Summary
Background
Von Willebrand factor (VWF) is a multimeric coagulation factor that tethers platelets to injured subendothelium. Type 2M von Willebrand Disease (VWD) is characterized by a qualitative defect in VWF with preserved multimer distribution.
Objectives
Through the Zimmerman Program for the Molecular and Clinical Biology for VWD, five VWF sequence variations were studied in subjects diagnosed with type 2M VWD.
Methods
Bleeding phenotype was assessed using the ISTH bleeding assessment tool. Full length VWF gene sequencing was performed for each subject. Each variant was placed into a recombinant VWF vector using site-directed mutagenesis and expressed in HEK293T cells as homozygous or heterozygous VWF. Variant expression, collagen binding, and platelet GPIbα binding were studied through ELISA assays. Multimer analysis was performed by gel electrophoresis.
Results
Bleeding scores were elevated for all subjects except for the p.P1162L and p.R1374C variants. Although all had reduced VWF ristocetin cofactor activity/VWF antigen ratios on plasma testing, recombinant VWF did not show a classic type 2M phenotype for any of the five variants. Homozygous expression of variants p.D1283Y, p.R1349C, p.R1374C, and p.I1453N was consistent with type 2A VWD, although all had normal expression as heterozygous recombinant VWF. Variant P1162L had normal VWF expression and function, consistent with the lack of bleeding symptoms.
Conclusions
Though originally classified as type 2M VWD, these homozygous recombinant VWF variants do not fulfill complete 2M VWD diagnostic criteria. A better classification schema and improved testing for putative type 2M variants is needed in order to effectively diagnose and treat affected patients.
Keywords: Von Willebrand Disease, Von Willebrand Factor, Platelets, Hemorrhage, Clinical laboratory techniques
Introduction
Von Willebrand factor (VWF) is a multimeric coagulation factor comprised of high molecular weight multimers (HMWM) that connect platelets to damaged subendothelium through collagen and platelet ligand sites. VWF binds to platelet glycoprotein Ib (GPIb) via the A1 domain [1]. VWF binds to collagens 1 and 3 via the A3 domain, and collagens 4 and 6 via the A1 domain [2,3]. Von Willebrand disease (VWD) is an inherited defect of VWF structure or quantity [4].
Multiple subtypes of VWD exist that include qualitative and quantitative anomalies [4]. Type 2M VWD is a qualitative variant of VWD, characterized by a specific defect in platelet adhesion and/or collagen binding. Type 2M VWD is diagnosed by a marked decrease in VWF ristocetin cofactor activity (VWF:RCo) relative to VWF antigen (VWF:Ag) in the presence of a normal multimer distribution [5]. The type 2M VWD category also includes collagen binding defects [4]. A decreased VWF:RCo/VWF:Ag ratio (<0.6–0.7) is indicative of the presence of a dysfunctional VWF [6]. Type 2A VWD is also a qualitative defect in VWF. In type 2A VWD, mutations affect the multimer structure of VWF with a loss of HMWM, and concomitant loss of GPIbα binding. Some type 2A variants have altered ADAMTS13 cleavage of VWF [7]. Type 2A VWD is diagnosed by a decrease in VWF:RCo/VWF:Ag ratio, <0.6–0.7, with a loss of HMWM [5].
In a study by Castaman measuring gastrointestinal bleeding, 2A VWD subtypes had a higher bleeding tendency than 2M VWD subtypes [8]. Increased bleeding risk may be attributed to the lack of HMWM in type 2A VWD [8]. Accurate and efficient classification of VWD mutations can dictate therapeutic strategy for VWD patients, especially when in high-risk bleeding situations. Alternately, misdiagnosis of a normal individual as having VWD can lead to unnecessary treatment and expense. We examined five VWF sequence variants identified through the Zimmerman Program for the Molecular and Clinical Biology of VWD, all originally classified as type 2M.
Methods
Study subjects
Subjects for this research project were enrolled in the Zimmerman Program for the Molecular and Clinical Biology of VWD (Zimmerman Program) following informed consent. The study is comprised of healthy controls, subjects with any diagnosed type of VWD, and immediate family members from sites across the United States. Subjects were prompted with a questionnaire to assess bleeding severity and assign a bleeding score [9]. A panel of laboratory tests including factor VIII activity (FVIII), VWF antigen (VWF:Ag), ristocetin cofactor activity (VWF:RCo), and multimer distribution were performed, as previously described, on plasma samples obtained at time of enrollment from each subject [10].
Five subjects enrolled with type 2M VWD who had novel or atypical VWF variants were selected for further study. Novel and atypical variants were defined as 2M VWD variants that have not been described in depth throughout the literature. Criteria involving the selection of a 2M subject included a VWF:RCo/ VWF:Ag ratio <0.6, as well as a normal multimer distribution on plasma samples obtained from the index case. VWF sequencing was performed on all subjects as previously described [11]. All family members had targeted VWF sequencing performed to evaluate for the presence of the variant found in that family’s index case.
Synthesis of variant VWF constructs
QuikChange II XL Site- Directed Mutagenesis Kit (Agilent Technologies) was used to synthesize recombinant VWF constructs to include 1283Y, 1349C, 1374C, 1453N, and 1162L variants in the pCINeo plasmid. DNA containing the VWF constructs was purified and expressed in HEK293T cells for each variant, as well as a wild-type (WT) VWF construct for a positive control, and an empty pCINeo vector (mock) for a negative control. Supernatants of HEK293T cells that contained VWF were collected at 72 hours for analysis by ELISA as described below. Additional transfections were performed using a 1:1 ratio of variant DNA to wild-type human recombinant VWF (in the pCDNA3.1 myc his vector) to mimic the heterozygous state.
