Abstract
Recently, we developed a new paradigm in macromolecular therapeutics that avoids the use of low molecular weight drugs. The activity of the “drug-free macromolecular therapeutics” is based on the biorecognition of complementary motifs at cell surface resulting in receptor crosslinking and apoptosis induction. The system is composed of two nanoconjugates: (1) a single-stranded morpholino oligonucleotide (MORF1) attached to an anti-CD20 Fab′ fragment (Fab′-MORF1); (2) multiple copies of complementary oligonucleotide MORF2 grafted to a linear polymer of N-(2-hydroxypropyl)methacrylamide (HPMA) – P-(MORF2)x. The two conjugates crosslink CD20 antigens via MORF1-MORF2 hybridization at the surface of CD20+ malignant B-cells and induce apoptosis. Preclinical studies in a murine model of human non-Hodgkin’s lymphoma showed cancer cells eradication and long-term survivors. The aim of this study was to determine the relationship between the detailed structure of the nanoconjugates and apoptosis induction in Raji cells to allow system optimization. The factors studied include the length of the MORF sequence, the valence of P-(MORF2)x (varying x), molecular weight of P-(MORF2)x, incorporation of a miniPEG spacer between Fab′ and MORF1 and between polymer backbone and pendant MORF2, and comparison of two Fab′ fragments, one from 1F5 antibody (Fab′1F5), the other from Rituximab (Fab′RTX). The results of apoptosis induction in human Burkitt’s B-cell non-Hodgkin’s lymphoma (NHL) Raji cells as determined using three apoptotic assays (Annexin V, Caspase 3, and TUNEL) indicated that: a) An improvement of apoptotic activity was observed for a 28 base pair MORF sequence when compared to MORFs composed of 20 and 25 base pairs. The differences depended on type of assay, concentration and exposure schedule (consecutive vs. premixed). b) The higher the valence of P-(MORF2)x the higher the levels of apoptosis. c) Higher molecular weight of P-(MORF2)x induced higher levels of apoptosis. d) A miniPEG8 spacer was effective in enhancing apoptotic levels in contrast to a miniPEG2 spacer. e) There was not a statistically significant difference when comparing Fab′1F5-MORF1 with Fab′RTX-MORF1.
Keywords: Biorecognition, Drug-free macromolecular therapeutics, Receptor crosslinking, Apoptosis N-(2-hydroxypropyl)methacrylamide (HPMA), Morpholino oligonucleotide, CD20, B-cell lymphoma
1. Introduction
Molecular biorecognition is at the center of all biological processes. Many life activities are based on self-assembly and specific interaction between macromolecules, e.g. coiled-coil peptide/protein recognition, antibody-antigen binding and DNA hybridization. The high-fidelity self-assembling nature motifs and their mimics can be employed to label/functionalize cell surfaces [1], influence immune response [2], control cell signaling pathways [3–5], manipulate cell fate [6–7], and trigger cellular events [3,8–17]. Biorecognition forms the basis for the design of precisely defined smart systems, including targeted therapeutics [10,12], imaging agents, stimuli-sensitive and self-assembled biomaterials [18], and biosensors [19]. Incorporation of native biorecognition motifs as grafts attached to synthetic polymer chains results in hybrid macromolecules that have the potential for self-assembly. For example, two N-(2-hydroxypropyl)methacrylamide (HPMA) copolymers containing grafts of complementary coiled-coil forming peptides [20] or HPMA copolymers grafted with peptide nucleic acids self-assemble into 3D hydrogels [21]. Similarly, two poly(N,N-dimethylacrylamide)s grafted with complementary oligonucleotides (oligoT10 and oligoA10) self-assemble into hydrogels [22].
Cell surface receptor clustering (crosslinking) as a result of multivalent ligand biorecognition is a driving force of various cellular events such as growth factor signaling [23], immune system function [24,25], neuronal cell communication [26], hormone uptake [27], cell adhesion [28], activation [29], and apoptosis [30,31]. For example, EGF receptor crosslinking appears to be a necessary and sufficient signal for induction of DNA synthesis [32]. Crosslinking a variety of human plasmacytoid dendritic cell surface receptors leads to the regulation of interferon-α production [33]. Insulin-receptor crosslinking may enhance the biological activity of insulin [27]. When bound to antibodies CD20 receptors are hyper-crosslinked via Fc receptor-expressing immune effector cells (e.g., macrophages, natural killer cells), resulting in apoptosis induction in B-cells [34].
Conventional polymeric nanomedicines utilize polymers as delivery vehicles to modify the biodistribution of anticancer drugs. Recent designs of nanomedicines add another function – to trigger or improve therapeutic effects through natural biological responses [35]. Macromolecular therapeutics employ biomimetic strategies to stimulate or control specific cellular activities [36,37]. The research presented here is based on a novel paradigm in the nanomedicine research area – drug-free macromolecular therapeutics [9]. The basic idea is to induce apoptosis by crosslinking of cell-surface (slowly internalizing) receptors mediated by the biorecognition of high-fidelity natural binding motifs, such as antiparallel coiled-coil peptides [10] or complementary oligonucleotides [12].
Non-Hodgkin’s lymphoma (NHL) is a prevalent cancer in the United States with around 72,000 new cases in 2015 [38]. About 85% NHL originate from B-cells the remaining diseases are mostly of T-cell origin [39]. CD20 is one of the most reliable biomarkers of B-lymphocytes. It is highly expressed on the surfaces of most malignant B-cells, as well as normal B-cells; however, it is not expressed on stem cells or progenitor cells and mature or activated plasma cells [40]. Clustering of CD20 receptors at the surface of B-cells mediates the interaction of CD20 with Src-family kinases and triggers apoptotic signaling [41]. The therapeutic efficacy of anti-CD20 mAb (Rituximab) is ascribed to three cellular events: antibody-dependent cellular cytotoxicity (ADCC), complement-dependent cytotoxicity (CDC), and CD20-mediated apoptosis [30,34]. All of these mechanisms require immune effector cells to function [42]. The clinical non-responsiveness and adverse effects of Rituximab or other therapeutic mAb has been partly attributed to the Fc fragment-related biological events [43,44].
In contrast, drug-free macromolecular therapeutics trigger direct and specific apoptosis of B-cell lymphomas without the help of effector cells [10]. This is achieved by the design of synthetic effectors that reproduce the function of immune effector cells. Two nanoconjugates are being used: the first is an anti-CD20 Fab′ fragment conjugated to one natural motif (peptide 1, oligonucleotide 1). B cells exposed to this nanoconjugate are decorated with the natural motif. Since the CD20 receptor is very slowly internalizing a new biorecognition site is created. Further exposure of decorated cells to an HPMA copolymer grafted with multiple copies of the complementary motif (peptide 2, oligonucleotide 2) results in crosslinking of CD20 receptors and initiation of apoptosis [9]. This design was validated using a complementary pair of coiled-coil forming peptides CCE and CCK [20]. Apoptosis was initiated by CD20 receptor crosslinking both in vitro [10,17,45] and in vivo [11].
Our recent studies use a pair of complementary morpholino oligonucleotides (MORF1, MORF2) due to their fast hybridization, excellent binding affinity, and stability in plasma as well as water-solubility. The system is composed of two nanoconjugates: (1) a single-stranded morpholino oligonucleotide MORF1 attached to an anti-CD20 Fab′ fragment, (2) multiple copies of complementary oligonucleotide MORF2 grafted to a linear HPMA copolymer – P-(MORF2)x. The two conjugates crosslink CD20 antigens via MORF1-MORF2 hybridization at the surface of CD20+ malignant B-cells and induce apoptosis [12–17]. Preclinical studies in a murine model of human non-Hodgkin’s lymphoma showed cancer cells eradication and long-term survivors [12,14].
The aim of this study is to determine the relationship between the detailed structure of nanoconjugates and apoptosis induction in Raji cells to allow system optimization. The factors studied include the length of the MORF sequence, the valence of P-(MORF2)x (varying x), molecular weight of P-(MORF2)x, incorporation of a miniPEG spacer between Fab′ and MORF1 and between polymer and MORF2, and comparison of two Fab′ fragments, one from 1F5 antibody (Fab′1F5), the other from Rituximab (Fab′RTX).
2. Materials and methods
2.1. Materials
N-(3-Aminopropyl)methacrylamide hydrochloride (APMA) was purchased from Polysciences (Warrington, PA). 2,2′-Azobis(2,4-dimethylvaleronitrile) (V-65), 2,2′-azobis[2-(2-imidazolin-2-yl) propane]dihydrochloride (VA-044), and 4,4-azobis(4-cyanopentanoic acid) (V-501) were from Wako Chemicals (Rich-mond, VA). Succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) was from Soltec Ventures (Beverly, MA), cross-linkers SM(PEG)2 (succinimidyl-[(N-maleimidopropionamido)-diethyleneglycol] ester) and SM(PEG)8 (succinimidyl-[(N-maleimidopropionamido)-octaethyleneglycol] ester), were from Thermo Fisher Scientific. N-(2-Hydroxypropyl)methacrylamide (HPMA) [46] and 4-cyanopentanoic acid dithiobenzoate (CPDB) [47] were prepared as previously described. Pepsin, 1-amino-2-propanol, and cysteamine were from Sigma-Aldrich and tris(2-carboxyethyl)phosphine (TCEP) from Thermo Scientific. All solvents were obtained from Sigma-Aldrich as the highest purity available.
The complementary 3′-amine-derivatized (20, 25 and 28)-mer phosphorodiamidate morpholino oligonucleotides and 3′-disulfide amide 25-mer phosphorodiamidate morpholino oligomers (MORF2-SSR) were from Gene Tools (Philomath, OR). MORF1 (20 bp, Mw = 6926 Da; 25 bp, Mw = 8631 Da; 28 bp, Mw = 9639 Da); MORF2-SSR (25 bp, Mw = 8585); MORF2, (20 bp, Mw = 6792 Da; 25 bp, Mw = 8437 Da; 28 bp, Mw = 9438 Da). For the design of base sequences, a sequence scrambling software (http://www.sirnawizard.com/scrambled.php) and a sequence analysis software (http://www.basic.northwestern.edu/biotools/oligocalc.html) were used [12].
2.2. Methods
2.2.1. Synthesis of Fab′1F5-MORF1, Fab′1F5-PEG2-MORF1 and Fab′RTX-MORF1 conjugates
The 1F5 mAb (IgG2a) was prepared from a mouse hybridoma cell subclone 1F5 (ATCC) in a CellMax® bioreactor (Spectrum Laboratories) and purified on a Protein G Sepharose 4 Fast Flow column (GE Healthcare). Rituximab (RTX) was purchased from Biogen Idec and Genentech USA. Preparation of Fab′1F5 (or Fab′RTX) followed a previously reported procedure [12,14]. Briefly, antibodies were digested into F(ab′)2 with 10% (w/w) pepsin in citric buffer (pH 4.0). Immediately before conjugation, F(ab′)2 was reduced to Fab′-SH by 10 mM TCEP.
