (pH 5.3 and 10 mM GSH) led to the quickest siRNA release (81.4% of siRNA released after 48 h). Taken together, these observations suggest that siRNA can be efficiently decomplexed from siRNA-complexed NPs inside of cells.
3.3. Enhanced endosomal/lysosomal escape capability of NPs
Another big challenge of nanocarriers for siRNA delivery is their poor endosomal/lysosomal escape capabilities. siRNA needs to be released into the cytoplasm for efficient gene silencing. Therefore, it is of great importance to functionalize the siRNA nanocarriers with good endosomal/lysosomal escape capabilities. As mentioned above, the imidazole groups in the cationic segment promote endosomal/lysosomal escape through the proton-sponge effect, thereby facilitating the release of siRNA to the cytosol. To verify endosomal/lysosomal escape, fluorescence microscopy was used to study the intracellular localizations of siRNA. The cells (MDA-MB-468 TNBC cell line) were treated with pure medium (control) or media containing free siRNA or siRNA-complexed nanoparticles. siRNA was labeled with Cy5.5 (red) for detection. After 2 h incubation, the nucleus and endosomes/lysosomes of cells were stained with Hoechst (blue) and Lysotracker (green), respectively. As shown in Fig. 5, siRNA complexed with NPs were taken up efficiently as signified by the strong red signal. More importantly, the red signals barely overlapped with the green ones (endosomes/lysosomes), and they were distributed relatively uniformly in the cytosol, demonstrating that the majority of the siRNA escaped from the endosomes/lysosomes. The z-stack images shown in Fig. 6 also confirmed the excellent endososomal/lysosomal escape capabilities of the siRNA-complexed NPs, which, thereby would lead to efficient gene silencing.
Fig. 5.
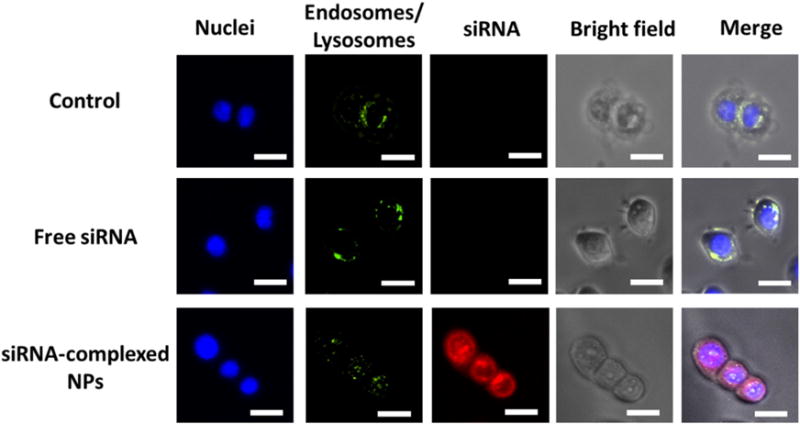
Assessment of the endosomal/lysosomal escape of siRNA-complexed NPs in MDA-MB-468 cells after 2 h incubation. Endosomes/lysosomes were stained with Lysotracker (green). siRNA was labeled with Cy5.5 (red). The nuclei were stained with Hoechst (blue). Scale bar: 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
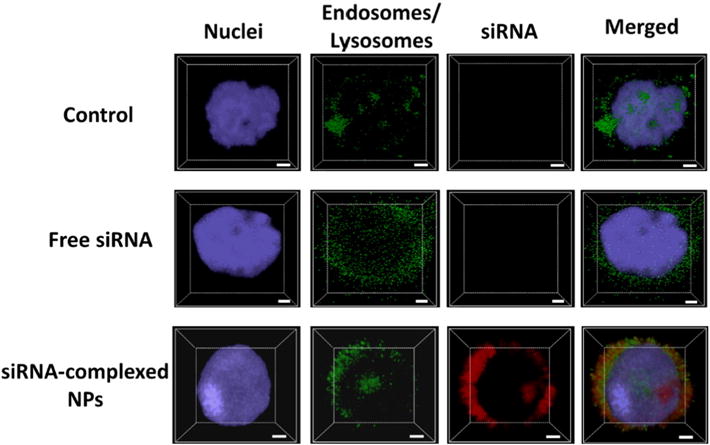
Z-stack CLSM images for the assessment of the endosomal/lysosomal escape of siRNA-complexed NPs in MDA-MB-468 cells. Scale bar: 2 μm.