VWF assays
Expression of variants was analyzed by ELISA for VWF:Ag as described previously [10]. The wild-type, mock, and variant constructs were serially diluted in ELISA block buffer and tested in triplicate at three different dilutions. Biotinylated anti-VWF monoclonal antibodies (AVW-4 and VWF-15, Blood Research Institute) were added at 1 mcg mL−1 for detection of bound VWF. Retention of variants was measured by performing VWF:Ag on the cell lysates as previously described [12]. To measure collagen 4 binding, the ELISA plate was coated with collagen type 4 (Southern BioTech) diluted to 1 mcg mL−1, as previously described [13]. Presence of VWF was measured by a combination of biotinylated anti-VWF monoclonal antibodies (AVW-1 and AVW-15, Blood Research Institute). To measure collagen 3 binding, the ELISA plate was coated with collagen type 3 (Southern BioTech) diluted to 1 mcg mL−1 as previously described [14]. Platelet GPIbα binding (VWF:GPIbM) was tested using a 96 well Immulon-4 HBX plate (Thermo Scientific, Rochester, NY, USA) as previously described using an anti-GPIb monoclonal antibody for capture of the recombinant GPIbα and a combination of biotinylated anti-VWF monoclonal antibodies (AVW-1 and AVW-15, Blood Research Institute) for detection in the absence of ristocetin [15]. VWF multimers were analyzed using sodium dodecyl sulfate agarose gel electrophoresis for all variant constructs. Multimer protein products were then transferred to a PVDF membrane. Immuno detection of multimers was performed as previously described using a combination of anti-VWF monoclonal antibodies (105.4, AVW-1, and AVW-5, Blood Research Institute) [16].
Results
Type 2M VWD subjects
Five subjects enrolled in the Zimmerman Program with a pre-existing diagnosis of type 2M VWD were evaluated. At the time of evaluation, 14 subjects in the study carried a diagnosis of type 2M and had DNA sequencing results available, but many of the sequence variants found had been previously characterized [17,18]. Baseline laboratory characteristics, including VWF levels, bleeding scores, and sequence variants, are detailed in table 1. These were similar to the values seen for other type 2M subjects enrolled in the Zimmerman Program (mean VWF:Ag 50 IU/dL and mean VWF:RCo 24 IU/dL for type 2M subjects recruited to date). Each subject had a decreased VWF:RCo/VWF:Ag ratio less than 0.6, but normal multimer distribution consistent with a clinical diagnosis of type 2M VWD. No subject had an elevated VWF propeptide/VWF:Ag ratio. Genetic variants in 4 subjects were located in the A1 domain of the VWF gene, with one subject possessing a mutation in the D3 domain (figure 1).
Table 1.
Clinical data for subjects with type 2M VWD.
| Variant | p.P1162L | p.D1283Y | p.Y1349C | p.R1374C | p.I1453N |
|---|---|---|---|---|---|
| Age (years) | 52 | 85 | 29 | 39 | 25 |
| Race | African American | Caucasian | Caucasian | Caucasian | Caucasian |
| Sex | Female | Female | Female | Male | Male |
| Bleeding score | 2 | 12 | 8 | 2 | 11 |
| VWF:Ag (IU/dL) | 144 | 35 | 49 | 22 | 20 |
| VWF:RCo (IU/dL) | 72 | 12 | 17 | 12 | 11 |
| VWF:RCo/VWF:Ag ratio | 0.50 | 0.34 | 0.35 | 0.55 | 0.55 |
| Multimer distribution | Normal | Normal | Normal | Normal | Normal |
| VWF:GPIbM (U/dL) | 98 | 6 | 11 | 18 | 23 |
| VWF:GPIbM/VWF:Ag ratio | 0.68 | 0.17 | 0.22 | 0.82 | 1.15 |
| VWF:CB3(IU/dL) | 156 | 35 | 48 | 19 | 19 |
| VWF:CB3/VWF:Ag ratio | 1.08 | 1.00 | 0.98 | 0.86 | 0.95 |
| VWF:CB4(U/dL) | 179 | 24 | 34 | 14 | 18 |
| VWF:CB4/VWF:Ag ratio | 1.24 | 0.69 | 0.69 | 0.64 | 0.90 |
VWF:Ag = VWF antigen; VWF:RCo = VWF ristocetin cofactor activity; VWF:GPIbM = VWF platelet binding using gain-of-function GPIb; VWF:CB3 = VWF collagen binding to type III collagen; VWF:CB4 = VWF collagen binding to type IV collagen
Figure 1. Type 2M variants cluster in the VWF A1 domain.
Spheres in the VWF A1 domain crystal structure [30] indicate variant locations. Note that variant P1162L is located in the D3 domain and is thus not depicted in the crystal structure.
Subjects with p.P1162L and p.R1374C exhibited normal bleeding scores of 2, while subjects with p.Y1349C, p.I1453N, and p.D1283Y exhibited elevated bleeding scores of 8, 11, and 12, respectively. The median bleeding score for the entire type 2M Zimmerman cohort was 11. Values of >2 are abnormal for children, with the cut-off increasing to >3 for adult males and >5 for adult females. Interestingly, subjects had varied bleeding scores but a similar range of VWF:RCo/VWF:Ag ratios. The VWF:RCo/VWF:Ag ratio alone did not predict bleeding symptoms in this group of subjects. Bleeding score measurements showed no correlation to VWF:Ag, VWF:RCo, or VWF:GPIbM.