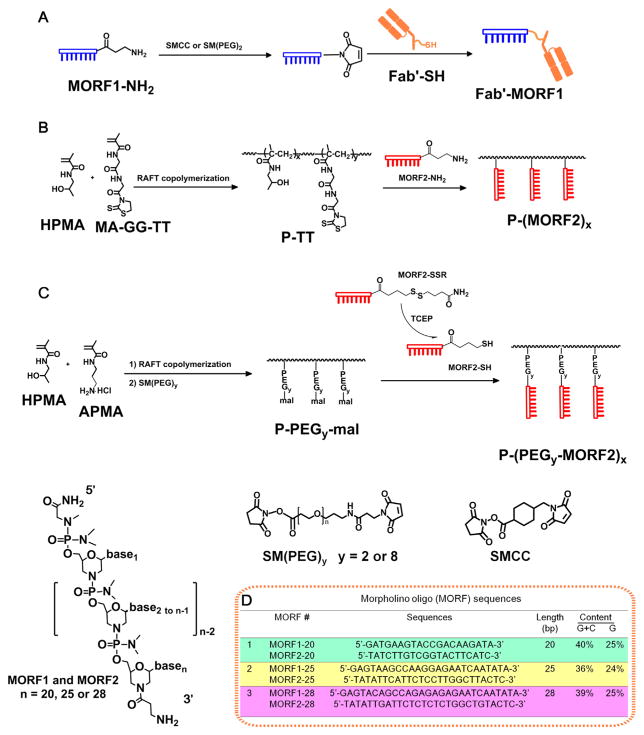
The Fab′1F5-MORF1, Fab′1F5-PEG2-MORF1 and Fab′RTX-MORF1 conjugates were synthesized similarly as previously described [12,14]. A typical procedure (Scheme 1) was as follows: First, 200 nmol MORF1-NH2 (20, 25 or 28 bps) was reacted with 1.34 mg (4 μmol) SMCC (or SM(PEG)2) in 200 μL DMSO to produce the MORF1-mal (containing a 3′-maleimide). The reaction was performed at room temperature (RT) for 24 h. The excess of SMCC (or SM(PEG)2) was removed by ultrafiltration (3000 Da cut-off) with PBS (pH 6.5) four times to yield MORF1-mal.
Scheme 1.
Synthesis of Fab′-MORF1 and P-(MORF2)X conjugates. (A) Scheme of Fab′-MORF1 synthesis. SMCC: succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate heterobifunctional linker. SM(PEG)n: NHS- and maleimide-activated miniPEG. (B) Scheme of the synthesis of P-(MORF2)x. (C) Scheme of synthesis of P-(PEGy-MORF2)x. MA-GG-TT: N-methacryloylglycylglycine thiazolidine-2-thione, APMA: N-(3-aminopropyl)methacrylamide hydrochloride. (D) Structure of morpholino oligonucleotide sequences.
Second, the MORF1-mal was mixed with 60 nmol (3 mg) freshly reduced Fab′-SH in 600 μL PBS (pH 6.5). The reaction was performed at 4 °C for 24 h. Finally, the Fab′-MORF1 conjugate was purified using size exclusion chromatography (SEC) to remove free, unconjugated Fab′ and MORF1. An ÄKTA FPLC system equipped with Sephacryl S-100 HR16/60 column (GE Healthcare) eluted with PBS (pH 7.2) was used. To determine Fab′ equivalent concentration of the Fab′-MORF1 conjugate, a bicinchoninic acid (BCA) protein assay (Pierce) was used. UV–visible spectroscopy (Varian Cary 400, Agilent Technologies) was used for quantification of the MORF1 equivalent concentration; the molar absorptivity of MORF1-20 bp, MORF1-25 bp and MORF1-28 bp were 220,470, 278,000, and 309,980 M−1 cm−1, respectively (at 265 nm, in 0.1 N HCl).
2.2.2. Synthesis of P1-MORF2 conjugates
2.2.2.1. Synthesis of multivalent P1-(MORF2)x
The multivalent P1-(MORF2)x (25 bp) conjugates and conjugates with different MORF2 sequences were synthesized as previously described [12,14]. First, the polymer precursor (P1-TT), a copolymer of HPMA and N-methacryloylglycylglycine thiazolidine-2-thione (MA-GG-TT) was synthesized by RAFT copolymerization. Second, P1-TT was reacted with MORF2-NH2 to produce P1-(MORF2)x conjugates (Scheme 1B).
2.2.2.1.1. Synthesis of P1-TT
In RAFT copolymerization, CPDB was used as the chain transfer agent (CTA), and 2,2′-azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride (VA-044) as the initiator. The reaction was carried out in DMSO and methanol containing 0.3% (v/v) acetic acid (MeOH/H+). A typical procedure was as follows: HPMA (122 mg, 0.85 mmol) and MA-GG-TT (53 mg, 0.15 mmol) were added into an ampoule attached to a Schlenk line. After three vaccum/nitrogen cycles to remove oxygen, 600 μL degassed DMSO was added to dissolve monomers, followed by addition of CPDB solution (0.28 mg in 70 μL DMSO) and VA-044 solution (0.11 mg in 55 μL MeOH/H+) via syringe. The mixture was bubbled with nitrogen for 15 min before sealing the ampoule; the copolymerization was performed at 40 °C for 40 h. The copolymer was isolated by precipitation into acetone and purified by dissolution-precipitation in methanol-acetone twice and dried under vacuum. Yield of P1-TT was 100 mg (51%). The number average molecular weight (Mn) and molecular weight distribution (polydispersity, PDI) of P1-TT were determined by SEC using ÄKTA FPLC equipped with miniDAWN and OptilabREX detectors (GE Healthcare). Superose 6 HR10/30 column (GE Healthcare) was used, with sodium acetate buffer (pH 6.5) and 30% acetonitrile (v/v) as the mobile phase. To remove the terminal (active) dithiobenzoate groups, P1-TT copolymers were reacted with 2,2′-azobis(2,4-dimethyl valeronitrile) (V-65). Briefly, P1-TT (100 mg, Mn = 86 kDa, 1.16 μmol) and V-65 (40× excess, 11 mg, 46 μmol) were added into an ampoule. After three vacuum-nitrogen cycles to remove oxygen, 0.6 mL MeOH/H+ was added. The solution was bubbled with nitrogen for 15 min, sealed and reacted at 55 °C for 2 h. The end-modified copolymer was purified by precipitation into acetone twice and then dried under vacuum (yield 94 mg, 94%). The content of TT groups in the copolymers was determined by UV absorbance at 305 nm (molar absorptivity = 10,900 M−1 cm−1 in methanol) [48].
2.2.2.1.2. Attachment of MORF2-NH2 to P1-TT to produce P1-(MORF2)x
The P1-TT described above was reacted with MORF2-NH2 to produce multivalent P1-(MORF2)x (25 bp) or different sequences (20 or 28 bp). P1-TT (1 mg; Mn = 86 kDa; containing 770 nmol TT groups) was mixed with MORF2-NH2-25 bp (1.1 mg, 130 nmol; 2.1 mg, 250 nmol; 5.1 mg, 600 nmol), MORF2-NH2-20 bp (4.1 mg, 600 nmol) or MORF2-NH2-28 bp (5.8 mg, 600 nmol) in 200 μL PBS (pH 7.4). The solution mixture in an ampoule was stirred at RT for 24 h; then 1 μL 1-amino-2-propanol was added and stirred for another 15 min to aminolyze unreacted TT groups on the polymer chains, if any. After reaction, unreacted MORF2-NH2 was removed by ultrafiltration (30,000 Da cut-off) four times with DI H2O. P1-(MORF2)x was characterized by UV absorbance at 265 nm. To quantify the content of MORF2 and determine the valence (number of MORF2 per polymer chain), the P1-(MORF2)x conjugates were freeze-dried and dissolved in 0.1 N HCl prior to UV–Vis analysis. The molar absorptivity of MORF2-20 bp, MORF2-25 bp and MORF2-28 bp were 201,090 M−1 cm−1, 252,120 M−1 cm−1 and 280,910 M−1 cm−1, respectively (at 265 nm, in 0.1 N HCl). The valences of the P1-(MORF2)x conjugates were calculated based on the resulting MORF2 contents and the Mn of the polymer backbones (as previously determined by SEC).
2.2.2.2. Synthesis of multivalent P2-PEG2-MORF2, P2-PEG8-MORF2 and P3-PEG2-MORF2 conjugates
These conjugates were prepared by the reaction of HPMA copolymer containing maleimido groups at side chain termini with 3′-thiol modified MORF2 (Scheme 1C). Briefly:
2.2.2.2.1. Synthesis of P2-PEG2-mal, P2-PEG8-mal and P3-PEG2-mal
In RAFT copolymerization, CPDB was used as the CTA and V-501 as the initiator. The reaction was carried out in H2O. A typical procedure was as follows: HPMA (129 mg, 0.9 mmol) and APMA (17.8 mg, 0.1 mmol) were added into an ampoule attached to a Schlenk line. After three vacuum nitrogen cycles to remove oxygen, 600 μL degassed H2O was added to dissolve monomers, followed by addition of 0.33 mg CPDB (or 0.16 mg, in 33 μL MeOH) and 0.17 mg V501 solution (or 0.08 mg, in 17 μL MeOH) via syringe. The mixture was bubbled with nitrogen for 30 min before sealing the ampoule; the copolymerization was performed at 70 °C for 24 h (or 40 h). The copolymers were purified by dialysis in DI H2O and the products isolated by freeze-drying. Yield of P2-NH2 was 115 mg, 78% and of P3-NH2 118 mg, 79%. Mn and PDI of P2-NH2 and P3-NH2 were 112 kDa and 1.17 or 291 kDa and 1.17, respectively as determined by SEC, using ÄKTA FPLC equipped with Superose 6 HR10/30 column with PBS buffer as the mobile phase. After end-modification with 40-times excess of V-65, the amino content of the copolymers was determined by ninhydrin assay [49]. P2-NH2: [NH2]: 75 per copolymer chain, Mn = 112 kDa; P3-NH2: [NH2]: 151 per copolymer chain, Mn = 291 kDa.
P2-NH2 was mixed with 2 eq. SM(PEG)2 (or SM(PEG)8) and stirred at RT for 6 h to prepare P2-PEG2-mal (or P2-PEG8-mal). The excess SM(PEG)2 (or SM(PEG)8) was removed by ultrafiltration (30 kDa cutoff) four times with DI H2O/MeOH (9/1). Maleimide group content was determined by modified Ellman’s assay [50]: ([mal]: 54 per polymer, 72% conversion for SM(PEG)2; [mal]: 33 per polymer, 44% conversion for SM(PEG)8).
P3-NH2 was mixed with 2 eq. SM(PEG)2 and stirred at RT overnight to prepare P3-PEG2-mal. After removing excess SM(PEG)2, maleimide group content was determined by modified Ellman’s assay: ([mal]: 125 per polymer, 71% conversion for SM(PEG)2).