3.4. In vitro cellular uptake studies
EGFR is overexpressed in many common types of cancer. In this study, an EGFR targeting peptide, GE11, was used as an active-tumor-targeting ligand to enhance cellular uptake. MDA-MB-468, a TNBC cell line that overexpresses EGFR, was used as the model cell line. Cells were incubated with either non-targeted (i.e., NPs without GE11 conjugation) or targeted (i.e., GE11 conjugated) NPs for 2 h. Cells without any treatment were used as a negative control. Cy5 was conjugated onto the NPs for the studies. Fluorescence imaging analysis was performed to compare cellular uptake. As shown in Fig. 7, the targeted NPs showed a markedly higher Cy5 fluorescence intensity than non-targeted ones. In the blocking experiment (co-incubated cells with free GE11 and targeted NPs), after the EGFR was saturated with free GE11, the cellular uptake of the targeted NPs returned to the level of the non-targeted ones, thereby demonstrating the targeting ability of GE11. Taken together, the targeted NPs increased the cellular uptake of NPs through EGFR-mediated endocytosis.
Fig. 7.
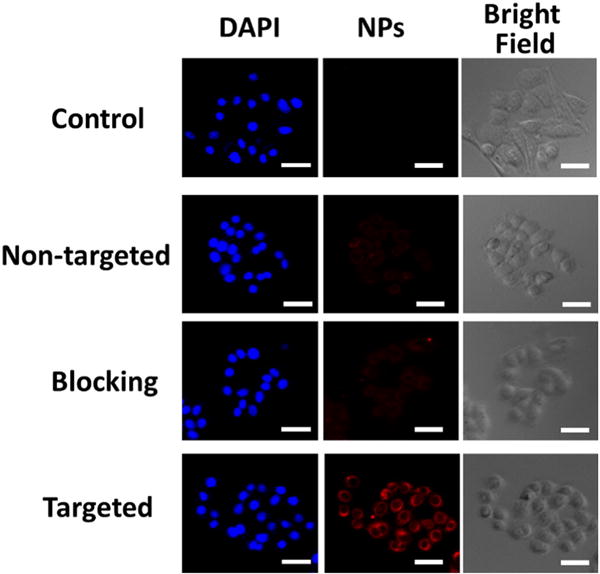
In vitro cellular uptake studies. Fluorescence images of MDA-MB-468 TNBC cells incubated with pure medium (control), Cy5-labeled non-targeted (without GE11 conjugation) NPs, targeted (GE11-conjugated) NPs with a blocking dose (2 μM) of GE11 (i.e., blocking), and targeted NPs at 37 °C for 2 h (NP concentration: 100 μg/mL). Targeted NPs significantly enhanced the cellular uptake in EGFR-overexpressing TNBC cells. Scale bar: 50 μm.
3.5. In vitro gene silencing efficiency and cell viability studies
To determine the potential of NPs to deliver siRNA, gene silencing was assessed in vitro. The gene silencing capacity of GFP-siRNA toward MDA-MB-468 cells stably expressing green fluorescent protein was evaluated for siRNA-complexed non-targeted and targeted NPs. Pure medium was used as the negative control. RNAiMAX, a commercially available transfection agent, was used as the positive control. As shown in Fig. 8 (A) and (B), both targeted and non-targeted groups induced GFP reduction compared to the negative control. As expected based on the cellular uptake studies, the extent of knockdown was dependent on GE11 functionalization. Relative to the negative control group, the siRNA-complexed non-targeted NPs produced a 47% GFP down-regulation. In contrast, the targeted NPs induced a 79% GFR reduction, which is comparable to that of RNAiMAX treatment (81%). However, the assessment of cell viability on these treatments revealed that siRNA-complexed RNAiMAX exhibited significant cytotoxicity, inducing >25% cell death (Fig. 8 (C)), which is consistent with the previous report [50]. However, no apparent cytotoxicity associated with NPs was observed. In fact, no significant cytotoxicity was observed for NPs alone up to 500 μg/mL (Fig. S2 in the Supporting Information). Taken together, these findings reveal that GE11-conjugated NPs are suitable nanocarriers for siRNA delivery targeted at TNBC cells and, potentially, other EGFR-overexpressing cells.
Fig. 8.
