Abstract
Adhesion molecules play a key role in cancer progression and tumorigenesis. Genetic polymorphism of adhesion molecules may alter the normal functioning thereby leading to bladder cancer susceptibility. Hence we aimed to evaluate three SNPs of CD166 gene (CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G) in bladder cancer patients and normal controls of North Indian population. A total of 270 healthy controls and 240 confirmed bladder cancer patients were recruited for this study. Three SNPs of CD166 gene viz. CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G were selected for this study. CD166rs6437585 C/T and CD166rs10511244 C/T were genotyped by Taqman allelic discrimination assay and CD166rs1157 A/G was genotyped by PCR–RFLP. The statistical analysis was done using the SPSS software, version 16.0 (SPSS, Chicago, IL), and p < 0.05 was considered statistically significant. Haplotypic analysis was done by using SNP analyzer version 1.2A. CD166rs6437585 C/T and CD166rs10511244 C/T showed significant association with reduced risk in bladder cancer while CD166rs1157 A/G showed significant high risk along with association at genotypic and allelic levels. Haplotypic analysis showed 1.8-folds risk in CCG combination, whereas CTA and TCG showed significant association with reduced risk. Further stratification on the basis of smoking, tumor grade/stage and BGC therapy revealed no association of these three polymorphic sites of CD166. Our study suggests that CD166rs6437585 C/T and CD166rs10511244 C/T are predictive for the reduced risk of bladder cancer, whereas CD166rs1157 A/G had shown significant association with high risk of bladder cancer in North Indians. This somehow suggests that CD166rs1157 A/G can be used as a marker for risk prediction of bladder cancer.
Keywords: CD166 gene (ALCAM), Bladder cancer, PCR–RFLP, BCG immunotherapy
Introduction
Bladder cancer is the 9th most common cancer worldwide, with an estimated 74,000 new cases expected to occur every year [1]. Bladder cancer incidence is about 4 times higher in men than in women. Bladder cancer incidence rates decreased from 2007 to 2011 by 1.6 % per year in men and by 1.1 % per year in women. An estimated 16,000 deaths will occur in 2015, 72 % of which will be in men [1]. In males, it is the fourth most common cancer (4 % of male total), whilst it is the 13th most common cancer in females (2 % of female total) [2]. As a general prevalence, in India, out of 1,00,000 people 3.0 male and 1 female develop BC each year [3].
Cancer stem cell is a recent theory in cancer study which is being established and extensively studied [4, 5]. Cancer stem cells are the population of TICs (Tumor Initiating Cells) which have the potential of generating a whole new tumor. Studies suggest their involvement in tumor initiation, progression, relapse and metastasis [6]. One of the best ways of using CSCs in novel treatment modality is to target their surface markers. For which the surface marker needs to be extensively studied. Out of various CSC markers, CD166 is identified as a purported stem cell marker in various cancers [7–10].
CD166 gene, also known as activated leukocyte cell adhesion molecule (ALCAM) is located on chromosome 3(3q13.1q13.2) containing 16 exons. CD166 is a 110-kDa multi-domain type 1 transmembrane glycoprotein of the immunoglobulin super family and is highly conserved. Hanahan and Weinberg [11], described various hallmarks of cancer like limitless replicative potential, uninterrupted angiogenesis, evasion of apoptosis, self-sufficiency with respect to growth signals, resistance to anti-growth signals, tissue invasion and metastasis [11]. Adhesion molecules are involved in the hallmark processes of cancer development. They also found to play role in various physiological functions like development of different tissues during embryogenesis, it is also expressed in varied class of malignancies such as melanoma and esophageal, gynecologic, prostate, and pancreatic cancers, and its expression is associated with diverse outcomes in different tumors [12]. Therefore, modulation of the function of adhesion molecules must be studied in various cancers.