2.2.2.2.2. Attachment of MORF2-SSR to P2-PEG2-mal or P3-PEG2-mal to produce P2-PEG2-MORF2 and P3-PEG2-MORF2 conjugates
MORF2-SSR (2.6 mg, 300 nmol) was reduced with TCEP (10 mM) in 1 mL PBS (RT, 2 h). The excess TCEP was removed by ultrafiltration (3000 cut-off) four times with PBS. 1.4 mg P2-PEG2-mal (or 3.6 mg P2-PEG8-mal, or 1.8 mg P3-PEG2-mal) in 200 uL PBS was added and incubated at RT overnight, then 10× excess of cysteamine (10 mM × 54 μL in PBS) was added and stirred for another 2 h to block unreacted mal groups on the polymer chains. The excess cysteamine and free MORF2-SH were removed by ultrafiltration (50,000 cut-off) four times with DI H2O. The valences of the P2-(PEG2-MORF2)x, P2-(PEG8-MORF2)x and P3-(PEG2-MORF2)x conjugates were calculated based on MORF2 contents and Mn of the polymer backbones (as previously determined by SEC).
2.3. Detection of hybridization by UV–visible spectroscopy
Analysis of the hypochromic effect upon MORF1-MORF2 hybridization was performed using a Varian Cary 400 Bio UV–visible spectrophotometer (Agilent Technologies, Santa Clara, CA). Fab′-MORF1 and P1-(MORF2)x conjugates were dissolved in 1 mL PBS (pH 7.4) each at a concentration of 1.2–1.5 μM (MORF equivalent) and then mixed in different ratios. The final concentrations of MORF oligos (MORF1 + MORF2) in every solution mixture were kept constant. For example, the mixture containing 75% MORF1 (or 25% MORF2 20 bp) was done by mixing 0.75 mL of 1.5 μM MORF1 solution with 0.25 mL of 1.5 μM MORF2 solution. Samples were placed in a 1-cm quartz cuvette for measurement. The optical density (OD) at 260 nm (contributed by bases) was recorded. All measurements were performed in triplicate.
2.4. Dynamic light scattering (DLS)
The hydrodynamic effective diameters of the conjugates, Fab′-MORF1 and P-(MORF2)x in PBS (pH 7.4) were analyzed by DLS using a Wyatt DynaPro Plate Reader II and analyzed using Dynamics 7 software. The conjugates at a concentration of about 1 mg/mL were filtered through a 0.22 μm filter prior to measurement. The mean particle diameters were recorded. Furthermore, DLS was used to characterize the change of particle size upon the binding of Fab′-MORF1 and P-(MORF2)x. The analysis was performed at different times (5, 10, 30, and 60 min) after mixing the two conjugates at equimolar MORF1/MORF2 concentrations. All samples contained a major population of particles indicating the hybridized conjugates, as well as minor populations indicating unbound Fab′-MORF1 and P-(MORF2)x. The mean effective diameter of the major population was recorded. All measurements were performed in triplicate.
2.5. Apoptosis assays
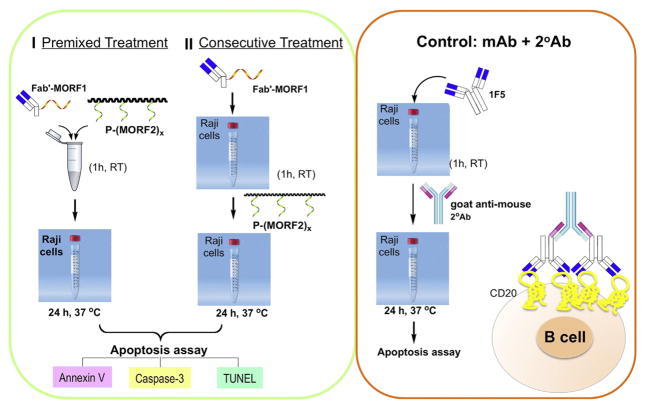
In vitro apoptosis induction of human Burkitt’s B-cell non-Hodgkin’s lymphoma (NHL) Raji cells by co-treatment with Fab′-MORF1 and P-(MORF2)x was evaluated by three assays: Annexin V/propidium iodide (PI) binding assay, Caspase-3 activation assay, and TUNEL (terminal deoxynucleotide mediated-dUTP nick-end labeling) assay. Two cell exposure protocols were used (Scheme 2). Consecutive exposure: the cells were incubated with Fab′-MORF1 conjugate first. After 1 h the cells decorated with MORF1 were exposed to P-(MORF2)x for 24 h. Premixed exposure: The nanoconjugates Fab′-MORF1 and P-(MORF2)x were mixed at 37 °C and after 1 h Raji cell were exposed to a preformed, self-assembled multivalent conjugate for 24 h. In all experiments, 1F5 or Rituximab (RTX) mAb hyper-crosslinked with a goat antimouse (GAM) secondary antibody (2° Ab) (KPL, Gaithersburg, MD) was used as a positive control (molar ratio 1F5:GAM = 2:1). Non-treated cells (in culture media) were used as negative controls (Scheme 2). The procedures of each assay are described below:
Scheme 2.
Apoptosis induction in Raji cells. Premixed exposure, consecutive exposure, and positive control.
2.5.1. Annexin V/PI assay
Annexin V-FITC and PI staining were performed following the RAPID™ protocol provided by the manufacturer (Oncogene Research Products, Boston, MA). For the consecutive treatment, 2 × 105 Raji cells were suspended in 0.4 mL fresh growth medium containing 0.2 or 1 μM Fab′-MORF1. The cells were incubated for 1 h in a humidified atmosphere at 37 °C with 5% CO2, and then washed twice with PBS + 1% bovine serum albumin (BSA), followed by resuspension in 0.4 mL medium containing 0.2 or 1 μM (MORF2-eqv.) of P-(MORF2)x. The cell suspension was incubated for 24 h. For the premixed treatment, first, 0.2 or 1 μM Fab′-MORF1 was mixed with 0.2 or 1 μM (MORF2-eqv.) P-(MORF2)x in culture medium at RT for 1 h, and then 2 × 105 Raji cells were suspended in 0.4 mL of the premixed solution. The cell suspension was incubated for 24 h. For the positive control, cells were first incubated with 0.5 or 1 μM of 1F5 mAb or RTX in culture medium for 1 h, and then washed twice with PBS + 1% BSA, followed by resuspension in 0.4 mL of fresh growth medium containing 0.25 or 0.5 μM GAM. The cells were incubated for another 24 h at 37 °C. Prior to staining, cells were washed twice with PBS. All experiments were carried out in triplicate.
2.5.2. Caspase-3 assay
A Phi-PhiLux kit (OncoImmunin, Gaithersburg, MD) was used. For the consecutive treatment, 2 × 105 Raji cells were suspended in 0.4 mL fresh growth medium S7 containing 1 or 0.2 μM Fab′-MORF1. The cells were incubated for 1 h in a humidified atmosphere at 37 °C with 5% CO2, and then washed twice with PBS + 1% BSA, followed by resuspension in 0.4 mL medium containing 1 or 0.2 μM (MORF2-eqv.) P-(MORF2)x. The cell suspension was incubated for 24 h. For the premixed treatment, first, 1 or 0.2 μM Fab′-MORF1 was mixed with 1 or 0.2 μM (MORF2-eqv.) P1-(MORF2)x in culture medium at 37 °C for 1 h, and then 2 × 105 Raji cells were suspended in 0.4 mL of the premixed solution. The cell suspension was incubated for 24 h. For the positive control, cells were firstly incubated with 0.4 mL 0.5 or 1 μM of 1F5 mAb or RTX in culture medium for 1 h, and then washed twice with PBS + 1% BSA, followed by resuspension in 0.4 mL of fresh growth medium containing 0.25 or 0.5 μM GAM. The cells were incubated for another 24 h at 37 °C. After the treatment, cells were washed twice with PBS and analyzed for caspase-3 activity following the manufacturer’s protocol. All experiments were carried out in triplicate.
2.5.3. TUNEL assay
An Apo Direct TUNEL kit (Phoenix Flow Systems, San Diego, CA) was used. For the consecutive treatment, 106 Raji cells were suspended in 1 mL fresh growth medium containing 1 μM Fab′-MORF1. The cells were incubated for 1 h in a humidified atmosphere at 37 °C with 5% CO2, and then washed twice with PBS + 1% BSA, followed by resuspension in 0.5 mL medium containing 1 μM (MORF2-eqv.) P-(MORF2)x. The cell suspension was incubated for 24 h. For the premixed treatment, 1 μM Fab′-MORF1 was mixed with 1 μM (MORF2-eqv.) P-(MORF2)x in culture medium at 37 °C for 1 h, and then 106 Raji cells were suspended in 1 mL of the premixed solution. The cell suspension was incubated for 24 h. For the positive control, cells were firstly incubated with 1 mL 1 μM of 1F5 mAb or RTX in culture medium for 1 h, and then washed twice with PBS + 1% BSA, followed by resuspension in 1 mL of fresh growth medium containing 0.5 μM GAM. The cells were incubated for another 24 h at 37 °C. After the treatments, cells were washed twice with PBS and fixed with 2% paraformaldehyde in PBS for 1 h at RT. Cells were then permeabilized in 70% ethanol overnight at 4 °C. Prior to analysis, nick-end labeling was performed following the manufacturer’s protocol. All experiments were carried out in triplicate.
2.6. Statistical analysis
Statistical analyses were performed by Student’s t-test to compare between two groups, or one-way analysis of variance (ANOVA) to compare three or more groups (with p value < 0.05 indicating statistically significant difference).
3. Results
3.1. Influence of the structure and length of MORF strands on hybridization and apoptosis induction in Raji B-cells
All our previous studies were done with 25 bp MORFs [12–17]. Here two additional pairs of MORFs (20 bp and 28 bp) were employed to evaluate the effect of length on hybridization and apoptosis induction. The 20 and 28 bp MORFs (see structure in Scheme 1D and Table S1) were designed using the same approach as previously described [12]. For the design of base sequences, a sequence scrambling software (http://www.sirnawizard.com/scrambled.php) and a sequence analysis software (http://www.basic.northwestern.edu/biotools/oligocalc.html) were used. To avoid self-complementarity, the “minimum” base pairs required for self-dimerization and “minimum base pairs required for a hairpin” were set to “3” (for 20 bp) and “4” (for 28 bp). The properties of 20 and 28 bp conjugates were compared with the 25 bp conjugates.