Affected family members
Through the Zimmerman program, family members of each subject were investigated. The index case and two family members with p.P1162L had a normal clinical phenotype with normal bleeding scores in the setting of decreased VWF:RCo/VWF:Ag ratios. The index case subject with variant p.R1374C had a normal bleeding score. Twenty-five family members were tested, some affected with VWD. With the exception of one p.R1374C family member having a high bleeding score, all other twenty-four family members had normal bleeding scores. There were six pediatric (under 18 years old) members in this family at time of draw, all of whom had normal bleeding scores. Other family members were older than 18 years old. The index case subject with variant p.Y1349C had a high bleeding score and had five affected family members with varied bleeding scores, ranging from 4–14. The index case with variant p.D1283Y also had a high bleeding score, and a single affected family member with a comparable high bleeding score of 13. The index case subject with variant p.I1453N had a high bleeding score but had no affected family members enrolled to analyze.
VWF secretion
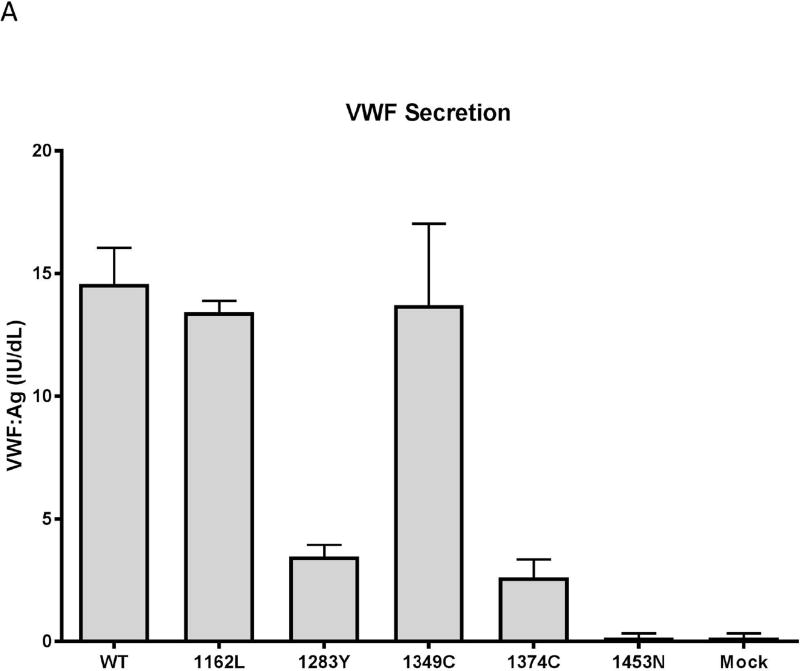
Classic type 2M VWD is thought to have normal VWF secretion with a specific defect in VWF function, typically with deficient platelet binding. ELISA assays on the secreted VWF protein showed that three of the five VWF variants did not secrete as well as the wild-type model. Variants 1283Y and 1374C showed significant decrease in the secretion of VWF as measured by VWF:Ag, approximately three times less than the wild-type secretion. 1453N did not secrete at all. 1162L secretion was similar to that seen for wild-type VWD (figure 2A).
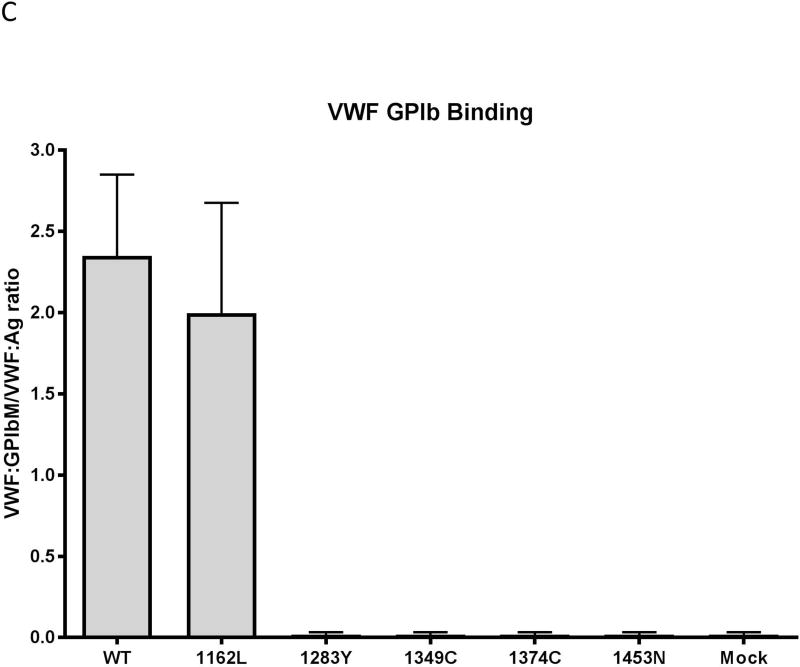
Figure 2. Variable in vitro secretion, collagen binding, and platelet GPIαb binding of type 2M variants.
Data shown represents averages of 3–4 different assays on 3–4 separate transfections. Error bars represent 1 standard deviation. Panel A: Expression. Variants 1283Y, 1374C and 1453N show a marked decrease in expression relative to wild-type. Variants 1162L and 1349C have normal expression. Panel B: Collagen binding. Binding to collagen 3 is shown in light gray and binding to collagen 4 is shown in dark gray. Variants 1283Y, 1349C, and 1374C have decreased binding to collagen 3 and 4. 1453N has absent binding to both collagen 3 and 4. 1162L has normal binding to collagen 3 and 4. Panel C: Platelet GPIαb binding. Variants 1283Y, 1349C, 1374C, and I1453N have absent binding to platelet GPIb. 1162L has normal binding. Data represents the average of three different VWF:GPIbM ELISA assays performed on three separate transfections.