Bladder cancer is the second most common among all forms of Urogenital cancer The prevalence of BC worldwide is estimated at over a million and is increasing steadily [13]. Despite its low incidence in western countries, it is still a major problem in India. The prevalence of BC is 3:1 in men: women [14]. More than 90 % of all newly diagnosed BC cases are transitional cell carcinomas. Zhou and group reported CD166 rs6437585 to be associated with increased risk of breast cancer among Chinese population [15]. CD166rs10511244 is very least reported, it showed no association with gall bladder cancer risk among North Indians [16]. CD166rs1157 is widely reported in many cancers viz. positive association with bladder cancer risk in Swedish population [17], No association with bladder cancer in Polish population [17], significant association with colon cancer risk [18] etc. Zhang et al. [19] in their review article have compiled various studies of genetic variants to be significantly associated with colon cancer risk and CD166rs1157 is among them. The presented case–control study was performed on North Indian population and focused on the effects of putative functionally relevant SNPs in candidate gene with a strong probability to be involved in BC risk. We investigated the effects of SNPs in CD166 gene and their contribution to BC susceptibility, tumour stage/grade and outcome after BCG immunotherapy with the aim to identify possible clinical markers. Here, we present the study of three unique genetic variants of CD166 gene i.e. CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G in a case–control study from North India in BC.
Materials and Methods
Study Subjects
A total of 240 confirmed bladder cancer patients and 270 healthy controls were recruited in the present study. All subjects in this study were of similar ethnicity, North India. All the patients enrolled in this study were histologically confirmed bladder cancer. Those with a previous history of other cancer, cancer metastasized to other site of body from another origin and previous radiotherapy was excluded. At the same time, 270 healthy controls (Mean age = 54.5 years, M:F = 249:21) were recruited from volunteers who came to the hospital for their routine checkups, unrelated to patients and to each other were also age and ethnicity matched. The criteria for selecting controls included no evidence of any personal history of cancer or other malignant conditions or any other chronic diseases. 240 confirmed bladder cancer patients were employed in this study. The ratio of male: female among 240 patients is 211:29, with the mean age of 56.9 years. The patients were subjected to detailed demographic, clinical and pathological investigations, which contained the details of age, stage, disease history, family history and other relevant details such as smoking history, occupational history and other lifestyle factors. 5-ml blood sample was drawn into coded tubes from every subject. Informed and written consent were taken from all subjects when interviewing for the demographic details and blood sample collection. The Ethical Review Board of the Institute approved the study.
Epidemiology
An epidemiological questionnaire was designed for the participants of this study to obtain data of demographic characteristics such as occupation, smoking and other lifestyles. Individuals who smoked once a day for more than 5 years were defined as smokers. The individuals who had never smoked in their lifetime were regarded as non smokers. At the conclusion of the interview, 5 ml of blood sample was drawn into coded EDTA vials. The demographic and clinical characteristics of the patients are demonstrated in Table 1.
Table 1.
Baseline demographic and clinical characteristics of bladder cancer patients and healthy controls
| Variables | Cases n = 240 n (%) |
Controls n = 270 n (%) |
Chi square p value# |
|---|---|---|---|
| Sex | |||
| Female | 29 (12.1) | 21 (7.8) | 0.105 |
| Male | 211 (87.9) | 249 (92.2) | |
| Age (years) | |||
| Mean age ± SD | 56.96 ± 13.86 | 54.50 ± 10.23 | 0.138 |
| Smokinga | |||
| Non smokers | 48 (29.6) | 214 (79.3) | <0.001 |
| Smokers | 116 (70.4) | 56 (20.7) | |
| Tumor grade stage | |||
| TaG1 | 48 (20.0) | – | – |
| TaG2-3 + T1G1-3 | 128 (53.3) | – | |
| T2+ | 64 (26.7) | – | |
| Intravesical therapy | |||
| Non treated | 83 (47.7) | – | – |
| BCG induction (BCG i + m) | 86 (52.3) | – | |
| Event | |||
| Recurrence | 74 (43.9) | – | – |
| Non-recurrence | 95 (56.1) | – | |
BCG i + m, Bacillus calmette-guerin induction + maintenance
Statistically significant values are shown in bold
aThe sum could not add up to the total due to some missing values
#Student t-test was used to determine the p value
Clinical Data Collection
The clinical information about tumor stage and grade, intravesical therapy and dates of recurrence, radical cystectomy and pathological findings at cystectomy were provided by the urologists in our department. The classification tumor stages were as per the American Joint Committee on Cancer’s TNM staging system [20]. Of the 240 total patients enrolled in the study, 180 patients had non muscle invasive bladder cancer (NMIBC) while the rest 60 had muscle invasive bladder cancer (MIBC). Patients with NMIBC at high risk (high grade, multiple and large tumor) were treated with intravesical Bacillus Calmette-Guerin (BCG) (n = 94). The patients with NMI cancer of low risk (low grade and single small tumor) were kept on cystoscopic surveillance and considered as non-BCG patients. Subsequently, all the patients were examined by cystoscopy after every 3 months in first and second years and later at 6 monthly intervals as long as there was no tumor recurrence. BCG treatment consisted of 6 weekly instillation induction BCG (n = 94). Since the number of patients receiving maintenance BCG was too low, we did not categorize the patients according to BCG regime for statistical analysis. The end point of study included tumor recurrence, defined as a newly found bladder tumor following a previous negative follow-up cystoscopy, or end of study time (60 months). Patients with invasive BC (n = 60) were treated with radical cystectomy with or without adjuvant chemotherapy, which included cisplatin, gemcitabine followed by periodical cystoscopy.