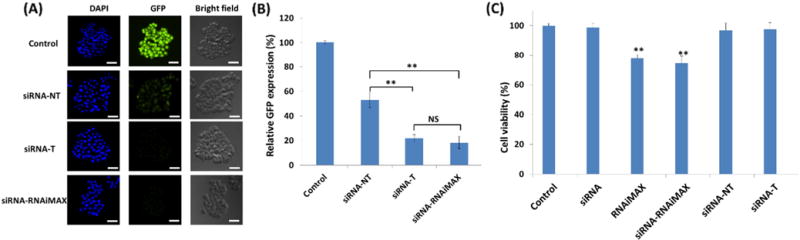
In vitro assessment of gene silencing efficiency using (A) fluorescence microscope and (B) flow cytometry. GFP-expressing MDA-MA-468 cells treated with pure medium (control), siRNA-complexed non-targeted NPs (siRNA-NT), siRNA-complexed targeted NPs (siRNA-T), and siRNA-complexed RNAiMAX (siRNA-RNAiMAX) for 24 h (40 nM of GFP-siRNA). (C) Cell viability studies. MDA-MA-468 cells treated with pure medium (control), siRNA-NT, siRNA-T, siRNA-RNAiMAX, and pure RNAiMAX for 24 h (40 nM of GFP-siRNA). All values are presented as a mean ± SD (n = 5); **: p < 0.01; NS: not significant. Scale bar: 100 μm.
4. Conclusion
We have developed a smart unimolecular NP for tumor-targeted delivery of siRNA. The unimolecular NPs demonstrated excellent endosomal/lysosomal escape capabilities via a proton sponge effect owing to the imidazole groups. Moreover, the pH/redox dual-sensitive characteristics of the unimolecular NPs facilitated the release of siRNA from the NPs inside the cells. GE11-targeting ligand enhanced the cellular uptake of NPs in EGFR-overexpressing TNBC cells. Furthermore, GE11-conjugated GFP-siRNA-complexed NPs exhibited excellent GFP down-regulation capabilities that were comparable to RNAiMAX, a commercially available agent. Meanwhile, RNAiMAX induced significantly higher cytotoxicity compared to the unimolecular NPs. Thus, GE11-conjugated unimolecular NPs could be a promising nanoplatform for targeted siRNA delivery in EFGR-overexpressing cancers.
Supplementary Material
Acknowledgments
This project was financially supported by grants from the NIH (K25CA166178 and R21CA196653). We would also like to express our gratitude to Professor Wei Xu for kindly providing us with the GFP-expressing MDA-MB-468 cells.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jconrel.2017.01.042.
References
- 1.Guo W, Chen W, Yu W, Huang W, Deng W. Small interfering RNA-based molecular therapy of cancers. Chin J Cancer. 2013;32:488–493. doi: 10.5732/cjc.012.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 3.Möller K, Müller K, Engelke H, Bräuchle C, Wagner E, Bein T. Highly efficient siRNA delivery from core–shell mesoporous silica nanoparticles with multifunctional polymer caps. Nanoscale. 2016;8:4007–4019. doi: 10.1039/c5nr06246b. [DOI] [PubMed] [Google Scholar]
- 4.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banan M, Puri N. The ins and outs of RNAi in mammalian cells. Curr Pharm Biotechnol. 2004;5:441–450. doi: 10.2174/1389201043376643. [DOI] [PubMed] [Google Scholar]
- 6.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 8.Saraswathy M, Gong S. Recent developments in the co-delivery of siRNA and small molecule anticancer drugs for cancer treatment. Mater Today. 2014;17:298–306. [Google Scholar]
- 9.Gary DJ, Puri N, Won Y-Y. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13:1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 11.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wadhwa R, Kaul S, Miyagishi M, Taira K. Vectors for RNA interference. Curr Opin Mol Ther. 2004;6:367–372. [PubMed] [Google Scholar]
- 13.Huang P-I, Lo W-L, Cherng J-Y, Chien Y, Chiou G-Y, Chiou S-H. Non-viral delivery of RNA interference targeting cancer cells in cancer gene therapy. Curr Gene Ther. 2012;12:275–284. doi: 10.2174/156652312802083576. [DOI] [PubMed] [Google Scholar]
- 14.Glover DJ, Lipps HJ, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 15.Ngamcherdtrakul W, Morry J, Gu S, Castro DJ, Goodyear SM, Sangvanich T, Reda MM, Lee R, Mihelic SA, Beckman BL. Cationic polymer modified mesoporous silica nanoparticles for targeted siRNA delivery to HER2+ breast cancer. Adv Funct Mater. 2015;25:2646–2659. doi: 10.1002/adfm.201404629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aliabadi HM, Uludağ H. Nanoparticle carriers to overcome biological barriers to siRNA delivery. Nanomedicines. 2016:46–105. [Google Scholar]