Cell lysates were also examined to look at intracellular retention of VWF. 1283Y, 1349C, and 1374C all had VWF:Ag in the cell lysate at or higher than the level seen for WT VWF, suggesting intracellular retention. 1453N had lysate levels that were approximately half that seen for the WT VWF, suggesting a defect in VWF formation as well as secretion (data not shown).
VWF collagen binding
Variant 1162L demonstrated normal binding to collagen 3 and 4 that was similar to wild-type (figure 2B). Conversely, variants 1283Y and 1374C showed decreased VWF:CB3/VWF:Ag with over a four-fold decrease in collagen 3 and collagen 4 binding compared to WT (p<0.05). Variant 1349C showed a non-significant decrease in collagen 4 binding. Since 1453N was not secreted, no collagen binding was observed.
GPIbα binding
GPIbα binding was absent in variants 1283Y, 1349C, and 1374C on VWF:GPIbM ELISA assay. Variant 1162L had platelet GPIb binding similar to that seen for wild-type VWF (Figure 2C). As expected given the lack of expression, no binding was observed for 1453N.
Heterozygous expression
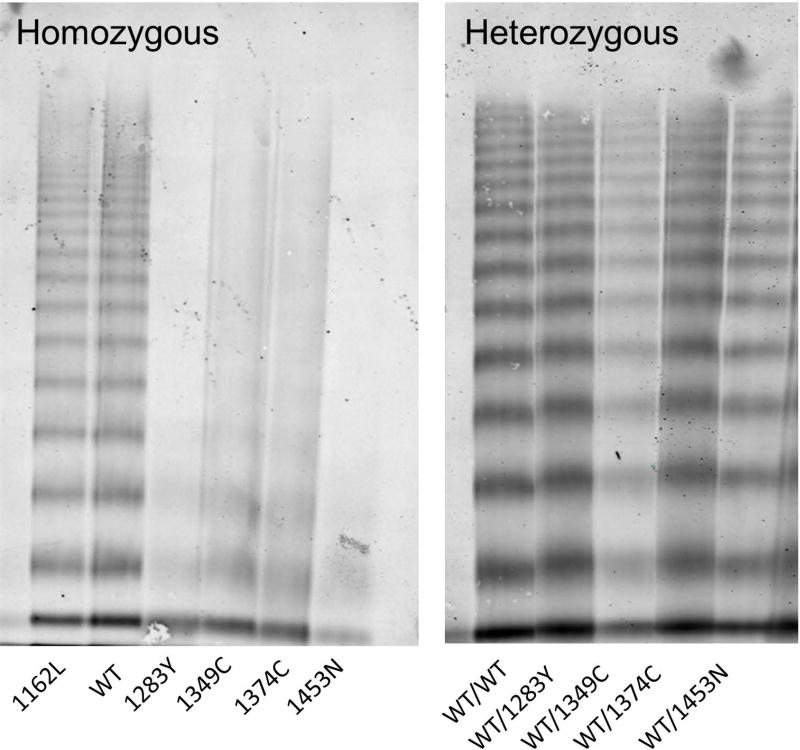
To mimic the heterozygous state seen in patients, 1283Y, 1349C, 1374C, and 1453N, were co-transfected with a wild-type VWF. Secretion was reduced to half of the wild-type VWF for 1283Y, 1374C, and 1453N. All heterozygous variants had normal binding to collagen 3, 4, and platelet GPIb. Figure 3 shows the comparison of homozygous VWF multimer distribution to the heterozygous VWF multimer distribution. The WT VWF and variant VWF were labeled in different colors, which demonstrated the predominance of multimers from the WT construct with minimal contribution of variant VWF for 12873Y, 1374C, and 1453N. For 1349C, both WT and variant constructs contributed to the normal multimer distribution (supplemental figure 1). Table 2 shows relative laboratory comparisons of the VWF variants to its heterozygous wild-type counterpart. The normal results may therefore reflect the predominance of WT VWF in the heterozygous expression.
Figure 3. Some homozygous variants have defective multimer structure that is improved when expressed in heterozygous form.
Variants 1283Y, 1349C, 1374C, and 1453N have loss of HMWM when expressed as homozygotes. Variants 1283Y, 1349C, 1374C, and 1453N have improved multimer distribution when expressed as heterozygous variant/wild-type. The multimer distribution for homozygous 1162L is similar to wild-type VWF.
Table 2.
Summary of VWF Expression Data.
| Secretion | VWF:GPIbM | VWF:CB3 | VWF:CB4 | Multimer distribution |
|
|---|---|---|---|---|---|
| WT | Normal | Normal | Normal | Normal | Normal |
| 1162L | Normal | Normal | Normal | Normal | Normal |
| 1283Y | ↓↓ | ↓↓↓ | ↓ | ↓↓ | ↓↓ |
| WT/1283Y | ↓↓ | Normal | Normal | Normal | Normal |
| 1349C | Normal | ↓↓↓ | ↓ | ↓ | ↓ |
| WT/1349C | ↓↓ | Normal | Normal | Normal | Normal |
| 1374C | ↓↓ | ↓↓↓ | ↓↓ | ↓↓ | ↓ |
| WT/1374C | ↓↓ | Normal | Normal | Normal | Normal |
| 1453N | ↓↓↓ | ↓↓↓ | ↓↓↓ | ↓↓↓ | ↓↓↓ |
| WT/1453N | ↓↓ | Normal | Normal | Normal | Normal |
Discussion
Each of the five variants studied here was originally classified as Type 2M VWD based on a low plasma VWF:RCo/VWF:Ag ratio in the setting of normal VWF multimer distribution. However in vitro studies of homozygous 1283Y, 1349C, 1374C, 1453N, and 1162L demonstrate behavior inconsistent with a diagnosis of classical type 2M VWD.