Candidate SNPs
SNP selection was based on previous studies on association between CD166 gene and cancer in different populations [4–7, 9–13] as well as on the basis of functional properties of the gene. The functional polymorphisms within the CD166 gene were selected by using the HapMap Project database (www.hapmap.org) based on the GIH population data of hapmap. We used certain criteria for the candidate gene polymorphisms viz., a minor allele frequency (MAF) greater than 10 % in Caucasian population; situated in the 3′UTR, 5′UTR, intronic and exonic regions of the tested genes which shows some biological significance according to the location within the gene. The LD Plot with SNPs is furnished in Fig. 1.
Fig. 1.
Linkage disequilibrium (LD) plot of CD166 gene in Hapmap-GIH population
Three SNPs of CD166 viz. CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G were selected for the presented study. rs6437585 is present in 5′-UTR of CD166 gene and is found to be potentially functional in tumor progression. rs10511244 is present in intronic region of CD166 gene and it also contributes in cancer progression. Third candidate SNP rs1157 is present in 3′-UTR and is also associated with enhancement of cancer.
Genotyping
Genomic DNA was extracted from venous blood by following standard salting out method [21]. The isolated DNA was qualified and quantified by using Nanodrop spectrophotometer (Thermo Fisher Scientific/Nanodrop Products, Wilmington, Delaware, USA). Genotyping of CD166 rs6437585 C/T and CD166 rs10511244 C/T was done by using Taqman allelic discrimination assay. For the assay primers and probes were provided as predesigned assays by Applied Biosystems (Foster City, CA). Genotyping was performed with ABI 7500HT Fast Sequence Detection System (Applied Biosystems, Foster City, CA) using 96-well plates. Positive and negative controls were used in each genotyping assay plate, and 10 % of the samples were randomly selected and run in duplicates with 100 % concordance. The results were reproducible with no discrepancy in genotyping. CD166 rs1157 A/G was genotyped by PCR-based restriction fragment length polymorphism (PCR–RFLP) analysis. The primer sequence used for CD166rs1157 A/G were adopted from a previous study [18]. Genotyping was done on 10 % Poly-Acrylamide Gel using molecular weight markers and visualized after staining with ethidium bromide. Positive and negative controls were used in each genotyping assay, and 10 % of the samples were randomly selected and run in duplicates with 100 % concordance. The results were reproducible with no discrepancy in genotyping. About 5 % of the randomly selected samples were validated by sequencing.
Statistical Analysis
Hardy–Weinberg equilibrium (HWE) test of SNP was performed using Michael H. Court’s (2005–2008) online calculator (http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls). Tests in bladder cancer patients and healthy unrelated controls did not show any significant deviation from HWE for any of the SNPs.
The power of the study was calculated using Quanto software, version 1.0 (available from: http://hydra.usc.edu/gxe). The present study achieved 80 % of the statistical power. The goodness-of-fit Chi square test was used to analyze any deviation from the Hardy–Weinberg equilibrium in controls. A binary logistic regression model was used to estimate the risk as the OR at the 95 % confidence interval. The statistical analysis was done using the Statistical Package for Social Sciences software, version 16.0 (SPSS, Chicago, IL), and p < 0.05 was considered statistically significant. Haplotypic analysis was done by using SNP analyzer version 1.2A.
Results
Demographic Characteristics of Study Subject
Of the 510 samples (patients and controls), there was no significant difference between the patients and controls in age (p = 0.138), sex (p = 0.105). The cases had significantly higher percentage of smokers (70.4 %) than the controls (20.7 %) (p < 0.001). The demographic details of the study subjects and clinical characteristics of the patients are described in Table 1.
Frequency Distribution of CD166 Gene Polymorphism (CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G)
The genotypic distributions of CD166 gene polymorphisms in controls were in Hardy–Weinberg equilibrium. The genotypic and allelic frequencies of CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G gene polymorphism and their association with BC risk is demonstrated in Table 2.