3.1.1. Fab′1F5-MORF1 conjugates
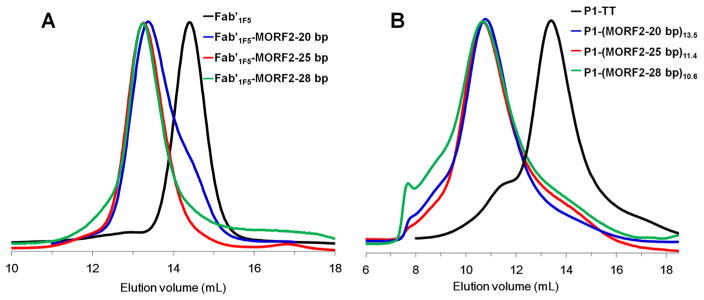
Fab′1F5-MORF1 conjugates (20 and 28 bp) were synthesized by the same methods as previously described except that MORF1-25 bp was replaced by MORF1-20 bp or MORF-28 bp. FPLC analysis confirmed the successful synthesis and purity of Fab′1F5-MORF1-20 bp and Fab′1F5-MORF1-28 bp conjugates (Fig. 1A). Fab′1F5-MORF1 conjugates with different MORF1 sequences are characterized in Table 1.
Fig. 1.
SEC analysis of (A) Fab′1F5 fragment (Fab′1F5-SH) of 1F5 mAb and the Fab′1F5-MORF1 conjugates with different MORF1 sequences. Sephacryl S-100 HR16/60 column with PBS as the mobile phase; (B) P1-TT and P1-(MORF2)x conjugates with different MORF2 sequences. Superose 6 HR10/30 column with sodium acetate buffer (pH 6.5) and 30% acetonitrile (v/v) as the mobile phase.
Table 1.
Characterization of Fab′-MORF1 conjugates.
| No. | Fab′-MORF1 conjugatea | Fab′/MORF1 |
|---|---|---|
| 1a | Fab′1F5-MORF1-20 bp | 1/1.1 |
| 1b | Fab′1F5-MORF1 | 1/1.1 |
| 1c | Fab′1F5-MORF1-28 bp | 1/1 |
| 2 | Fab′1F5-PEG2-MORF1 | 1/1.2 |
| 3 | Fab′RTX-MORF1 | 1/1.1 |
If not otherwise stated, MORF1 oligos were 25 bp.
3.1.2. P1-(MORF2)x conjugates
MORF2-NH2 (20 or 28 bp) was attached to polymer precursor P1-TT to synthesize P1-MORF2-20 bp and P1-MORF2-28 bp conjugates. After removing free MORF2 by ultrafiltration (50 kDa cut-off), P1-MORF2 (20 and 28 bp) conjugates were characterized by FPLC (Fig. 1B). The valences of two P1-(MORF2)x conjugates were 13.5 and 10.6 for 20 bp and 28 bp conjugates, respectively. P1-(MORF2)x conjugates with different MORF2 sequences are characterized in Table 2.
Table 2.
Characterization of P-MORF2 conjugates.
| No. | Polymer-MORF2 conjugatea | Polymer precursor | Conjugate | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Mn (kDa) | PDI | Mn (kDa)b | PDI | Valencec | |||
| 1a | P1-(MORF2)2.6 | P1-TT | 87 | 1.18 | 109 | 1.36 | 2.6 |
| 1b | P1-(MORF2)4.0 | P1-TT | 87 | 1.18 | 121 | 1.34 | 4.0 |
| 1c | P1-(MORF2)11.4 | P1-TT | 87 | 1.18 | 183 | 1.34 | 11.4 |
| 2a | P1-(MORF2-20 bp)13.5 | P1-TT | 87 | 1.18 | 179 | 1.31 | 13.5 |
| 2b | P1-(MORF2-28 bp)10.6 | P1-TT | 87 | 1.18 | 187 | 1.41 | 10.6 |
| 3a | P2-(PEG2-MORF2)12.7 | P2-mal | 112 | 1.17 | 219 | 1.36 | 12.7 |
| 3b | P2-(PEG8-MORF2)11.2 | P2-mal | 112 | 1.17 | 207 | N/D | 11.2 |
| 4a | P3-(PEG2-MORF2)13.7 | P3-mal | 291 | 1.17 | 407 | N/D | 13.7 |
| 4b | P3-(PEG2-MORF2)25.1 | P3-mal | 291 | 1.17 | 503 | N/D | 25.1 |
If not otherwise stated, MORF2 oligos were 25 base pairs (bp).
Number average molecular weight (Mn) was calculated from the molecular weight of the polymer precursor and the content of MORF2 as determined by UV–visible spectrophotometry.
MORF2 per polymer chain as determined by UV–visible spectrophotometry.
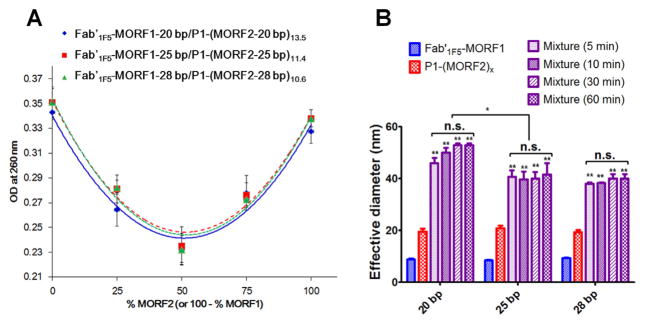
Hybridization of the conjugates via MORF1-MORF2 biorecognition was evaluated by UV–visible spectroscopy as previously described [12]. Two complementary conjugates were mixed in different ratios, and the optical density at 260 nm (contributed by bases) was measured. Upon mixing Fab′-MORF1 and P1-(MORF2)x, a “hypochromic effect” was observed (Fig. 2). The OD260nm reached a minimum when a molar ratio of 1:1 (MORF1:MORF2) was used. Such decrease was due to hydrogen bonding between complementary bases that limited the resonance of the aromatic rings. Similar data were obtained for all (20 bp, 25 bp and 28 bp) conjugates (Fig. 2A). These results indicated that the MORF1-MORF2 hybridization was preserved after attachment to Fab′ or to HPMA polymer. The biorecognition of all three pairs evaluated was high.
Fig. 2.
In vitro hybridization of Fab′1F5-MORF1 and P1-(MORF2)x with different sequences. (A) Hypochromic effect upon hybridization of Fab′1F5-MORF1 and P1-(MORF2)x as analyzed by UV–vis. The optical density (OD) at 260 nm decreased when the two conjugates were mixed (in different ratios). (B) Effective hydrodynamic diameters of the two conjugates and their mixture (equimolar MORF1/MORF2; tested at different times after mixing) as characterized by dynamic light scattering. Statistics, unless otherwise indicated, was performed by comparing the mixture with P1-(MORF2)x (*p < 0.05, **p < 0.005, n.s.: no significant difference).
Furthermore, the binding of Fab′1F5-MORF1 with P1-(MORF2)x containing MORF pairs of different length was characterized by DLS (Fig. 2B). A rapid increase of hydrodynamic size upon mixing the conjugates (at equimolar ratio of MORF1 and MORF2) was observed for all three pairs. However, the kinetics of the 20 bp seems to be a little slower than the kinetics of binding of the conjugate containing MORFs with 25 and 28 bp. Also, the size of the 20 bp hybridized conjugates was some larger than those of the other two pairs. It is likely due to a higher number of MORF2 grafts per polymer chain (13.5 for 20 bp vs. 11.4 for 25 bp and 10.6 for 28 bp).
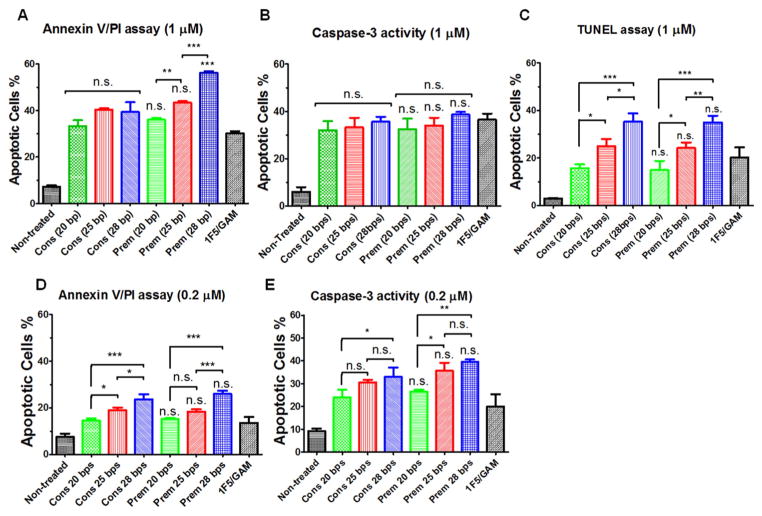
3.1.3. Apoptosis induction
The impact of the structure and length of MORF strands on apoptosis levels in Raji B cells was evaluated by three assays: Annexin V/propidium iodide (PI) binding, Caspase 3 activation, and TUNEL. (1) Consecutive treatment (Cons): Raji cells were exposed to Fab′1F5-MORF1 for 1 h, followed by exposure to P1-(MORF2)x for 24 h; (2) Premixture treatment (Prem): Fab′1F5-MORF1 and P1-(MORF2)x were mixed first. After 1 h Raji cells were exposed to the self-assembled multivalent conjugate (Scheme 2).
The apoptotic activity was evaluated at two concentrations 0.2 and 1 μM (Fig. 3). As expected, increased concentration of nanoconjugates resulted in higher levels of apoptosis. The dose-dependent trends were observed in both consecutive and premixed treatment regimens as well as in the positive control (mAb + 2° Ab). The impact of MORF length was clearly demonstrated. Generally, the longer the MORF sequences, the higher the apoptotic levels. This was observed in four out of five treatment protocols (Fig. 3A, C, D, and E). The exception was the caspase 3 assay at 1 μM concentration (Fig. 3B). There was no statistically significant difference between consecutive and premixed treatm –ents. Only the early apoptotic Annexin V/PI assay at 1 μM concentration showed a difference for 28 pb conjugates. The premixed apoptotic level was about 54%, whereas the consecutive level was about 42%. All nanoconjugates were more effective than the positive control – 1F5/GAM.
Fig. 3.
Effect of the length of MORF sequences on apoptosis induction of Raji B-cells analyzed by different assays. (A, D) Annexin V/PI binding assay. (B, E) Caspase-3 activation assay. (C) TUNEL assay. Incubation time was 24 h. For data in figures A, B, D and E, 2 × 105 cells were used. For data in figure C, 1 × 106 cells were used. Non-treated, cells in culture medium; 1F5/GAM, 1F5 mAb (1 μM or 0.2 μM) followed (1 h later) by goat anti-mouse secondary Ab (0.5 μM or 0.1 μM); Cons, Consecutive, Fab′1F5-MORF1 (1 μM or 0.2 μM) followed (1 h later) by P1-(MORF2)x (1 μM or 0.2 μM, MORF2-eqv); Prem, Premixture of Fab′1F5-MORF1 (1 μM or 0.2 μM) and P1-(MORF2)x (1 μM or 0.2 μM, MORF2-eqv). The percentage of apoptotic cells was quantified by flow cytometry. Statistics, unless otherwise indicated, was performed by comparing each premixed treatment with the corresponding consecutive treatment (***p < 0.0001, **p < 0.005, *p < 0.05, n.s.: no significant difference). All data are presented as mean ± SD (n = 3). The valences of three different P1-(MORF2)x are 13.5 for P1-(MORF2-20 bp)x, 11.4 for P1-(MORF2-25 bp)x, 10.6 for P1-(MORF2-28 bp)x.
