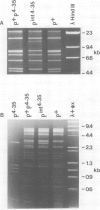
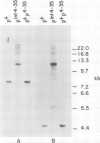
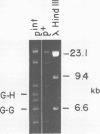
Abstract
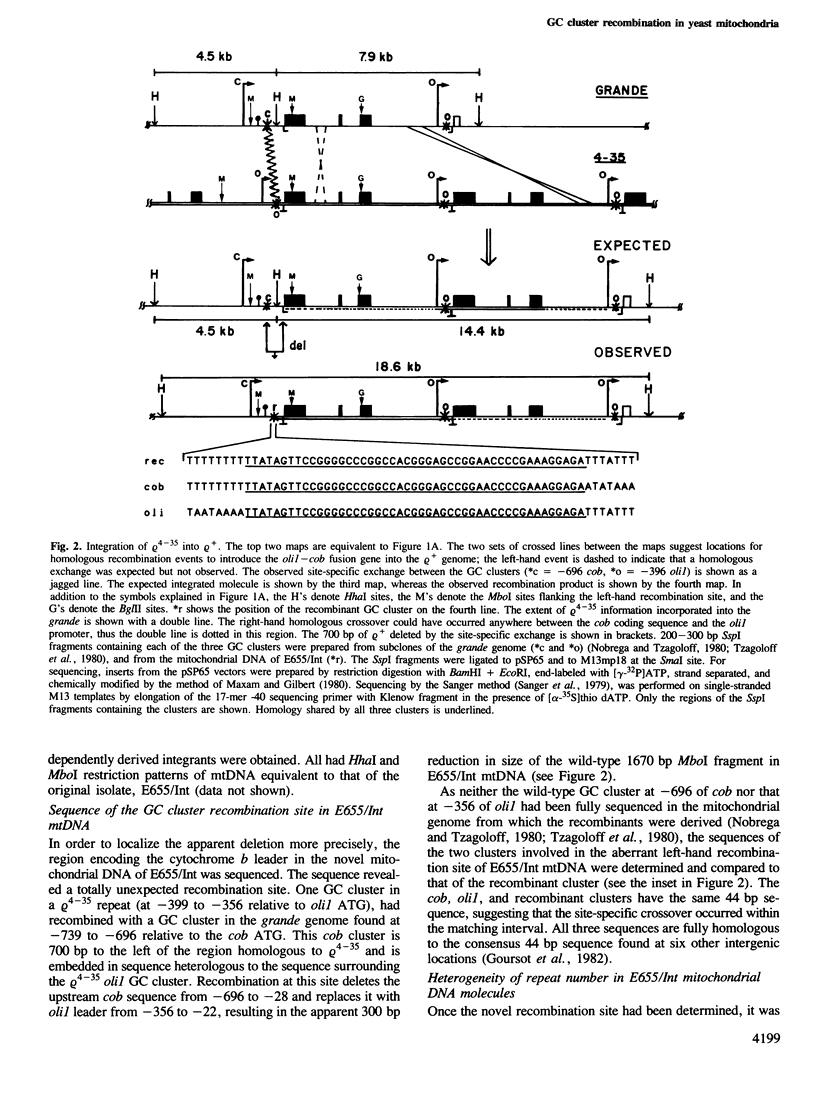
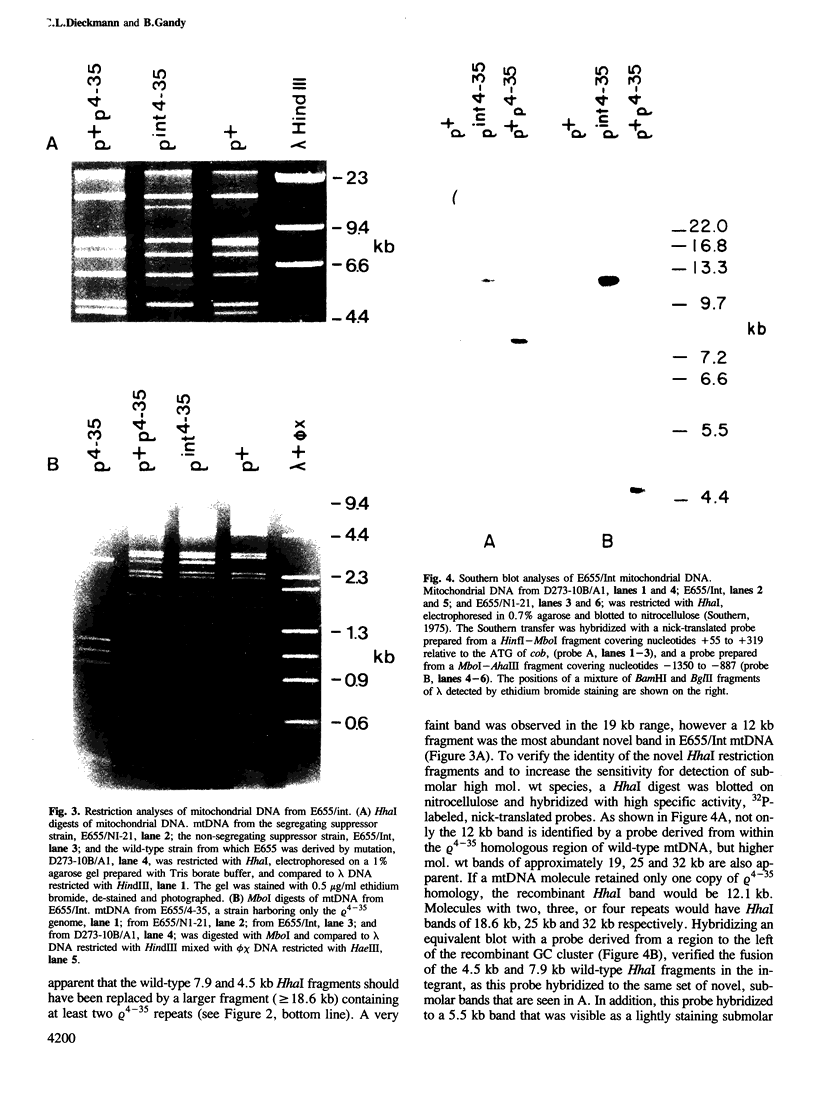
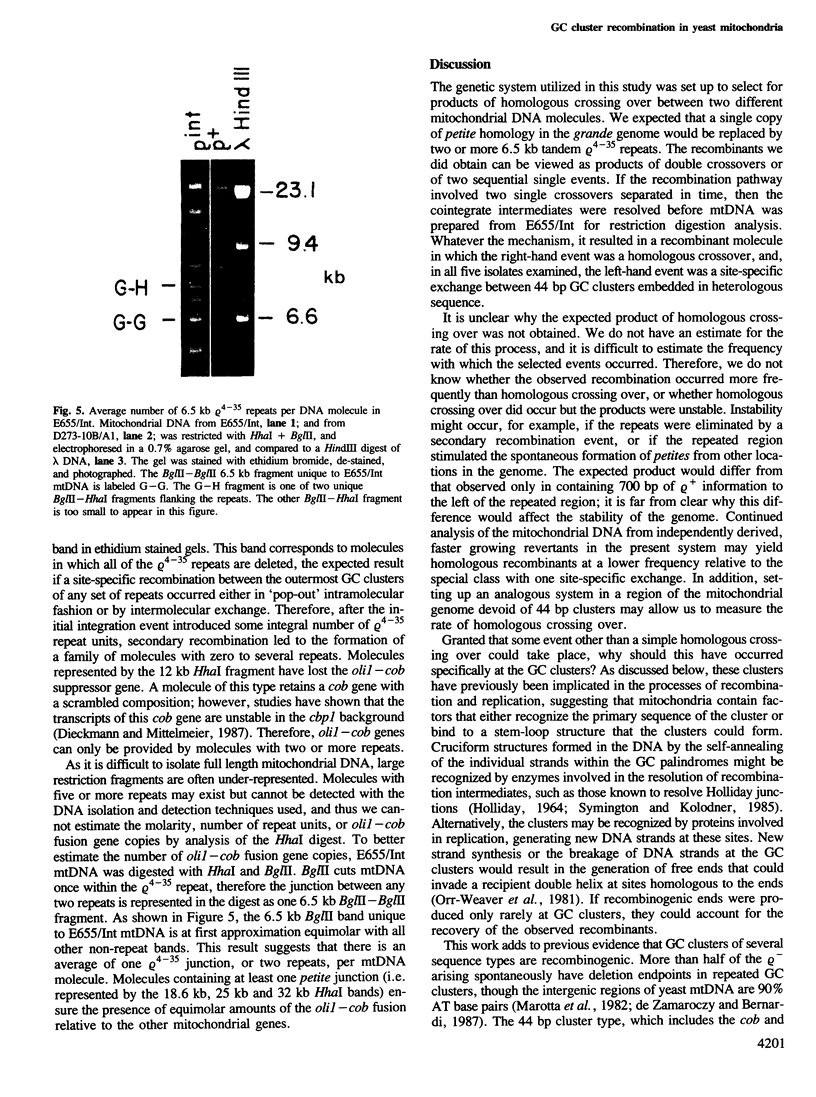
Yeast mitochondrial DNA molecules have long, AT-rich intergenic spacers punctuated by short GC clusters. GC-rich elements have previously been characterized by others as preferred sites for intramolecular recombination leading to the formation of subgenomic petite molecules. In the present study we show that GC clusters are favored sites for intermolecular recombination between a petite and the wild-type grande genome. The petite studied retains 6.5 kb of mitochondrial DNA reiterated tandemly to form molecules consisting of repeated units. Genetic selection for integration of tandem 6.5 kb repeats of the petite into the grande genome yielded a novel recombination event. One of two crossovers in a double exchange event occurred as expected in the 6.5 kb of matching sequence between the genomes, whereas the second exchange involved a 44 bp GC cluster in the petite and another 44 bp GC cluster in the grande genome 700 bp proximal to the region of homology. Creation of a mitochondrial DNA molecule with a repetitive region led to secondary recombination events that generated a family of molecules with zero to several petite units. The finding that 44 bp GC clusters are preferred as sites for intermolecular exchange adds to the data on petite excision implicating these elements as recombinational hotspots in the yeast mitochondrial genome.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldacci G., Bernardi G. Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J. 1982;1(8):987–994. doi: 10.1002/j.1460-2075.1982.tb01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacci G., Chérif-Zahar B., Bernardi G. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 1984 Sep;3(9):2115–2120. doi: 10.1002/j.1460-2075.1984.tb02099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Physical map of the Oxi3 locus of yeast mitochondrial DNA. J Biol Chem. 1980 Dec 25;255(24):11922–11926. [PubMed] [Google Scholar]