Table 2.
Frequency distribution of CD166 gene variants CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G among bladder cancer patients and controls
| Genetic model | Genotypes | Controls n = 270 n (%) |
Patients n = 240 n (%) |
p value | ORa (95 % CI) |
|---|---|---|---|---|---|
| CD166rs6437585 C/T | |||||
| Additive | CC | 165 (61.1) | 174 (72.5) | Ref | Ref |
| CT | 81 (30.0) | 61 (25.4) | 0.095 | 0.714 (0.481–1.060) | |
| TT | 24 (8.9) | 5 (2.1) | 0.001 | 0.198 (0.074–0.530) | |
| Dominant | CC | 165 (61.1) | 174 (72.5) | Ref | Ref |
| CT + TT | 105 (38.9) | 66 (27.5) | 0.007 | 0.596 (0.410–0.867) | |
| Multiple | C | 411 (76.1) | 409 (85.2) | Ref | Ref |
| T | 129 (23.9) | 71 (14.8) | <0.001 | 0.553 (0.401–0.762) | |
| CD166rs10511244 C/T | |||||
| Additive | CC | 129 (47.8) | 138 (57.5) | Ref | Ref |
| CT | 109 (40.4) | 86 (35.8) | 0.108 | 0.738 (0.509–1.069) | |
| TT | 32 (11.9) | 16 (6.7) | 0.021 | 0.467 (0.245–0.892) | |
| Dominant | CC | 129 (47.8) | 138 (57.5) | Ref | Ref |
| CT + TT | 141 (52.2) | 102 (42.5) | 0.028 | 0.676 (0.477–0.960) | |
| Multiple | C | 367 (68.0) | 362 (75.4) | Ref | Ref |
| T | 173 (32.0) | 118 (24.6) | 0.009 | 0.692 (0.525–0.911) | |
| CD166rs1157 A/G | |||||
| Additive | AA | 132 (48.9) | 91 (37.9) | Ref | Ref |
| AG | 109 (40.4) | 114 (47.5) | 0.029 | 1.517 (1.043–2.206) | |
| GG | 29 (10.7) | 35 (14.6) | 0.050 | 1.751 (1.000–3.065) | |
| Dominant | AA | 132 (48.9) | 91 (37.9) | Ref | Ref |
| AG + GG | 138 (51.1) | 149 (62.1) | 0.013 | 1.566 (1.100–2.230) | |
| Multiple | A | 373 (69.0) | 296 (61.7) | Ref | Ref |
| G | 167 (30.9) | 184 (38.3) | 0.013 | 1.388 (1.071–1.799) | |
Statistically significant values are shown in bold
aOR (95 % CI) age, gender adjusted odds ratio and 95 % confidence interval
A significant association with reduced risk was found in additive model (TT; p = 0.001: Adjusted OR = 0.198, 95 % CI = 0.074–0.530), dominant model (CT + TT; p = 0.007: Adjusted OR = 0.596, 95 % CI = 0.410–0.867) and allelic model (T allele; p < 0.001: Adjusted OR = 0.553: 95 % CI = 0.401–0.762) of CD166rs6437585 C/T. Similar kind of results were seen in CD166rs10511244 C/T, at genotypic level in additive (TT; p = 0.021) as well as dominant model (CT + TT; p = 0.028), at allelic level, the variant allele, T showed reduced risk for BC (p = 0.009). Whereas, in CD166rs1157 A/G we found significant association with risk in heterozygous genotype, AG (p = 0.029, Adjusted OR = 1.517, 95 % CI = 1.043–2.206) and a marginal association in variant genotype, GG of the additive model (p = 0.050, Adjusted OR = 1.751, 95 % CI = 1.000–3.065). In the dominant model of CD166rs1157 A/G, AG + GG, significant risk for BC was seen (p = 0.031, Adjusted OR = 1.566, 95 % CI = 1.100–2.230). Also, at allelic level the variant allele, G showed reduced risk to BC (p = 0.013, Adjusted OR = 1.388, 95 % CI = 1.071–1.799) (Table 2).
Association of CD166 Gene Polymorphisms CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G at Genotypic Level with Smoking
We correlated CD166 gene polymorphism (CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G) with smoking habits among patients. For this we stratified patients in two groups: Smokers and non-smokers. This was analyzed by using Fischer’s exact test. No association was seen in any variants of CD166 gene with respect to smoking (Table 3).