3.2. Effect of P-(MORF2)x valence on apoptosis induction in Raji cells
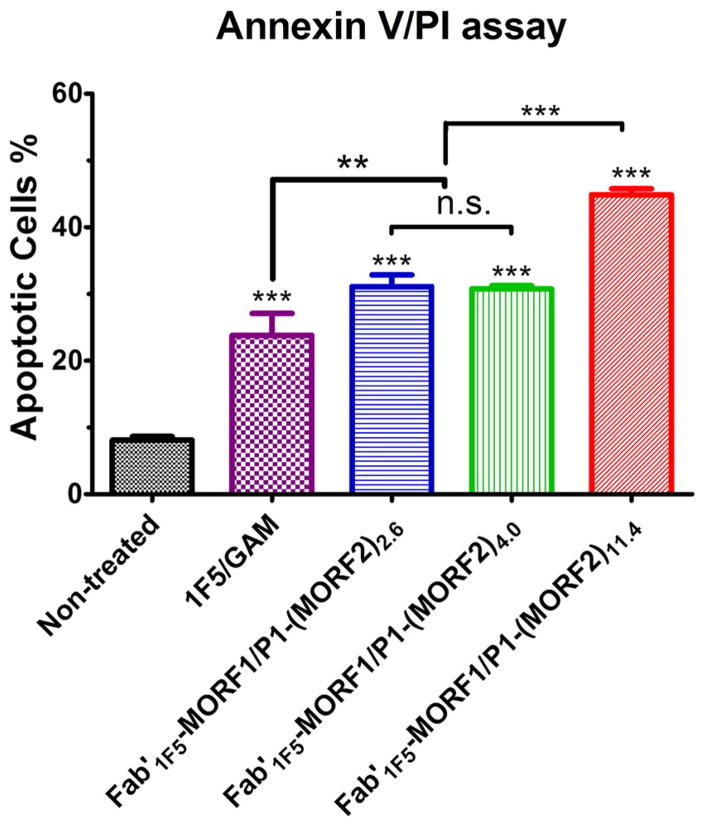
To evaluate the effect of valence on apoptosis induction, multivalent P1-(MORF2)x (25 bp) conjugates were synthesized from the same polymer precursor P1-TT (Mn 87 kDa; PDI 1.18) by polymeranalogous reaction with varying amounts of MORF2-NH2 to produce P1-(MORF2)2.6, P1-(MORF2)4.0 and P1-(MORF2)11.4. The conjugates characterization is summarized in Table 2 and Fig. S1. The conjugate Fab′-MORF1 is characterized in Table 1 and Figs. S2 and S3.
The results of apoptosis induction as determined by Annexin V/PI assay are shown in Fig. 4. Apoptotic levels obtained following incubation of Raji cells with premixed nanoconjugates Fab′1F5-MORF1 and P1-(MORF2)x indicate a relationship between the valence of the P1-(MORF2)x and levels of apoptosis. Increase of valence from 2.6 to 4.0 did not have a statistically significant effect. Both conjugates created about 33% apoptotic cells. However, the conjugate P1-(MORF2)11.4 with a valence of 11.4 possessed a higher apoptosis induction efficacy and produced 45% of apoptotic cells (Fig. 4). All three conjugates were more active than the positive control – treatment with the whole 1F5 Ab followed by secondary goat anti-mouse Ab.
Fig. 4.
Effect of valence on apoptosis induction of Raji B-cells. Percentage of apoptotic cells were analyzed by Annexin V/PI binding and quantified by flow cytometry. Incubation time was 24 h. Untreated, cells in culture medium; 1F5/GAM, 1F5 mAb (0.5 μM) followed (1 h later) by goat anti-mouse secondary Ab (0.25 μM); Fab′1F5-MORF1/P1-(MORF2)x, premixture of Fab′1F5-MORF1 (1 μM) and P1-(MORF2)x with different valences (1 μM, MORF2-eqv). The percentage of apoptotic cells was quantified by flow cytometry. Statistics, unless otherwise indicated, was performed by comparing each treatment with the non-treated (***p < 0.0001, **p < 0.005, n.s.: no significant difference). All data are presented as mean ± SD (n = 3).
3.3. Impact of P-(PEG2-MORF2)x molecular weight on apoptosis induction in Raji B cells
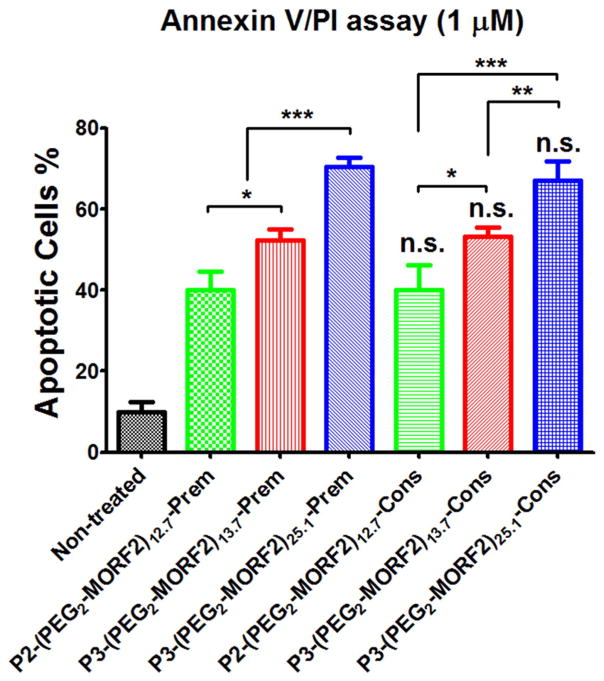
To estimate the impact of molecular weight of the MORF2 containing HPMA graft copolymers on apoptosis induction we compared P2-(PEG2-MORF2)12.7 (112 kDa; 12.7 MORF2 per macromolecule) with P3-(PEG2-MORF2)13.7 (291 kDa; 13.7 MORF2 per macromolecule). In addition, we synthesized a high molecular weight, high valence conjugate P3-(PEG2-MORF2)25.1 (291 kDa; 25.1 MORF2 per macromolecule) to evaluate a conjugate with both high molecular weight and high valence (Table 2). As the complementary conjugate, Fab′1F5-PEG2-MORF1 (Table 1) was used.
P2 and P3 based conjugates were synthesized using modified chemistry. Whereas in P1 based conjugates the MORF2 was attached to the P1-TT precursor by reaction of MORF2’s amino group with the TT groups at P1-TT side chain termini (Scheme 1B), in P2 and P3 based conjugates thiolene reaction was used. The side-chain amino groups of the HPMA + APMA copolymer were converted to maleimido groups by reaction with heterobifunctional reagent SM(PEG)y followed by reaction with thiol derivatized MORF2 (Scheme 1C).
Hybridization of Fab′1F5-PEG2-MORF1 with P3-(PEG2-MORF2)13.7 (or P3-(PEG2-MORF2)25.1) was evaluated by UV–visible spectroscopy as described above. Maximum hypochromic effect occurred when mixing Fab′1F5-PEG2-MORF1 with P3-(PEG2-MORF2)x at 1:1 mole ratio MORF1:MORF2 (Fig. S4). This indicates that polymer molecular weight did not impair the hybridization of the two nanoconjugates. The kinetics of Fab′1F5-PEG2-MORF1 binding with P3-(PEG2-MORF2)13.7 or P3-(PEG2-MORF2)25.1 was characterized by DLS (Fig. S5). The results showed a significant and rapid biorecognition of MORF1/MORF2 as shown by increase of hydrodynamic size for both pairs of conjugates.
The apoptosis induction activity of the three pairs of nanoconjugates is depicted on Fig. 5. AnnexinV/PI assay was used and Raji cells incubated both in consecutive and premixed arrangements. Comparison of two nanoconjugate pairs with similar number of grafts per macromolecule but different molecular weights, namely Fab′1F5-PEG2-MORF1 + P2-(PEG2-MORF2)12.7 (Mn 112; valence 12.7) vs. Fab′-PEG2-MORF1 + P3-(PEG2-MORF2)13.7 (Mn 291; valence 13.7) clearly indicated enhanced apoptotic activity of the higher molecular weight conjugate (Fig. 5). As expected, increasing the valence of the high molecular weight conjugate enhanced the efficacy further; the pair Fab′1F5-PEG2-MORF1 + P3-(PEG2-MORF2)25.1 (Mn 291; valence 25.1) possessed the highest activity of the three pairs evaluated. There was no statistically significant difference between the consecutive or premixed exposure of Raji cells to the nanoconjugates.
Fig. 5.
Effect of molecular weight on apoptosis induction of Raji B-cells. Percentage of apoptotic cells were analyzed by Annexin V/PI binding and quantified by flow cytometry. Incubation time was 24 h. Non-treated, cells in culture medium; Prem, premixture of Fab′1F5-PEG2-MORF1 (1 μM) and conjugates with different valences (P2-(PEG2-MORF2)12.7 or P3-(PEG2-MORF2)13.7 or P3-(PEG2-MORF2)25.1; 1 μM, MORF2-eqv). Cons, consecutive, Fab′1F5-PEG2-MORF1 (1 μM) followed (1 h later) by conjugates with different valences (P2-(PEG2-MORF2)12.7 or P3-(PEG2-MORF2)13.7 or P3-(PEG2-MORF2)25.1; 1 μM, MORF2-eqv). Percentage of apoptotic cells was quantified by flow cytometry. Statistics, unless otherwise indicated, was performed by comparing each consecutive treatment with premixed treatment (***p < 0.0001, **p < 0.005, *p < 0.05, n.s.: no significant difference). All data are presented as mean ± SD (n = 3).
3.4. Effect of spacer length on apoptosis induction
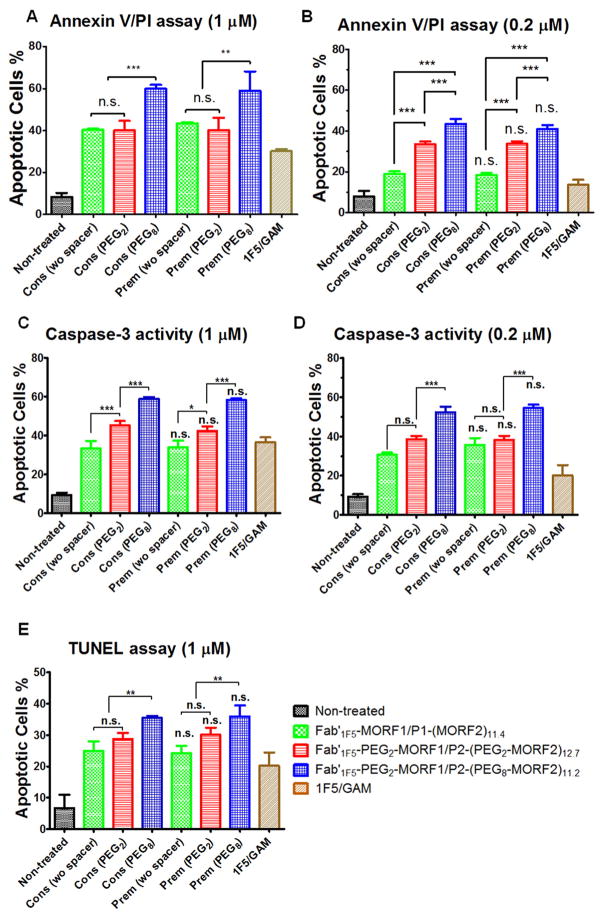
To evaluate the effect of spacer length on the efficacy of apoptosis induction we have compared three nanoconjugate pairs. The traditional conjugate pair (no spacer in Fab′1F5-MORF1; GG spacer in P-(MORF2)x) was compared with conjugates that contained PEG2 spacer between Fab′1F5 and MORF1 and PEG2 or PEG8 spacers between the 3-aminopropyl side chains of the HPMA copolymer and MORF2.