Variant p.P1162L: normal variant
When first discovered, VWF variant p.P1162L was categorized as Type 2M. This classification was due to the decreased VWF:RCo/VWF:Ag of 0.5, noted in Table 1. However in vitro, 1162L presented phenotypically identical to the wild-type VWF. In figure 2A, 1162L is secreted in the HEK293T cells to a similar degree as wild- type VWF. Furthermore, figure 2B and 2C depict binding to collagen 3, collagen 4, and platelet GPIbα at the same capacity as wild-type VWF. The p.P1162L variant presents as a wild-type phenotype both in vitro as shown above and in vivo with a normal bleeding score of 2 in the index case and affected family members. The index case also had a VWF:RCo well above the lower limit of normal.
Variant p.P1162L is located in the D3 binding domain. Perhaps because p.P1162L is not located within the A1 domain, this does not hinder platelet binding. The most likely explanation for the low VWF:RCo/VWF:Ag ratio is due to the presence of a homozygous p.D1472H VWF A1 domain variant found both in this subject and the affected family members. This variant has previously been linked to low VWF:RCo/VWF:Ag ratios without causing clinical bleeding [10]. Based on this evidence, we would suggest that p.P1162L should not be considered a pathologic 2M VWD variant but rather included as a genetic variant of normal VWF. Variant p.P1162L has previously been identified in other Zimmerman Program index cases, including type 1, 1C, 2A, and 2B VWD, all of whom had another causative variant explaining their phenotype.
Variants p.D1283Y, p.R1349C, p.R1374C, p.I1453N :2A or 2M?
Type 2A VWD is characterized by a loss of HMWM and intermediate molecular weight multimers. This arises from either a defect in synthesis or vulnerable cleavage to ADAMTS-13 [7]. A defect in HMWM could severely affect VWF’s functional role of binding platelets and collagen. Though categorized as 2M VWD, variants p.D1283Y, p.R1349C, and p.R1374C are most consistent with a 2A phenotype in vitro when expressed in homozygous form. Interestingly, p.I1453N had no detectable expression in vitro, which appears more consistent with a type 3 VWD defect. Despite this lack of expression, the patient phenotype showed a functional defect with decreased VWF:RCo more consistent with type 2 VWD. Unfortunately no family members were available to study this variant further. In heterozygous form, decreased secretion was seen, but binding to platelet GPIb and collagen was normal. Multimer distribution was primarily composed of wild-type multimers, supporting a synthetic defect as opposed to a preferential destruction of abnormal VWF variants. The normal multimers seen on plasma testing could reflect the VWF synthesized from the normal allele. Previous work has shown variable expression for the p.M1304R variant [19]. It is certainly possible that in vivo the “good” allele is dominant in some VWD patients.
Interestingly, the subject with variant p.R1374C had a normal bleeding score despite similar functional defects observed in vitro. Of note, there are other index cases of p.R1374C in the Zimmerman program with elevated bleeding scores that were classified as type 1C and 2A. This variant has been classified alternately as type 1, type 2M and type 2A in the literature [20,21,22]. There is recent evidence for variable phenotype seen with p.1374C, with patients in the same family evidencing variable bleeding as assessed by bleeding score [23].
2M Classification Schema
Though all subjects were diagnosed as Type 2M VWD, VWF variants 1283Y, 1349C, 1374C, 1162L, and 1453N do not necessarily behave in vitro as type 2M. A 2M VWD variant has qualitative defects in VWF with a normal multimer distribution, but variants 1283Y, 1349C, 1374C, and 1453N all had abnormal multimer distribution when expressed in homozygous form (Figure 3). However, our study is limited by the analysis of recombinant VWF in heterozygous and homozygous form, which may not reflect the plasma VWF in affected patients.
Previous research has suggested that VWF:RCo to measure VWF function is not the most informative schema to assess VWF’s in vivo function [24]. Given the high coefficient of variation with the VWF:RCo [25,26], relying on this assay to diagnose type 2 variants can be problematic. The VWF:GPIbM assay has been considered a potential replacement for the VWF:RCo [27]. In this group, the VWF:GPIbM assay did not uniformly identify a decrease in VWF function. Only the subjects with p.D1283Y and p.Y1349C had decreased VWF:GPIbM/VWF:Ag ratios. While the ratio would be expected to be normal for the subject with p.P1162L, the normal ratios in the subjects with p.R1374C and p.I1453N may have been secondary to variable secretion [19]. It is also possible that the VWF:RCo is an aberrant result and p.I1453N is more akin to a type 3 variant.