Table 3.
Association of CD166 gene polymorphisms on the basis of smoking
| Genotype | Patients non smokers n = 48 n (%) |
Patients smoker n = 116 n (%) |
p value | ORa (95 % CI) |
|---|---|---|---|---|
| CD166rs6437585C/T | ||||
| CC | 33 (68.8) | 80 (69.0) | Ref | Ref |
| CT | 14 (29.2) | 33 (28.4) | 0.941 | 0.972 (0.462–1.048) |
| TT | 1 (2.1) | 3 (2.6) | 0.856 | 1.237 (0.124–1.833) |
| CD166rs10511244C/T | ||||
| CC | 25 (52.1) | 64 (55.2) | Ref | Ref |
| CT | 19 (39.6) | 46 (39.7) | 0.877 | 0.946 (0.467–1.217) |
| TT | 4 (8.3) | 6 (5.2) | 0.437 | 0.586 (0.152–0.953) |
| CD166rs1157A/G | ||||
| AA | 17 (35.4) | 41 (35.3) | Ref | Ref |
| AG | 28 (58.3) | 55 (47.4) | 0.579 | 0.814 (0.394–1.183) |
| GG | 3 (6.3) | 20 (17.2) | 0.137 | 2.764 (0.725–3.043) |
aOR (95 % CI) age, gender adjusted odds ratio and 95 % confidence interval
Association of CD166 Gene Variants CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G Genotypes with Tumor Stage/Grade of BC Patients
For this study the BC patients were stratified into three groups based on their tumor stage/grade [TaG1 (low risk NMIBC), TaG2–3 + T1G1–3 (High risk NMIBC) and T2+ (muscle invasive)]. TaG1 was taken as a reference. The patients with similar stage but with different grades respond to treatment differently. However, no association was found in any of CD166 gene variants CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G with any of the tumor stage/grade of BC patients (Table 4).
Table 4.
Association of CD166 gene variants CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G genotypes with tumor stage/grade of BC patients
| Genotypes | (a) n = 48 n (%) |
(b) n = 128 n (%) |
(c) n = 64 n (%) |
p value (a–b) | ORa (95 % CI) | p value (a–c) | ORa (95 % CI) | p value (b–c) | ORa (95 % CI) |
|---|---|---|---|---|---|---|---|---|---|
| CD166rs6437585C/T | TaG1 | TaG2-3,T1G1-3 | T2+ | ||||||
| CC | 38 (79.2) | 93 (72.7) | 43 (67.2) | Reference | Reference | Reference | |||
| CT | 10 (20.8) | 32 (25.0) | 19 (29.3) | 0.513 | 1.308 (0.585–1.922) | 0.249 | 1.679 (0.696–2.053) | 0.466 | 1.284 (0.655–2.517) |
| TT | NA | 3 (2.3) | 2 (3.1) | NA | NA | NA | NA | 0.694 | 1.442 (0.232–1.947) |
| CD166rs10511244C/T | TaG1 | TaG2-3,T1G1-3 | T2+ | ||||||
| CC | 29 (60.4) | 78 (60.9) | 31 (48.4) | Reference | Reference | Reference | |||
| CT | 16 (33.3) | 42 (32.8) | 28 (43.8) | 0.947 | 0.976 (0.477–1.198) | 0.225 | 1.635 (0.739–2.029) | 0.110 | 1.677 (0.890–2.162) |
| TT | 3 (6.3) | 8 (6.3) | 5 (7.8) | 0.990 | 0.991 (0.246–1.395) | 0.566 | 1.559 (0.342–2.116) | 0.457 | 1.573 (0.477–2.181) |
| CD166rs1157A/G | TaG1 | TaG2-3,T1G1-3 | T2+ | ||||||
| AA | 26 (54.2) | 41 (32.0) | 24 (37.5) | Reference | Reference | Reference | |||
| AG | 17 (35.4) | 66 (51.6) | 31 (48.4) | 0.081 | 2.462 (1.192–3.083) | 0.100 | 1.975 (0.878–2.445) | 0.513 | 0.802 (0.415–1.253) |
| GG | 5 (10.4) | 21 (16.4) | 9 (14.1) | 0.079 | 2.663 (0.893–3.438) | 0.286 | 1.950 (0.572–2.444) | 0.511 | 0.732 (0.289–1.354) |
aOR (95 % CI) age, gender adjusted odds ratio and 95 % confidence interval
Association of CD166 Gene Variants CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G Haplotypes with Bladder Cancer Risk