- 17.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 18.Chen G, Jaskula-Sztul R, Esquibel CR, Lou I, Zheng Q, Dammalapati A, Harrison A, Eliceiri KW, Tang W, Chen H, Gong S. Neuroendocrine tumor-targeted upconversion nanoparticle-based micelles for simultaneous NIR-controlled combination chemotherapy and photodynamic therapy, and fluorescence imaging. Adv Funct Mater ( 2017 doi: 10.1002/adfm.201604671. 1604671-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uz M, Mallapragada SK, Altinkaya SA. Responsive pentablock copolymers for siRNA delivery. RSC Adv. 2015;5:43515–43527. [Google Scholar]
- 20.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 21.Shrestha R, Elsabahy M, Florez-Malaver S, Samarajeewa S, Wooley KL. Endosomal escape and siRNA delivery with cationic shell crosslinked knedel-like nanoparticles with tunable buffering capacities. Biomaterials. 2012;33:8557–8568. doi: 10.1016/j.biomaterials.2012.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pittella F, Zhang M, Lee Y, Kim HJ, Tockary T, Osada K, Ishii T, Miyata K, Nishiyama N, Kataoka K. Enhanced endosomal escape of siRNA-incorporating hybrid nanoparticles from calcium phosphate and PEG-block charge-conversional polymer for efficient gene knockdown with negligible cytotoxicity. Biomaterials. 2011;32:3106–3114. doi: 10.1016/j.biomaterials.2010.12.057. [DOI] [PubMed] [Google Scholar]
- 23.Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. Mechanistic investigation of poly (ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130:57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Wagner M, Rinkenauer AC, Schallon A, Schubert US. Opposites attract: influence of the molar mass of branched poly (ethylene imine) on biophysical characteristics of siRNA-based polyplexese. RSC Adv. 2013;3:12774–12785. [Google Scholar]
- 25.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S. Gold nanoparticles with a monolayer of doxorubicin-conjugated amphiphilic block copolymer for tumor-targeted drug delivery. Biomaterials. 2009;30:6065–6075. doi: 10.1016/j.biomaterials.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 26.Chen G, Wang L, Cordie T, Vokoun C, Eliceiri KW, Gong S. Multi-functional self-fluorescent unimolecular micelles for tumor-targeted drug delivery and bioimaging. Biomaterials. 2015;47:41–50. doi: 10.1016/j.biomaterials.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Jaskula–Sztul R, Harrison A, Dammalapati A, Xu W, Cheng Y, Chen H, Gong S. KE108-conjugated unimolecular micelles loaded with a novel HDAC inhibitor thailandepsin-A for targeted neuroendocrine cancer therapy. Biomaterials. 2016 doi: 10.1016/j.biomaterials.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, Siddiqui IA, Nihal M, Pilla S, Rosenthal K, Mukhtar H, Gong S. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials. 2013;34:5244–5253. doi: 10.1016/j.biomaterials.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Hong H, Chen G, Shi S, Zheng Q, Zhang Y, Theuer CP, Barnhart TE, Cai W, Gong S. Image-guided and tumor-targeted drug delivery with radiolabeled unimolecular micelles. Biomaterials. 2013;34:8323–8332. doi: 10.1016/j.biomaterials.2013.07.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaskula-Sztul R, Chen G, Dammalapati A, Harrison A, Tang W, Gong S, Chen H. AB3-loaded and tumor-targeted unimolecular micelles for medullary thyroid cancer treatment. J Mater Chem B Mater Biol Med. 2016;5:151. doi: 10.1039/C6TB02530G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Size-dependent endocytosis of nanoparticles. Adv Mater. 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putnam D, Gentry CA, Pack DW, Langer R. Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc Natl Acad Sci. 2001;98:1200–1205. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo S, Cheng R, Meng F, Park TG, Zhong Z. Water soluble poly (histamine acryl-amide) with superior buffer capacity mediates efficient and nontoxic in vitro gene transfection. J Polym Sci A Polym Chem. 2011;49:3366–3373. [Google Scholar]