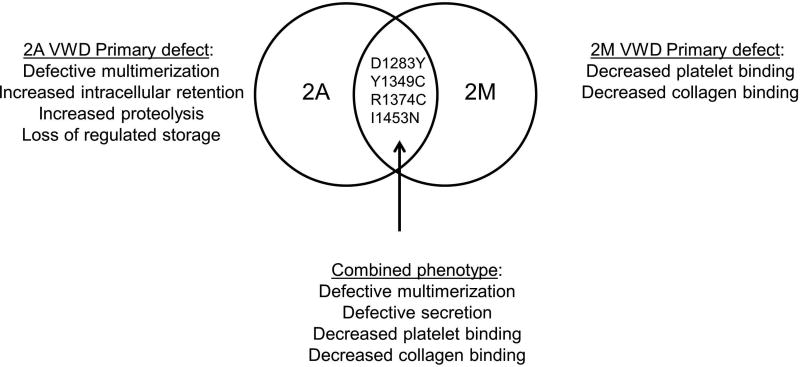
All subjects had normal plasma collagen binding with both collagen 3 and collagen 4, in addition to a normal multimer distribution in plasma, despite the lack of normal multimer distribution for the homozygous expressed variant constructs 1283Y, 1349C, 1374C and 1453N in vitro. This may be due in part to our inability to detect subtle multimer defects. It is possible that in patient plasma, the wild-type VWF allele is dominant. This would match with the normalization of results seen with heterozygous expression where normalization of the multimer distribution was seen for all variants. That explanation, however, fails to explain the decrease in VWF:RCo and/or VWF:GPIbM seen on plasma testing of the affected patients. This may be due to differences in secretion of VWF, additional VWF variants or modifier protein variants, or to the presence or absence of other coagulation defects that could modify phenotype. It may be that all type 2M variants have a qualitative defect affecting VWF function, with some also affecting VWF secretion, whereas all type 2A variants have loss of high molecular weight multimers with a corresponding qualitative defect in VWF function (figure 4). Clinical laboratory variability may reflect our imperfect assays, such that some variants may be alternately classified as either type 2A or type 2M. It should also be noted that collagen binding defects without a concomitant platelet binding defect fall into the category of type 2M VWD; however, none of the variants in this analysis showed collagen binding defects.
Figure 4. Intersecting phenotypes for types 2A and 2M VWD.
This diagram shows the overlap in phenotype for the variants described here, combining attributes of type 2A and type 2M VWD.
We propose that the current classification schema of type 2A VWD versus type 2M VWD does not completely encompass certain variants, whose laboratory phenotype may vary despite the presence of a unifying genetic mutation. In terms of laboratory diagnosis, multimer distribution may not be required in order to ensure appropriate treatment of affected patients. Clinically, classifying these as type 2A/2M is sufficient to recommend to clinicians the use of a von Willebrand factor containing concentrate for treatment, particularly if no clinical response to desmopressin is obtained. Some of these variants have previously been labeled “unclassified” which does not alleviate clinical confusion as to the phenotype. Classic type 2A VWD has a known association of increased gastrointestinal bleeding with type 2A VWD that is not seen in type 2M VWD [8]. Classic type 2M VWD is generally less symptomatic. The group with overlap in laboratory expression may also have an overlap in phenotype, but more work is required to characterize this interesting group of variants in flow based models.[28,29]
Supplementary Material
Heterozygous transfection of 50% normal and 50% variant DNA, with WT VWF labeled in green and variant VWF labeled in red for variants 1283Y, 1349C, 1374C, and 1453N. WT lane represents 50% green label and 50% red label, which scans as yellow. The 1349C variant also shows yellow color, representing contribution of both WT and variant DNA to the expressed protein. Heterozygous transfection of 1283Y, 1374C, and 1453N demonstrates predominance of the green WT VWF in the multimer distribution.
Essentials.
The pathophysiology of type 2M von Willebrand disease (VWD) is poorly understood.
Sequence variations in type 2M VWD subjects were characterized.
A high degree of clinical and laboratory variability exists within type 2M VWD variants.
Some type 2M variants may share features of type 2A VWD.
Acknowledgments
The authors would like to thank all the subjects, physicians, and staff involved in the Zimmerman Program. Funding from the National Heart, Lung, and Blood Institute provided support for the Zimmerman Program and multiple investigators (HL081588, HL102260, and HL126810). Additional support was provided by the MACC Fund (Midwest Athletes Against Childhood Cancer) and the BloodCenter Research Fund.
Footnotes
Addendum
A. L. Doruelo performed experiments, wrote the manuscript, and assisted in research design. Sandra L. Haberichter performed experiments, assisted in research design, and edited the manuscript. P. A. Christopherson collected subject data, edited the manuscript, and assisted in research design. L. N. Boggio collected subject data and edited the manuscript. S. Gupta collected subject data and edited the manuscript. S. R. Lentz collected subject data and edited the manuscript. A. D. Shapiro collected subject data and edited the manuscript. R. R. Montgomery assisted in research design and edited the manuscript. V. H. Flood supervised experiments, designed the research, and wrote the manuscript.
Disclosure of Conflicts of Interests
L. N. Boggio has served as a consultant for Alnylam, Bayer, Biogen, CSL Behring, Grifols, Novo Nordisk, Octapharma, OPKO, Pfizer, and Shire. S. R. Lentz. has received grants from NIH. V. H. Flood has received grants from NIH and has served as a consultant for CSL Behring and Shire.