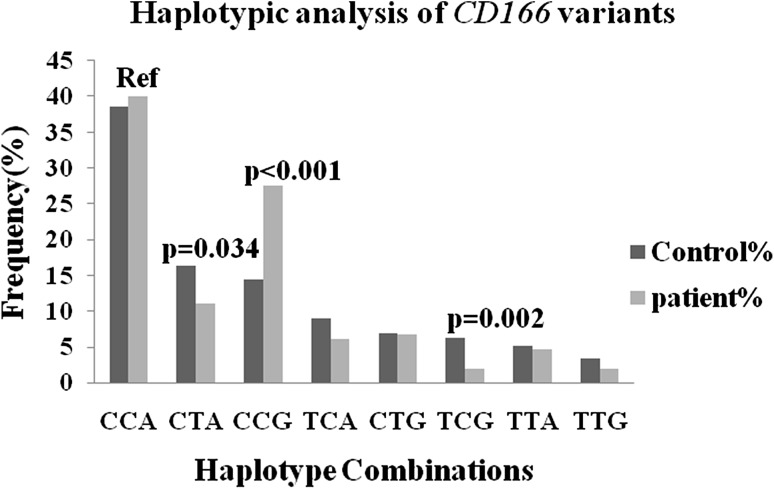
Haplotypic analysis could be more manifesting in predicting risk and finding the association of disease as compared to single nucleotide polymorphism analysis. Keeping this in mind, we examined the effects of CD166 gene variants by constructing haplotype sets, taking CCA as a reference as these three alleles are wild alleles from all three candidate SNPs.
Significant association was seen in three out of eight haplotypic combinations among these three sets one set i.e. CCG revealed significant association with about two fold risk for BC (CTA p = 0.034, OR = 0.653, 95 % CI = 0.440–0.967; CCG p < 0.001, OR = 1.844, 95 % CI = 1.308–2.599; and TCG p = 0.002, OR = 0.296, 95 % CI = 0.138–0.634), after applying Bonferroni correction (CCG pc = 0.008 and TCG pc = 0.016) (Fig. 2).
Fig. 2.
Haplotype analysis of CD166 gene variants
Modulation of CD166 Gene Variants, CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G Genotypes and Outcome After BCG Immunotherapy
To analyze the association of CD166rs6437585 C/T, CD166rs10511244 C/T, and CD166rs1157 A/G gene variants and the risk of recurrence in NMIBC patients, further analysis was trammeled only to NMIBC patients (n = 180). The median follow-up of NMIBC patients was 14 months. We analyzed the association of genotypes and risk of recurrence after BCG immunotherapy. We grouped patients into BCG treated (n = 94) and non-treated (n = 86) as these were patients of low grade tumors and did not require BCG immunotherapy. None of the polymorphisms were associated with risk of recurrence free survival (Data not shown).
Discussion
Adhesion molecules play an important role in the behavior of both malignant and benign cells. These molecules are thought to be involved in tumor growth and metastases, and are therefore important to prognosis. As a result, they may be targets for novel treatment modalities [22].
In this hospital based case–control study we found significant low risk to BC in case of CD166rs6437585 C/T polymorphism in the CD166 promoter. Also a reduced risk was seen in case of CD166rs10511244 C/T polymorphism of CD166 gene. However, we worked out a third variant of CD166 gene, CD166rs1157 A/G, and found significantly high risk at genotypic as well as allelic level.
CD166 is a member of the cell surface immunoglobulin super family which is involved in cell-to-cell interactions. In a previous study, which used dot blot analysis, it was found that CD166 transcript levels were higher than normal in the tissues of five out of eight breast cancers [23]. IHC based expression on human melanoma cells revealed CD166 expression with correlation to melanoma progression [24]. CD166 expression was observed in 68 % of esophageal squamous cell carcinoma at RNA and protein level which indicates that CD166 correlates with tumor invasion and metastasis [25]. Another IHC based expression study revealed strong association of CD166 with prognosis in high grade cervical cancer [26]. CD166 was shown to be upregulated in 86 % of prostate cancer patients, suggesting its role as a good prognostic as well as diagnostic marker for prostatic cancer [27].