The P2-(PEGy-MORF2)x conjugates were synthesized in three steps. First, a copolymer of HPMA and APMA was prepared by RAFT copolymerization (P2-NH2). Second, the amino groups at side chain termini were reacted with heterobifunctional reagents, SM(PEG)2 or SM(PEG)8 to convert amino groups into maleimido groups. Finally, MORF2-SH (freshly prepared by reducing MORF2-SSR with TCEP) was grafted to the copolymer via thioether bonds. The conjugates were purified and characterized by SEC (Fig. S6). Two P2-PEGy-MORF2 (y = 2 or 8) with the same polymer backbone (Mn 112 kDa; PDI 1.17) and similar valences were synthesized. The valences of P2-(PEG2-MORF2)12.7 and P2-(PEG8-MORF2)11.2 were 12.7 and 11.2, respectively.
To synthesize Fab′1F5-PEG2-MORF1 amine-derivatized MORF1 oligo (MORF1-NH2) was modified by SM(PEG)2 and attached to freshly prepared Fab′1F5-SH. The conjugate was purified by ultrafiltration (30,000 Da cut-off) with PBS and characterized by SEC (Fig. S7).
Hybridization of the Fab′1F5-PEG2-MORF1 with P2-(PEG2-MORF2)12.7 (or P2-(PEG8-MORF2)11.2) was characterized by UV spectrophotometry and DLS as described above. The hybridization was effective (Fig. S8) and the kinetics of binding fast (Fig. S9).
The apoptotic induction activity in Raji cells was evaluated by three assays: Annexin V/PI, Caspase 3, and TUNEL (Fig. 6). For AnnexinV/PI and caspase 3 assays two concentrations (0.2 and 1 μM) were used. The TUNEL assay was evaluated at 1 μM. Generally, the insertion of a PEG2 spacer resulted in no (Fig. 6A,E) or minor (Fig. 6B, C, and D) enhancement of apoptosis. In contrast, insertion of a PEG8 spacer resulted in enhancement of apoptosis in all three assays and both concentrations – compare Fab′1F5-PEG2-MORF1 + P2-(PEG8-MORF2)11.2 with Fab′1F5-PEG2-MORF1 + P2-(PEG2-MORF2)12.7 and Fab′1F5-MORF1 + P1-(MORF2)11.4.
Fig. 6.
Effect of spacer length on apoptosis induction of Raji B-cells analyzed by different assays. (A, B) Annexin V/PI binding assay. (C, D) Caspase-3 activation assay. (E) TUNEL assay. Incubation time was 24 h. For figures A, B, C, and D, 2 × 105 cells were used for each test. For figure E, 1 × 106 cells were used. Non-treated, cells in culture medium; Cons, Consecutive exposure, Fab′1F5-MORF1 or Fab′1F5-PEG2-MORF1 (0.2 μM or 1 μM) followed (1 h later) by P1-(MORF2)11.4, P2-(PEG2-MORF2)12.7 or P2-(PEG8-MORF2)11.2 (0.2 μM or 1 μM, MORF2-eqv); Prem, Cells were exposed to the premixture of Fab′1F5-MORF1 or Fab′1F5-PEG2-MORF1 (0.2 μM or 1 μM) and P1-(MORF2)11.4, P2-(PEG2-MORF2)12.7 or P2-(PEG8-MORF2)11.2 (0.2 μM or 1 μM, MORF2-eqv); 1F5/GAM, 1F5 mAb (1 μM) followed (1 h later) by goat anti-mouse secondary Ab (0.5 μM). Percentage of apoptotic cells was quantified by flow cytometry. Statistics, unless otherwise indicated, was performed by comparing each premixed treatment with the corresponding consecutive treatment (***p < 0.0001, n.s.: no significant difference). All data are presented as mean ± SD (n = 3). Statistical analyses were performed by Student’s t-test to compare between two groups, or one-way analysis of variance (ANOVA) to compare three or more groups (with p value < 0.05 indicating statistically significant difference). (wo spacer): Fab′1F5-MORF1 with P1-(MORF2)11.4; (PEG2): Fab′1F5-PEG2-MORF1 with P2-(PEG2-MORF2)12.7; (PEG8): Fab′1F5-PEG2-MORF1 with P2-(PEG8-MORF2)11.2.
3.5. Effect of structure of Fab′ fragment on apoptosis induction
The experiments described above were performed with nanoconjugates containing the Fab′ fragment from the 1F5 antibody. This antibody was used previously in four NHL patients and minor toxicities and immunogenicity was detected [51]. In animal experiments minor immunogenicity of Fab′ fragment from 1F5 Ab was detectable too [52]. Consequently, in preparation for clinical translation, we compared the efficacy of the system based on Fab′ from 1F5 with nanoconjugates based on Fab′ from FDA approved Rituximab [53]. Fab′RTX-MORF1 was synthesized from Rituximab and MORF1 by the same method as Fab′1F5-MORF1. The SEC analysis of Fab′RTX-MORF1 is shown in Fig. S11. The mole ratio Fab′RTX: MORF1 in the conjugate was close to 1:1 as determined by BCA assay and UV–visible spectroscopy.
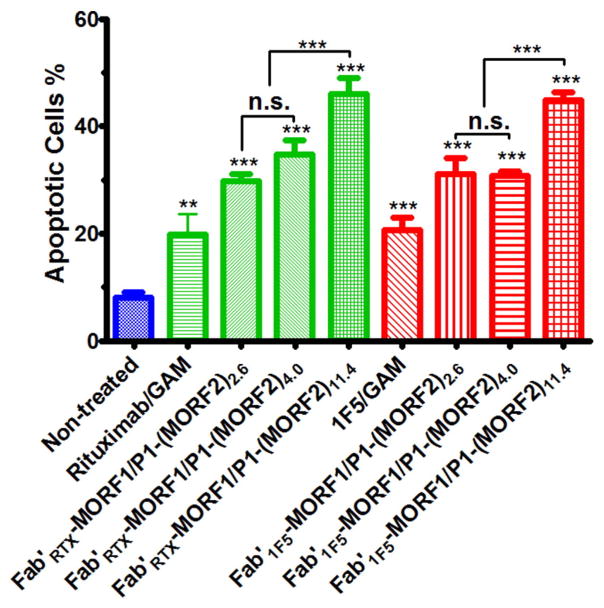
Apoptosis induction in Raji cells was determined by Annexin V/PI binding assay (Fig. 7). Results indicated that Fab′RTX-MORF1 in combination with three graft copolymers, P1-(MORF2)2.6, P1-(MORF2)4.0, and P1-(MORF2)11.4 produced apoptotic levels fully comparable with the similar combinations based on Fab′ fragment from 1F5 antibody, i.e. Fab′1F5-MORF1 in combination with P1-(MORF2)2.6, P1-(MORF2)4.0, and P1-(MORF2)11.4. This bodes well for further translational development of this technology.
Fig. 7.
Effect of structure of antibody fragment on apoptosis induction of Raji B-cells. Percentage of apoptotic cells was analyzed by Annexin V/PI binding and quantified by flow cytometry. Incubation time was 24 h. Non-treated, cells in culture medium; 1F5/GAM, 1F5 mAb (0.5 μM) followed (1 h later) by goat anti-mouse secondary Ab (GAM; 0.25 μM); RTX/GAM, RTX mAb (0.5 μM) followed (1 h later) by GAM (0.25 μM); Fab′RTX-MORF1/P1-(MORF2)x (or Fab′1F5-MORF1/P1-(MORF2)x), premixture of Fab′RTX-MORF1 (or Fab′1F5-MORF1, 1 μM) and P1-(MORF2)x with different valences (1 μM, MORF2-eqv). Percentage of apoptotic cells was quantified by flow cytometry. Statistics, unless otherwise indicated, was performed by comparing each treatment with the non-treated. (***p < 0.0001, **p < 0.005, n.s.: no significant difference). All data are presented as mean ± SD (n = 3).
4. Discussion
The excellent biorecognition of the complementary domains and the self-assembly of domain-containing graft copolymers was an inspiration for the design of new nanomedicines. It created a bridge between the design of biomaterials and the design of nanomedicines [54]. Complementary CCK and CCE peptides that self-assemble into antiparallel coiled-coil heterodimers were engaged in the design of a new CD20+ cell apoptosis induction system, called drug-free macromolecular therapeutics [10,11]. Indeed, the biorecognition of CCE/CCK peptide motifs at the cellular surface was able to crosslink CD20 receptors and control apoptosis of B cells. Multiple fluorescence imaging studies, including 2-channel FMT, 3D confocal microscopy, and 4-color FACS demonstrated co-localization of both nanoconjugates, Fab′-CCE and P-(CCK)9, on NHL Raji cell surface, indicating “two-step” targeting specificity. The fluorescent images also revealed that these two conjugates could disrupt normal membrane lipid distribution and form lipid raft clusters, leading to cancer cell apoptosis. This “two-step” biorecognition capacity was further demonstrated in a NHL xenograft model, using fluorescent images at whole-body, tissue and cell levels [45].
Recent designs of drug-free macromolecular therapeutics use complementary morpholino oligonucleotides as biorecognition units. The CCE-CCK system worked well. However, these peptides do not have a strong secondary structure at neutral pH. They first interact by hydrophobic and electrostatic interactions and subsequently fold into precise coiled-coils [20]. To be efficient in vivo, an excess of the second nanoconjugate needs to be used [11]. Morpholino oligonucleotides were selected due to their fast hybridization, excellent binding affinity, stability in plasma, and water solubility [9,12,14]. The MORF oligonucleotides are charge neutral, resulting in significantly stronger binding than natural DNA and RNA [55]. This new therapeutic system was composed of two hybrid conjugates: (1) anti-CD20 Fab′ linked to MORF1 (Fab′-MORF1), and (2) HPMA copolymer grafted with multiple copies of MORF2 (P-(MORF2)x). The two conjugates self-assemble via MORF1-MORF2 hybridization at the surface of CD20+ B-cells, which crosslinks CD20 and initiates apoptosis in vitro and in vivo [12,14–17]. Following crosslinking the CD20 receptors tend to cluster as dimers or tetramers, redistribute and become localized into lipid rafts. This mediates the interaction between clustered CD20 and Src-family kinases (which are also located in lipid rafts), and triggers apoptotic signaling.