- Dieckmann C. L., Koerner T. J., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4722–4731. [PubMed] [Google Scholar]
- Dieckmann C. L., Pape L. K., Tzagoloff A. Identification and cloning of a yeast nuclear gene (CBP1) involved in expression of mitochondrial cytochrome b. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1805–1809. doi: 10.1073/pnas.79.6.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Dujon B. Yeast mitochondrial genomes consisting of only A.T base pairs replicate and exhibit suppressiveness. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7156–7160. doi: 10.1073/pnas.81.22.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goursot R., Mangin M., Bernardi G. Surrogate origins of replication in the mitochondrial genomes of ori-zero petite mutants of yeast. EMBO J. 1982;1(6):705–711. doi: 10.1002/j.1460-2075.1982.tb01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker J., Rabinowitz M., Getz G. S. Tandem inverted repeats in mitochondrial DNA of petite mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1366–1370. doi: 10.1073/pnas.71.4.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta R., Colin Y., Goursot R., Bernardi G. A region of extreme instability in the mitochondrial genome of yeast. EMBO J. 1982;1(5):529–534. doi: 10.1002/j.1460-2075.1982.tb01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Morimoto R., Rabinowitz M. Physical mapping of the yeast mitochondrial genome: derivation of the fine structure and gene map of strain D273-10B and comparison with a strain (MH41-7B) differing in genome size. Mol Gen Genet. 1979 Feb 16;170(1):25–48. [PubMed] [Google Scholar]
- Müller P. P., Reif M. K., Zonghou S., Sengstag C., Mason T. L., Fox T. D. A nuclear mutation that post-transcriptionally blocks accumulation of a yeast mitochondrial gene product can be suppressed by a mitochondrial gene rearrangement. J Mol Biol. 1984 Jun 5;175(4):431–452. doi: 10.1016/0022-2836(84)90178-5. [DOI] [PubMed] [Google Scholar]