Our study suggested significant association of CD166rs6437585 C/T and T/T of CD166 gene. CD166rs6437585 C/T hetero and variant genotype were found to have OR = 1.38 for developing bladder cancer as compared to the wild genotype. Expression studies showed that the variant allele i.e. T allele was associated with high transcriptional activity of CD166 gene in breast cancer [15]. Whereas Jiang et al. showed CD166rs6437585 C/T to be associated with increased risk of breast cancer in Chinese population. They also checked CD166rs6437585 C/T in various in vitro assays and found its variant allele to be involved in enhanced transcription rates of CD166 gene. Their study also suggested involvement of CD166rs6437585 T, variant allele in tumor progression.
CD166rs10511244 C/T, in our study showed reduced risk for bladder cancer. Although the association and risk factors vary from ethnicity to ethnicity as well as in case of different diseases. The third SNP of CD166 gene CD166rs1157 A/G, demonstrated significant high risk for bladder cancer in the present study. Supporting our study, rs1157 had shown a hazard ratio of 3.42 in Swedish population for breast cancer [17]. Being located in the 3′UTR region of CD166 gene and having a functional role in miRNA binding [17] any aberration in rs1157 can be said to be involved in a discrepancy in normal functioning of the cell, which may lead to cancer. CD166 is a ligand for CD6, it is a cell surface scavenger receptor involved in T cell activation and proliferation, as well as in thymocyte differentiation. Chappell and group reported the binding sites on CD6 and CD166 and showed that a SNP in CD6 causes glycosylation that hinders the CD6/CD166 interaction and also suggested that how the interactions of consecutive domains can be perturbed by SNPS in ligands as well as receptors [28].
In our study, we showed that genetic variant in CD166 gene may be of significance for the prognosis of bladder cancer. Further analyses with explanation at functional consequences of these variants on mRNA and protein expression are warranted with more number of cases and in wide ethnicity variation.
Conclusion
Our study suggests that CD166rs6437585 C/T and CD166rs10511244 C/T are predictive for the reduced risk of bladder cancer, whereas CD166rs1157 A/G had shown significant association with high risk of bladder cancer in North Indians. This somehow suggests that CD166rs1157 A/G can be used as a marker for risk prediction of bladder cancer although some more studies with larger sample size, varied ethnicity and with more advanced techniques are suggested in support of this study. Our result suggests the importance of testing large sample sizes and of performing expression studies to validate genetic associations of CD166 in bladder cancer.
To the best of our knowledge, present study is the first to report a group of three SNPs of CD166 gene variants with bladder cancer risk in North Indian population.
Acknowledgments
This study was funded by Department of Science and Technology (DST) [SR/SO/HS-120/2007], New Delhi. The assistance of relevant clinical information of the patients by the Urologists and Pathologists are duly acknowledged.
Compliance with Ethical Standards
Conflict of interest
Authors have no conflicts of interest in this work.
References
- 1.American Cancer Society . Cancer facts and figures 2015. Atlanta: American Cancer Society; 2015. [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon: International Agency for Research on Cancer; 2013. http://globocan.iarc.fr. Accessed Dec 2013.
- 3.Murthy NS, Nandakumar BS, Pruthvish S, George PS, Mathew A. Disability adjusted life years for cancer patients in India. Asian Pac J Cancer Prev. 2010;11(3):633–640. [PubMed] [Google Scholar]
- 4.Kitamura H, Okudela K, Yazawa T, Sato H, Shimoyamada H. Cancer stem cell: implications in cancer biology and therapy with special reference to lung cancer. Lung Cancer. 2009;66(3):275–281. doi: 10.1016/j.lungcan.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Eramo A, Haas TL, De Maria R. Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene. 2010;29(33):4625–4635. doi: 10.1038/onc.2010.207. [DOI] [PubMed] [Google Scholar]