Our previous studies have shown the efficacy of the MORF-based drug-free macromolecular therapeutics in an animal model of NHL [12,14] and on patient cells [15]. The aim of this study was to determine the relationship between the detailed structure of the nanoconjugates and apoptosis induction in Raji cells to allow system optimization. The factors studied include the length of the MORF sequence, the valence of P-(MORF2)x (varying x), molecular weight of P-(MORF2)x, incorporation of a miniPEG spacer between Fab′ and MORF1 and between polymer and MORF2, and comparison of two Fab′ fragments, one from 1F5 antibody (Fab′1F5), the other from Rituximab (Fab′RTX).
To this end we designed complementary MORF sequences containing 20, 25, and 28 base pairs (Scheme 1, Table S1), prepared Fab′ fragments from 1F5 and Rituximab anti-CD20 antibodies and Fab′-MORF1 conjugates (Table 1), used RAFT polymerization to synthesize HPMA copolymer precursors and HPMA copolymer-MORF2 conjugates with different valences and varying molecular weight (Table 2). In addition, we inserted a PEG2 spacer between Fab′ fragment and MORF1 and PEG2 or PEG8 spacers between the HPMA copolymer side chains and MORF2 (Tables 1 and 2). Three apoptotic assays, hybridization assay and DLS were used to characterize the biological properties of the nanoconjugates.
The A/T/C/G content MORF sequences was selected to achieve optimal binding efficacy and specificity (G + C = 35–65%) [55], maintain aqueous solubility (G < 36%) [55], and potentially provide favorable pharmacokinetics (C < 7 to avoid rapid kidney uptake) [56]. The impact of the length and structure of MORF strands on the hybridization (Fig. 2) demonstrated high levels of recognition for all three MORF pairs (20, 25, and 28 bp). There was a slight indication that the kinetics of hybridization (as determined by DLS, Fig. 2B) was slower for 20 bp. However, apoptotic assays (Fig. 3) were considerably more sensitive than DLS to verify the differences in the properties of nanoconjugates containing MORFs of different length. The conjugates with 28 bp MORFs were more efficient than those with 20 or 25 bp. Conjugates with 25 bp were more efficient than those with 20 bp with the exception of caspase 3 assay at 1 μM concentration. Interestingly, there was no statistically significant difference between consecutive and premixed treatments. The translation of this technology will be based on the consecutive treatment. It permits pretargeting where the first nanoconjugate (Fab ′-MORF1) bound to CD20 remains at the B cell surface whereas off target bound Fab′-MORF1 will be internalized and degraded before the administration of the second nanoconjugate (P-(MORF2)x).
The effect of valence (Fig. 4) and molecular weight (Fig. 5) of the polymer backbone on the apoptotic activity was clearly defined. The higher the valence and the higher the molecular weight of the P-(MORF2)x conjugate the higher was the apoptotic activity. A multifunctional conjugate can interact with multiple CD20 receptors and enhance the biological activity. This is a well-known phenomenon – Dintzis et al. have demonstrated in their immunon model of immune response that only polyacrylamide molecules with long chains and a large number (12 to 16) of attached haptens per macromolecule could deliver immunogenic signal to the cell [57]. Our results demonstrate that for the system CD20 vs. ligand, there is no clear threshold for apoptosis induction, but multivalent systems produce dramatically higher apoptotic response. Multivalency leads to enhanced biorecognition [58], as shown in numerous examples related to the CD20 receptor. Multimeric Rituximab bound to activated dextran [59] or graphene oxide [60] enhanced apoptotic activity in CD20+ cells. To avoid Fc receptor interactions [61] and trogocytosis of CD20 receptors [62] our laboratory used only anti-CD20 Fab′ fragments attached to HPMA copolymers. Remarkably, HPMA copolymers with attached multiple Fab′ fragments also demonstrated a strong multivalency effect in apoptosis induction [13,63,64].
Effect of spacer/flexibility on apoptosis induction. Longer spacer imposes a higher apoptosis induction efficacy on the conjugates. Whereas a PEG2 spacer produced minimal changes, a longer, PEG8 spacer clearly enhanced the apoptotic levels (Fig. 6). The results obtained with the PEG8 spacer are due to a higher flexibility of MORF2 containing grafts attached to the HPMA copolymer backbone. Binding of receptors with ligands usually results in entropic penalty due to decrease of the molecule flexibility. As discussed in a recent study, keeping molecules flexible may reduce the entropy penalty and create stronger binding [65]. The HPMA copolymer backbone retains its flexibility when first MORF2 binds to MORF1 that is attached to CD20. The dynamic and flexible nature of HPMA copolymer backbone [66] would allow a conformational response within the nanoconjugate so that additional hybridizations may occur [63].
As mentioned in the Results section, Fab′1F5 may produce minor immune response [51,52]. Replacement with Fab′RTX, a fragment from the FDA approved Ab, produced nanocnjugates with equal apoptosis induction capacity. This bodes well for the translation potential of this technology.
In summary, we have evaluated the relationship between the structure of morpholino oligonucleotide-based drug-free macromolecular therapeutics and their efficacy in the induction of apoptosis in CD20+ Raji B cells. The results indicated that the efficacy of conjugates increases with increasing length of the MORF sequence, valence and molecular weight of the P-(MORF2)x conjugate, and longer spacer between the polymer backbone and MORF2. Fab′ fragments from two different Abs (1F5 and Rituximab) possessed equal activities. The results combined with previous in vivo studies provide design principles for nanoconjugates to be translated into clinical evaluation.
Acknowledgments
The research was supported in part by NIH grant GM095606, Huntsman Cancer Institute (project 160305) and Bastion Biologics (project 50503163).
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jconrel.2016.12.025.
Footnotes
Competing interests
J.Y. and J.K. are inventors on a pending US patent application (PCT/US2014/023784; assigned to the University of Utah and licensed to Bastion Biologics) related to this work. J.K. is Chief Scientific Advisor and J.Y. Scientific Advisor for Bastion Biologics. Otherwise, the authors declare no competing financial interests.
References
- 1.Krishnamachari Y, Pearce ME, Salem AK. Self-assembly of cell–microparticle hybrids. Adv Mater. 2008;20:989–993. [Google Scholar]
- 2.Koyfman AY, Braun GB, Reich NO. Cell-targeted self-assembled DNA nanostructures. J Am Chem Soc. 2009;131:14237–14239. doi: 10.1021/ja9015638. [DOI] [PubMed] [Google Scholar]
- 3.Cho MH, Lee EJ, Son M, Lee JH, Yoo D, Kim JW, Park SW, Shin JS, Cheon J. A magnetic switch for the control of cell death signaling in in vitro and in vivo systems. Nat Mater. 2012;11:1038–1043. doi: 10.1038/nmat3430. [DOI] [PubMed] [Google Scholar]
- 4.Lee SE, Liu GL, Kim F, Lee LP. Remote optical switch for localized and selective control of gene interference. Nano Lett. 2009;9:562–570. doi: 10.1021/nl802689k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 6.Kuang Y, Yuan D, Zhang Y, Kao A, Du XW, Xu B. Interactions between cellular proteins and morphologically different nanoscale aggregates of small molecules. RSC Adv. 2013;3:7704–7707. doi: 10.1039/C3RA41523F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuang Y, Xu B. Nanofibers of small hydrophobic molecules disrupt dynamics of microtubules and selectively inhibit glioblastoma cells. Angew Chem Int Ed. 2013;52:6944–6948. doi: 10.1002/anie.201302658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlsson M, Rebmann B, Lienemann PS, Sprossmann N, Ehrbar M, Radziwill G, Weber W. Pharmacologically controlled protein switch for ON-OFF regulation of growth factor activity. Sci Rep. 2013;3:2716–2721. doi: 10.1038/srep02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu TW, Kopeček J. Drug-free macromolecular therapeutics – a new paradigm in polymeric nanomedicines. Biomater Sci. 2015;3:908–922. doi: 10.1039/C4BM00442F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu K, Liu J, Johnson RN, Yang J, Kopeček J. Drug-free macromolecular therapeutics: induction of apoptosis by coiled-coil-mediated cross-linking of antigens on the cell surface. Angew Chem Int Ed. 2010;49:1451–1455. doi: 10.1002/anie.200906232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu K, Yang J, Liu J, Kopeček J. Coiled-coil based drug-free macromolecular therapeutics: in vivo efficacy. J Control Release. 2012;157:126–131. doi: 10.1016/j.jconrel.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu TW, Yang J, Zhang R, Sima M, Kopeček J. Cell surface self-assembly of hybrid nanoconjugates via oligonucleotide hybridization induces apoptosis. ACS Nano. 2014;8:719–730. doi: 10.1021/nn4053827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu TW, Yang J, Kopeček J. Anti-CD20 multivalent HPMA copolymer–Fab′ conjugates for the direct induction of apoptosis. Biomaterials. 2012;33:7174–7181. doi: 10.1016/j.biomaterials.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu TW, Zhang R, Yang J, Chao MP, Shami PJ, Kopeček J. A two-step pretargeted nanotherapy for CD20 crosslinking may achieve superior anti-lymphoma efficacy to rituximab. Theranostics. 2015;5:834–846. doi: 10.7150/thno.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu TW, Kosak KM, Shami PJ, Kopeček J. Drug-free macromolecular therapeutics induce apoptosis of patient chronic lymphocytic leukemia cells. Drug Deliv Transl Res. 2014;4:389–394. doi: 10.1007/s13346-014-0209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartley JM, Chu TW, Peterson EM, Zhang R, Yang J, Harris J, Kopeček J. Super-resolution imaging and quantitative analysis of membrane protein/lipid raft clustering mediated by cell surface self-assembly of hybrid nanoconjugates. ChemBioChem. 2015;16:1725–1729. doi: 10.1002/cbic.201500278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartley JM, Zhang R, Gudheti M, Yang J, Kopeček J. Tracking and quantifying polymer therapeutic distribution on a cellular level using 3D dSTORM. J Control Release. 2016;231:50–59. doi: 10.1016/j.jconrel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan W, Yang J, Kopečková P, Kopeček J. Smart hydrogels containing adenylate kinase: translating substrate recognition into macroscopic motion. J Am Chem Soc. 2008;130:15760–15761. doi: 10.1021/ja805634x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Mazumdar D, Lu Y. A simple and sensitive “dipstick” test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew Chem Int Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Xu C, Wang C, Kopeček J. Refolding hydrogels self-assembled from N-(2-hydroxypropyl)methacrylamide graft copolymers by antiparallel coiled-coil formation. Biomacromolecules. 2006;7:1187–1195. doi: 10.1021/bm051002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu TW, Feng J, Yang J, Kopeček J. Hybrid polymeric hydrogels via peptide nucleic acid (PNA)/DNA complexation. J Control Release. 2015;220:608–616. doi: 10.1016/j.jconrel.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagahara S, Matsuda T. Hydrogel formation via hybridization of oligonucleotides derivatized in water-soluble vinyl polymers. Polym Gels Netw. 1996;4:111–127. [Google Scholar]