References
- 1.Fujimura Y, Titani K, Holland LZ, Russell SR, Roberts JR, Elder JH, Ruggeri ZM, Zimmerman TS. Von willebrand factor. A reduced and alkylated 52/48-kDa fragment beginning at amino acid residue 449 contains the domain interacting with platelet glycoprotein ib. J Biol Chem. 1986;261:381–5. [PubMed] [Google Scholar]
- 2.Pareti FI, Niiya K, McPherson JM, Ruggeri ZM. Isolation and characterization of two domains of human von willebrand factor that interact with fibrillar collagen types I and III. J Biol Chem. 1987;262:13835–41. [PubMed] [Google Scholar]
- 3.Rand JH, Patel ND, Schwartz E, Zhou SL, Potter BJ. 150-kD von willebrand factor binding protein extracted from human vascular subendothelium is type VI collagen. J Clin Invest. 1991;88:253–9. doi: 10.1172/JCI115285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadler JE, Budde U, Eikenboom JC, Favaloro EJ, Hill FG, Holmberg L, Ingerslev J, Lee CA, Lillicrap D, Mannucci PM, Mazurier C, Meyer D, Nichols WL, Nishino M, Peake IR, Rodeghiero F, Schneppenheim R, Ruggeri ZM, Srivastava A, Montgomery RR, Federici AB. Working Party on von Willebrand Disease Classification. Update on the pathophysiology and classification of von willebrand disease: A report of the subcommittee on von willebrand factor. J Thromb Haemost. 2006;4:2103–14. doi: 10.1111/j.1538-7836.2006.02146.x. [DOI] [PubMed] [Google Scholar]
- 5.Nichols WL, Hultin MB, James AH, Manco-Johnson MJ, Montgomery RR, Ortel TL, Rick ME, Sadler JE, Weinstein M, Yawn BP. Von willebrand disease (VWD): Evidence-based diagnosis and management guidelines, the national heart, lung, and blood institute (NHLBI) expert panel report (USA) Haemophilia. 2008;14:171–232. doi: 10.1111/j.1365-2516.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 6.Favaloro EJ. Diagnosis and classification of von willebrand disease: A review of the differential utility of various functional von willebrand factor assays. Blood Coagul Fibrinolysis. 2011;22:553–64. doi: 10.1097/MBC.0b013e32834a7e01. [DOI] [PubMed] [Google Scholar]
- 7.Jacobi PM, Gill JC, Flood VH, Jakab DA, Friedman KD, Haberichter SL. Intersection of mechanisms of type 2A VWD through defects in VWF multimerization, secretion, ADAMTS-13 susceptibility, and regulated storage. Blood. 2012;119:4543–53. doi: 10.1182/blood-2011-06-360875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castaman G, Federici AB, Tosetto A, La Marca S, Stufano F, Mannucci PM, Rodeghiero F. Different bleeding risk in type 2 A and 2 M von willebrand disease: A two-year prospective study in 107 patients. J Thromb Haemost. 2012;10:632–638. doi: 10.1111/j.1538-7836.2012.04661.x. [DOI] [PubMed] [Google Scholar]
- 9.Rodeghiero F, Tosetto A, Abshire T, Arnold DM, Coller B, James P, Neunert C, Lillicrap D. ISTH/SSC joint VWF and Perinatal/Pediatric Hemostasis Subcommittees Working Group. ISTH/SSC bleeding assessment tool: A standardized questionnaire and a proposal for a new bleeding score for inherited bleeding disorders. J Thromb Haemost. 2010;8:2063–5. doi: 10.1111/j.1538-7836.2010.03975.x. [DOI] [PubMed] [Google Scholar]
- 10.Flood VH, Gill JC, Morateck PA, Christopherson PA, Friedman KD, Haberichter SL, Branchford BR, Hoffmann RG, Abshire TC, Di Paola JA, Hoots WK, Leissinger C, Lusher JM, Ragni MV, Shapiro AD, Montgomery RR. Common VWF exon 28 polymorphisms in african americans affecting the VWF activity assay by ristocetin cofactor. Blood. 2010;116:280–6. doi: 10.1182/blood-2009-10-249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellissimo DB, Christopherson PA, Flood VH, Gill JC, Friedman KD, Haberichter SL, Shapiro AD, Abshire TC, Leissinger C, Hoots WK, Lusher JM, Ragni MV, Montgomery RR. VWF mutations and new sequence variations identified in healthy controls are more frequent in the african-american population. Blood. 2012;119:2135–40. doi: 10.1182/blood-2011-10-384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haberichter SL, Allmann AM, Jozwiak MA, Montgomery RR, Gill JC. Genetic alteration of the D2 domain abolishes von willebrand factor multimerization and trafficking into storage. J Thromb Haemost. 2009;7:641–50. doi: 10.1111/j.1538-7836.2009.03290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flood VH, Schlauderaff AC, Haberichter SL, Slobodianuk TL, Jacobi PM, Bellissimo DB, Christopherson PA, Friedman KD, Gill JC, Hoffmann RG, Montgomery RR. Zimmerman Program Investigators. Crucial role for the VWF A1 domain in binding to type IV collagen. Blood. 2015;125:2297–304. doi: 10.1182/blood-2014-11-610824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flood VH, Lederman CA, Wren JS, Christopherson PA, Friedman KD, Hoffmann RG, Montgomery RR. Absent collagen binding in a VWF A3 domain mutant: Utility of the VWF:CB in diagnosis of VWD. J Thromb Haemost. 2010;8:1431–3. doi: 10.1111/j.1538-7836.2010.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flood VH, Gill JC, Morateck PA, Christopherson PA, Friedman KD, Haberichter SL, Hoffmann RG, Montgomery RR. Gain-of-function GPIb ELISA assay for VWF activity in the zimmerman program for the molecular and clinical biology of VWD. Blood. 2011;117:e67–74. doi: 10.1182/blood-2010-08-299016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flood VH, Gill JC, Friedman KD, Christopherson PA, Jacobi PM, Hoffmann RG, Montgomery RR, Haberichter SL. the Zimmerman Program Investigators. Collagen binding provides a sensitive screen for variant von willebrand disease. Clin Chem. 2013;59:684–91. doi: 10.1373/clinchem.2012.199000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancuso DJ, Kroner PA, Christopherson PA, Vokac EA, Gill JC, Montgomery RR. Type 2M:Milwaukee-1 von willebrand disease: An in-frame deletion in the Cys509-Cys695 loop of the von willebrand factor A1 domain causes deficient binding of von willebrand factor to platelets. Blood. 1996;88:2559–68. [PubMed] [Google Scholar]