- Nobrega F. G., Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence and organization of the cytochrome b gene in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1980 Oct 25;255(20):9828–9837. [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Strausberg R. L., Vincent R. D., Perlman P. S., Butow R. A. Asymmetric gene conversion at inserted segments on yeast mitochondrial DNA. Nature. 1978 Dec 7;276(5688):577–583. doi: 10.1038/276577a0. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Foury F. Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett. 1976 Jun 15;65(3):391–395. doi: 10.1016/0014-5793(76)80154-8. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Zassenhaus H. P., Butow R. A. Expression of GC clusters in the yeast mitochondrial var 1 gene. Transcription into stable RNAs. J Biol Chem. 1984 Jul 10;259(13):8417–8421. [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. Sequence organization of the mitochondrial genome of yeast--a review. Gene. 1985;37(1-3):1–17. doi: 10.1016/0378-1119(85)90252-5. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The AT spacers and the var1 genes from the mitochondrial genomes of Saccharomyces cerevisiae and Torulopsis glabrata: evolutionary origin and mechanism of formation. Gene. 1987;54(1):1–22. doi: 10.1016/0378-1119(87)90342-8. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene. 1986;41(1):1–22. doi: 10.1016/0378-1119(86)90262-3. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The primary structure of the mitochondrial genome of Saccharomyces cerevisiae--a review. Gene. 1986;47(2-3):155–177. doi: 10.1016/0378-1119(86)90060-0. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty G., Baldacci G., Goursot R., Bernardi G. The ori sequences of the mitochondrial genome of a wild-type yeast strain: number, location, orientation and structure. Gene. 1984 Dec;32(3):439–457. doi: 10.1016/0378-1119(84)90019-2. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty G., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Gene. 1983 Mar;21(3):193–202. doi: 10.1016/0378-1119(83)90002-1. [DOI] [PubMed] [Google Scholar]