- 23.Heldin CH. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 24.Germain RN, Stefanova I. The dynamics of T cell receptor signaling: complex orchestration and the key roles of tempo and cooperation. Annu Rev Immunol. 1999;17:467–522. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- 25.Healy JI, Goodnow CC. Positive versus negative signaling by lymphocyte antigen receptors. Annu Rev Immunol. 1998;16:645–670. doi: 10.1146/annurev.immunol.16.1.645. [DOI] [PubMed] [Google Scholar]
- 26.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell-surface interactions. Curr Opin Chem Biol. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 27.Kahn CR, Baird KL, Jarrett DB, Flier JS. Direct demonstration that receptor crosslinking or aggregation is important in insulin action. Proc Natl Acad Sci U S A. 1978;75:4209–4213. doi: 10.1073/pnas.75.9.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu Y, van Seventer GA, Ennis E, Newman W, Horgan KJ, Shaw S. Crosslinking of the T cell-specific accessory molecules CD7 and CD28 modulates T cell adhesion. J Exp Med. 1992;175:577–582. doi: 10.1084/jem.175.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fourcin M, Chevalier S, Guillet C, Robledo O, Froger J, Pouplard-Barthelaix A, Gascan H. gp130 transducing receptor crosslinking is sufficient to induce interleukin-6 type responses. J Biol Chem. 1996;271:11756–11760. doi: 10.1074/jbc.271.20.11756. [DOI] [PubMed] [Google Scholar]
- 30.Shan D, Ledbetter JA, Press OW. Apoptosis of malignant human B cells by ligation of CD20 with monoclonal antibodies. Blood. 1998;91:1644–1652. [PubMed] [Google Scholar]
- 31.Vallat LD, Park Y, Li C, Gribben JG. Temporal genetic program following B-cell receptor cross-linking: altered balance between proliferation and death in healthy and malignant B cells. Blood. 2007;109:3989–3997. doi: 10.1182/blood-2006-09-045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber AB, Libermann TA, Lax I, Yarden Y, Schlessinger J. Biological role of epidermal growth factor-receptor clustering. J Biol Chem. 1983;25:846–853. [PubMed] [Google Scholar]
- 33.Fanning SL, George TC, Feng D, Feldman SB, Megjugorac NJ, Izaguirre AG, Fitzgerald-Bocarsly P. Receptor cross-linking on human plasmacytoid dendritic cells leads to the regulation of IFN-alpha production. J Immunol. 2006;177:5829–5839. doi: 10.4049/jimmunol.177.9.5829. [DOI] [PubMed] [Google Scholar]
- 34.Deans JP, Li H, Polyak MJ. CD20-mediated apoptosis: signalling through lipid rafts. Immunology. 2002;107:176–182. doi: 10.1046/j.1365-2567.2002.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couvreur P, Vauthier C. Nanotechnology: intelligent design to treat complex disease. Pharm Res. 2006;23:1417–1450. doi: 10.1007/s11095-006-0284-8. [DOI] [PubMed] [Google Scholar]
- 36.Callahan J, Kopeček J. Semitelechelic HPMA copolymers functionalized with triphenylphosphonium as drug carriers for membrane transduction and mitochondrial localization. Biomacromolecules. 2006;7:2347–2356. doi: 10.1021/bm060336m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aluri SR, Shi P, Gustafson JA, Wang W, Lin YA, Cui H, Liu S, Conti PS, Li Z, Hu P, Epstein AL, MacKay JA. A hybrid protein-polymer nanoworm potentiates apoptosis better than a monoclonal antibody. ACS Nano. 2014;8:2064–2076. doi: 10.1021/nn403973g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 39.Armitage JO, Weisenburger DD. New approach to classifying non-Hodgkin’s lymphomas: clinical features of the major histologic subtypes. Non-Hodgkin’s lymphoma classification project. J Clin Oncol. 1998;16:2780–2795. doi: 10.1200/JCO.1998.16.8.2780. [DOI] [PubMed] [Google Scholar]
- 40.Stashenko P, Nadler LM, Hardy R, Schlossman SF. Characterization of a human B lymphocyte-specific antigen. J Immunol. 1980;125:1678–1685. [PubMed] [Google Scholar]
- 41.Hoffmeister JK, Cooney D, Coggeshall KM. Clustered CD20 induces apoptosis: src-family kinase, the proximal regulator of tyrosine phosphorylation, calcium influx, and caspase 3-dependent apoptosis. Blood Cells Mol Dis. 2000;26:133–143. doi: 10.1006/bcmd.2000.0287. [DOI] [PubMed] [Google Scholar]
- 42.Okroj M, Österborg A, Blom AM. Effector mechanisms of anti-CD20 monoclonal antibodies in B cell malignancies. Cancer Treat Rev. 2013;39:632–639. doi: 10.1016/j.ctrv.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 43.Cartron G, Dacheux L, Salles G, Solala-Seligny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcγRIIIa gene. Blood. 2002;99:754–758. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 44.Lands LC. New therapies, new concerns: rituximab-associated lung injury. Pediatr Nephrol. 2010;25:1001–1003. doi: 10.1007/s00467-010-1476-3. [DOI] [PubMed] [Google Scholar]
- 45.Zhang R, Yang J, Chu TW, Hartley JM, Kopeček J. Multimodality imaging of coiled-coil mediated self-assembly in a “drug-free” therapeutic system. Adv Healthc Mat. 2015;4:1054–1065. doi: 10.1002/adhm.201400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopeček J, Bažilová H. Poly[N-(2-hydroxypropyl)methacrylamide]. 1. Radical polymerization and copolymerization. Eur Polym J. 1973;9:7–14. [Google Scholar]
- 47.Mitsukami Y, Donovan MS, Lowe AB, McCormick CL. Water-soluble polymers. 81. Direct synthesis of hydrophilic styrenic-based homopolymers and block copolymers in aqueous solution via RAFT. Macromolecules. 2001;34:2248–2256. [Google Scholar]
- 48.Šubr V, Ulbrich K. Synthesis and properties of new N-(2-hydroxypropyl)-methacrylamide copolymers containing thiazolidine-2-thione reactive groups. React Funct Polym. 2006;66:1525–1538. [Google Scholar]
- 49.Starcher B. A ninhydrin-based assay to quantitate the total protein content of tissue samples. Anal Biochem. 2001;292:125–129. doi: 10.1006/abio.2001.5050. [DOI] [PubMed] [Google Scholar]
- 50.Gergel D, Cederbaum AI. Inhibition of the catalytic activity of alcohol dehydrogenase by nitric oxide is associated with S nitrosylation and the release of zinc. Biochemistry. 1996;35:16186–16194. doi: 10.1021/bi962043r. [DOI] [PubMed] [Google Scholar]
- 51.Johnson TA, Press OW. Therapy of B-cell lymphomas with monoclonal antibodies and radioimmunoconjugates: the Seattle experience. Ann Hematol. 2000;79:175–182. doi: 10.1007/s002770050576. [DOI] [PubMed] [Google Scholar]
- 52.Kverka M, Hartley JM, Chu TW, Yang J, Heidchen R, Kopeček J. Immunogenicity of coiled-coil based drug-free macromolecular therapeutics. Biomaterials. 2014;35:5886–5896. doi: 10.1016/j.biomaterials.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith MR. Rituximab (monoclonal anti-CD20 antibody): mechanisms of action and resistance. Oncogene. 2003;22:7359–7368. doi: 10.1038/sj.onc.1206939. [DOI] [PubMed] [Google Scholar]
- 54.Yang J, Kopeček J. Polymeric biomaterials and nanomedicines. J Drug Deliv Sci Technol. 2015;30:318–330. doi: 10.1016/j.jddst.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 56.Liu G, He J, Dou S, Gupta S, Vanderheyden JL, Rusckowski M, Hnatowich DJ. Pretargeting in tumored mice with radiolabeled morpholino oligomer showing low kidney uptake. Eur J Nucl Med Mol Imaging. 2004;31:417–424. doi: 10.1007/s00259-003-1393-9. [DOI] [PubMed] [Google Scholar]
- 57.Dintzis H, Dintzis R, Vogelstein B. Molecular determinants of immunogenicity: the immunon model of immune response. Proc Natl Acad Sci U S A. 1976;73:3671–3675. doi: 10.1073/pnas.73.10.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mammen M, Choi S, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 59.Zhang N, Khawli LA, Hu P, Epstein AL. Generation of rituximab polymer may cause hyper-cross-linking – induced apoptosis in non-Hodgkin’s lymphomas. Clin Cancer Res. 2005;11:5971–5980. doi: 10.1158/1078-0432.CCR-05-0554. [DOI] [PubMed] [Google Scholar]
- 60.Luo C, Deng Z, Li L, Clayton F, Chen AL, Wei P, Miles R, Stephens DM, Glenn M, Wang X, Jensen PE, Chen X. Association of rituximab with grapheme oxide confers direct cytotoxicity for CD20-positive lymphoma cells. Oncotarget. 2016;7:12806–12822. doi: 10.18632/oncotarget.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okroj M, Österborg A, Blom AM. Effector mechanisms of anti-CD20 monoclonal antibodies in B cell malignancies. Cancer Treat Rev. 2013;39:632–639. doi: 10.1016/j.ctrv.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 62.Pham T, Mero P, Booth JW. Dynamics of macrophage trogocytosis of rituximabcoated B cells. PLoS One. 2011;6:e14498. doi: 10.1371/journal.pone.0014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson RN, Kopečková P, Kopeček J. Synthesis and evaluation of multivalent branched HPMA copolymer–Fab′ conjugates targeted to the B-cell antigen CD20. Bioconjug Chem. 2009;20:129–137. doi: 10.1021/bc800351m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson RN, Kopečková P, Kopeček J. Biological activity of anti-CD20 multivalent HPMA copolymer–Fab′ conjugates. Biomacromolecules. 2012;13:727–735. doi: 10.1021/bm201656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You W, Huang Y-MM, Kizhake S, Natarajan A, Chang CA. Characterization of promiscuous binding of phosphor ligands to breast-cancer-gene 1 (BRCA1) C-terminal (BRCT): molecular dynamics, free energy, entropy, and inhibition design. PLOS Comp Biol. 2016;12(8):e1005057. doi: 10.1371/journal.pcbi.1005057. http://dx.doi.org/10.1371/journal.pcbi.1005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bohdanecký M, Bažilová H, Kopeček J. Poly[N-(2-hydroxypropyl)methacrylamide] II. Hydrodynamic properties of dilute solutions. Eur Polym J. 1974;10:405–410. [Google Scholar]