- 34.Fleige E, Achazi K, Schaletzki K, Triemer T, Haag R. pH-responsive dendritic core– multishell nanocarriers. J Control Release. 2014;185:99–108. doi: 10.1016/j.jconrel.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 35.Xu S, Luo Y, Haag R. Water-soluble pH-responsive dendritic core-shell nanocarriers for polar dyes based on poly (ethylene imine) Macromol Biosci. 2007;7:968–974. doi: 10.1002/mabi.200700066. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Zhong P, Meng F, Cheng R, Deng C, Feijen J, Zhong Z. Redox and pH-responsive degradable micelles for dually activated intracellular anticancer drug release. J Control Release. 2013;169:171–179. doi: 10.1016/j.jconrel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Meng F, Cheng R, Deng C, Zhong Z. Intracellular drug release nanosystems. Mater Today. 2012;15:436–442. [Google Scholar]
- 38.Cheng R, Feng F, Meng F, Deng C, Feijen J, Zhong Z. Glutathione-responsive nanovehicles as a promising platform for targeted intracellular drug and gene delivery. J Control Release. 2011;152:2–12. doi: 10.1016/j.jconrel.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 39.Cheng R, Meng F, Deng C, Klok H-A, Zhong Z. Dual and multi-stimuli responsive polymeric nanoparticles for programmed site-specific drug delivery. Biomaterials. 2013;34:3647–3657. doi: 10.1016/j.biomaterials.2013.01.084. [DOI] [PubMed] [Google Scholar]
- 40.Phan UT, Arunachalam B, Cresswell P. Gamma-interferon-inducible lysosomal thiol reductase (GILT) maturation, activity, and mechanism of action. J Biol Chem. 2000;275:25907–25914. doi: 10.1074/jbc.M003459200. [DOI] [PubMed] [Google Scholar]
- 41.Arunachalam B, Phan UT, Geuze HJ, Cresswell P. Enzymatic reduction of disulfide bonds in lysosomes: characterization of a gamma-interferon-inducible lysosomal thiol reductase (GILT) Proc Natl Acad Sci. 2000;97:745–750. doi: 10.1073/pnas.97.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brinkman AM, Chen G, Wang Y, Hedman CJ, Sherer NM, Havighurst TC, Gong S, Xu W. Aminoflavone-loaded EGFR-targeted unimolecular micelle nanoparticles exhibit anti-cancer effects in triple negative breast cancer. Biomaterials. 2016;101:20–31. doi: 10.1016/j.biomaterials.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang H, Chen X, Rui M, Sun W, Chen J, Peng J, Xu Y. Effects of surface displayed targeting ligand GE11 on liposome distribution and extravasation in tumor. Mol Pharm. 2014;11:3242–3250. doi: 10.1021/mp5001718. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Zhao R, Wu X, Sun Y, Yao M, Li J, Xu Y, Gu J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005;19:1978–1985. doi: 10.1096/fj.05-4058com. [DOI] [PubMed] [Google Scholar]
- 45.Gumuskaya B, Alper M, Hucumenoglu S, Altundag K, Uner A, Guler G. EGFR expression and gene copy number in triple-negative breast carcinoma. Cancer Genet Cytogenet. 2010;203:222–229. doi: 10.1016/j.cancergencyto.2010.07.118. [DOI] [PubMed] [Google Scholar]
- 46.Sheng Q, Liu J. The therapeutic potential of targeting the EGFR family in epithelial ovarian cancer. Br J Cancer. 2011;104:1241–1245. doi: 10.1038/bjc.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tobita K, Kijima H, Dowaki S, Kashiwagi H, Ohtani Y, Oida Y, Yamazaki H, Nakamura M, Ueyama Y, Tanaka M. Epidermal growth factor receptor expression in human pancreatic cancer: significance for liver metastasis. Int J Mol Med. 2003;11:305–309. [PubMed] [Google Scholar]
- 48.Nicholson RI, Gee JMW, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 49.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S. Amphiphilic multi-arm-block copolymer conjugated with doxorubicin via pH-sensitive hydrazone bond for tumor-targeted drug delivery. Biomaterials. 2009;30:5757–5766. doi: 10.1016/j.biomaterials.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 50.Segovia N, Pont M, Oliva N, Ramos V, Borrós S, Artzi N. Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer. Adv Healthc Mater. 2015;4:271–280. doi: 10.1002/adhm.201400235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