- 6.Ni C, Zhang Z, Zhu X, Liu Y, Qu D, Wu P, et al. Prognostic value of CD166 expression in cancers of the digestive system: a systematic review and meta-analysis. PLoS ONE. 2013;8(8):e70958. doi: 10.1371/journal.pone.0070958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105(36):13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, Swain JR, Wong MH, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139(6):2072–2082. doi: 10.1053/j.gastro.2010.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, Lawson D, et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS ONE. 2012;7(8):e42564. doi: 10.1371/journal.pone.0042564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Tachezy M, Zander H, Wolters-Eisfeld G, Müller J, Wicklein D, Gebauer F, et al. Activated leukocyte cell adhesion molecule (CD166): an “inert” cancer stem cell marker for non-small cell lung cancer? Stem Cells. 2014;32(6):1429–1436. doi: 10.1002/stem.1665. [DOI] [PubMed] [Google Scholar]
- 13.Clauditz TS, von Rheinbaben K, Lebok P, Minner S, Tachezy M, Borgmann K, et al. Activated leukocyte cell adhesion molecule (ALCAM/CD166) expression in head and neck squamous cell carcinoma (HNSSC) Pathol Res Pract. 2014;2010(10):649–655. doi: 10.1016/j.prp.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 15.Zhou P, Du LF, Lv GQ, Yu XM, Gu YL, Li JP, et al. Functional polymorphisms in CD166/ALCAM gene associated with increased risk for breast cancer in a Chinese population. Breast Cancer Res Treat. 2011;128(2):527–534. doi: 10.1007/s10549-011-1365-x. [DOI] [PubMed] [Google Scholar]
- 16.Yadav A, Gupta A, Rastogi N, Agrawal S, Kumar A, Kumar V, Mittal B. Association of cancer stem cell markers genetic variants with gallbladder cancer susceptibility, prognosis, and survival. Tumour Biol. 2016;37(2):1835–1844. doi: 10.1007/s13277-015-3929-6. [DOI] [PubMed] [Google Scholar]
- 17.Varadi V, Bevier M, Grzybowska E, Johansson R, Enquist-Olsson K, Henriksson R, et al. Genetic variation in ALCAM and other chromosomal instability genes in breast cancer survival. Breast Cancer Res Treat. 2012;131(1):311–319. doi: 10.1007/s10549-011-1765-y. [DOI] [PubMed] [Google Scholar]
- 18.Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, Winder T, et al. Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res. 2011;17(21):6934–6943. doi: 10.1158/1078-0432.CCR-11-1180. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K, Civan J, Mukherjee S, Patel F, Yang H. Genetic variations in colorectal cancer risk and clinical outcome. World J Gastroenterol. 2014;20(15):4167–4177. doi: 10.3748/wjg.v20.i15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombel M, Soloway M, Akaza H, Böhle A, Palou J, Buckley R, Lamm D, Brausi M, Witjes JA, Persad R. Epidemiology, staging, grading and risk stratification of bladder cancer. Eur Urol Suppl. 2008;7(10):618–626. doi: 10.1016/j.eursup.2008.08.002. [DOI] [Google Scholar]
- 21.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang WG, Puntis MC, Hallett MB. Molecular and cellular basis of cancer invasion and metastasis: implications for treatment. Br J Surg. 1994;81(11):1576–1590. doi: 10.1002/bjs.1800811107. [DOI] [PubMed] [Google Scholar]
- 23.Kristiansen G, Pilarsky C, Wissmann C, Stephan C, Weissbach L, Loy V, et al. ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate. 2003;54(1):34–43. doi: 10.1002/pros.10161. [DOI] [PubMed] [Google Scholar]
- 24.Van Kempen LC, Meier F, Egeblad M, Kersten-Niessen MJ, Garbe C, Weidle UH, et al. Truncation of activated leukocyte cell adhesion molecule: a gateway to melanoma metastasis. J Investig Dermatol. 2004;122(5):1293–1301. doi: 10.1111/j.0022-202X.2004.22531.x. [DOI] [PubMed] [Google Scholar]
- 25.Verma A, Shukla NK, Deo SV, Gupta SD, Ralhan R. MEMD/ALCAM: a potential marker for tumor invasion and nodal metastasis in esophageal squamous cell carcinoma. Oncology. 2005;68(4–6):462–470. doi: 10.1159/000086989. [DOI] [PubMed] [Google Scholar]
- 26.Ihnen M, Kress K, Kersten JF, Kilic E, Choschzick M, Zander H, et al. Relevance of activated leukocyte cell adhesion molecule (ALCAM) in tumor tissue and sera of cervical cancer patients. BMC Cancer. 2012;4(12):140. doi: 10.1186/1471-2407-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weichert W, Knösel T, Bellach J, Dietel M, Kristiansen G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol. 2004;57(11):1160–1164. doi: 10.1136/jcp.2004.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chappell PE, Garner LI, Yan J, Metcalfe C, Hatherley D, Johnson S, Robinson V, Lea SM, Brown MH. Structures of CD6 and its ligand CD166 give insight into their interaction. Structure. 2015;23(8):1426–1436. doi: 10.1016/j.str.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]