- 18.Larsen DM, Haberichter SL, Gill JC, Shapiro AD, Flood VH. Variability in platelet- and collagen-binding defects in type 2M von willebrand disease. Haemophilia. 2013;19:590–4. doi: 10.1111/hae.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Hinckley JD, Haberichter S, Jacobi P, Montgomery R, Flood VH, Wong R, Interlandi G, Chung DW, Lopez JA, Di Paola J. Variable content of von willebrand factor mutant monomer drives the phenotypic variability in a family with von willebrand disease. Blood. 2015;126:262–9. doi: 10.1182/blood-2014-11-613935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penas N, Perez-Rodriguez A, Torea JH, Loures E, Noya MS, Lopez-Fernandez MF, Batlle J. Von willebrand disease R1374C: Type 2A or 2M? A challenge to the revised classification. high frequency in the northwest of spain (galicia) Am J Hematol. 2005;80:188–96. doi: 10.1002/ajh.20470. [DOI] [PubMed] [Google Scholar]
- 21.James PD, Notley C, Hegadorn C, Poon MC, Walker I, Rapson D, Lillicrap D. Association of Hemophilia Clinic Directors of Canada. Challenges in defining type 2M von willebrand disease: Results from a canadian cohort study. J Thromb Haemost. 2007;5:1914–22. doi: 10.1111/j.1538-7836.2007.02666.x. [DOI] [PubMed] [Google Scholar]
- 22.Goodeve A, Eikenboom J, Castaman G, Rodeghiero F, Federici AB, Batlle J, Meyer D, Mazurier C, Goudemand J, Schneppenheim R, Budde U, Ingerslev J, Habart D, Vorlova Z, Holmberg L, Lethagen S, Pasi J, Hill F, Hashemi Soteh M, Baronciani L, Hallden C, Guilliatt A, Lester W, Peake I. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von willebrand disease in the european study, molecular and clinical markers for the diagnosis and management of type 1 von willebrand disease (MCMDM-1VWD) Blood. 2007;109:112–21. doi: 10.1182/blood-2006-05-020784. [DOI] [PubMed] [Google Scholar]
- 23.Gupta S, Heiman M, Duncan N, Hinckley J, Di Paola J, Shapiro AD. Variable bleeding phenotype in an amish pedigree with von willebrand disease. Am J Hematol. 2016;91:E431–5. doi: 10.1002/ajh.24474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flood VH, Friedman KD, Gill JC, Morateck PA, Wren JS, Scott JP, Montgomery RR. Limitations of the ristocetin cofactor assay in measurement of von willebrand factor function. J Thromb Haemost. 2009;7:1832–9. doi: 10.1111/j.1538-7836.2009.03594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitchen S, Jennings I, Woods TA, Kitchen DP, Walker ID, Preston FE. Laboratory tests for measurement of von willebrand factor show poor agreement among different centers: Results from the united kingdom national external quality assessment scheme for blood coagulation. Semin Thromb Hemost. 2006;32:492–8. doi: 10.1055/s-2006-947863. [DOI] [PubMed] [Google Scholar]
- 26.Meijer P, Haverkate F. An external quality assessment program for von willebrand factor laboratory analysis: An overview from the european concerted action on thrombosis and disabilities foundation. Semin Thromb Hemost. 2006;32:485–91. doi: 10.1055/s-2006-947862. [DOI] [PubMed] [Google Scholar]
- 27.Graf L, Moffat KA, Carlino SA, Chan AK, Iorio A, Giulivi A, Hayward CP. Evaluation of an automated method for measuring von willebrand factor activity in clinical samples without ristocetin. Int J Lab Hematol. 2014;36:341–51. doi: 10.1111/ijlh.12218. [DOI] [PubMed] [Google Scholar]
- 28.Tischer A, Campbell JC, Machha VR, Moon-Tasson L, Benson LM, Sankaran B, Kim C, Auton M. Mutational constraints on local unfolding inhibit the rheological adaptation of von willebrand factor. J Biol Chem. 2016;291:3848–59. doi: 10.1074/jbc.M115.703850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colace TV, Fogarty PF, Panckeri KA, Li R, Diamond SL. Microfluidic assay of hemophilic blood clotting: Distinct deficits in platelet and fibrin deposition at low factor levels. J Thromb Haemost. 2014;12:147–58. doi: 10.1111/jth.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley J, Cruz M, Handin R, Liddington R. Crystal structure of the von willebrand factor A1 domain and implications for the binding of platelet glycoprotein ib. J Biol Chem. 1998;273:10396–401. doi: 10.1074/jbc.273.17.10396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heterozygous transfection of 50% normal and 50% variant DNA, with WT VWF labeled in green and variant VWF labeled in red for variants 1283Y, 1349C, 1374C, and 1453N. WT lane represents 50% green label and 50% red label, which scans as yellow. The 1349C variant also shows yellow color, representing contribution of both WT and variant DNA to the expressed protein. Heterozygous transfection of 1283Y, 1374C, and 1453N demonstrates predominance of the green WT VWF in the multimer distribution.