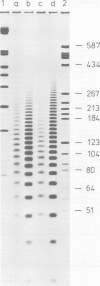
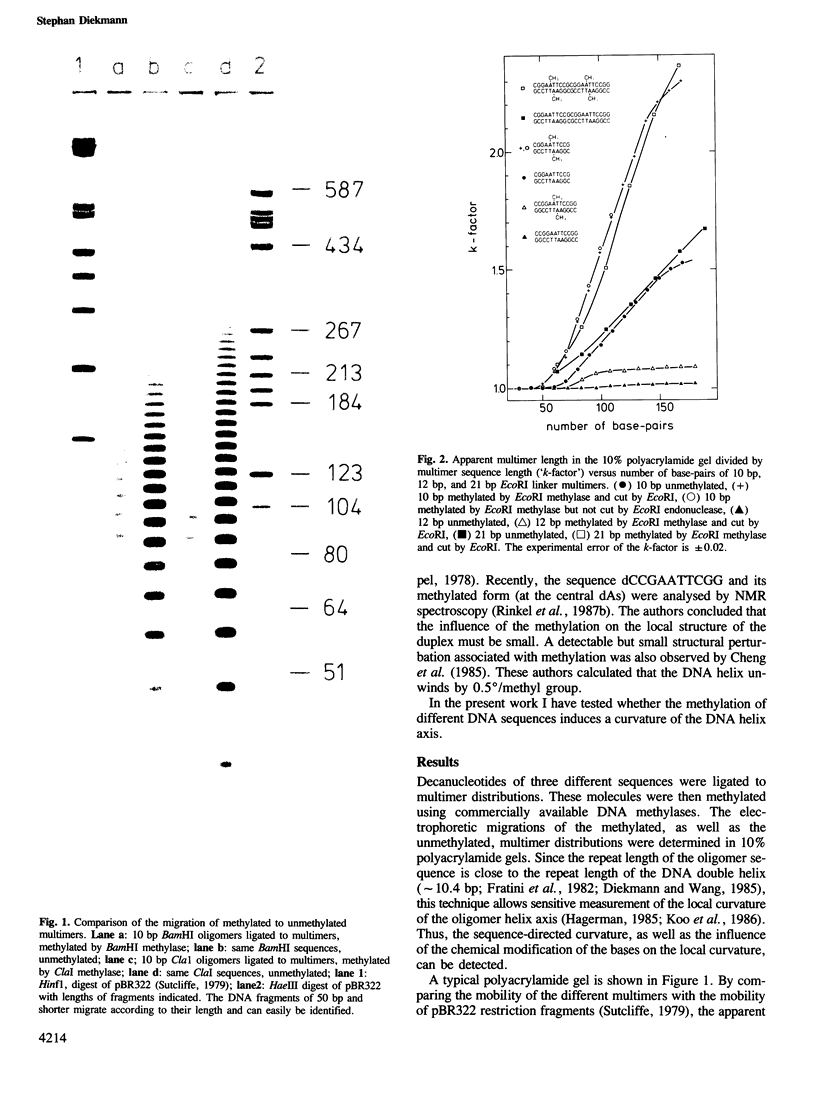
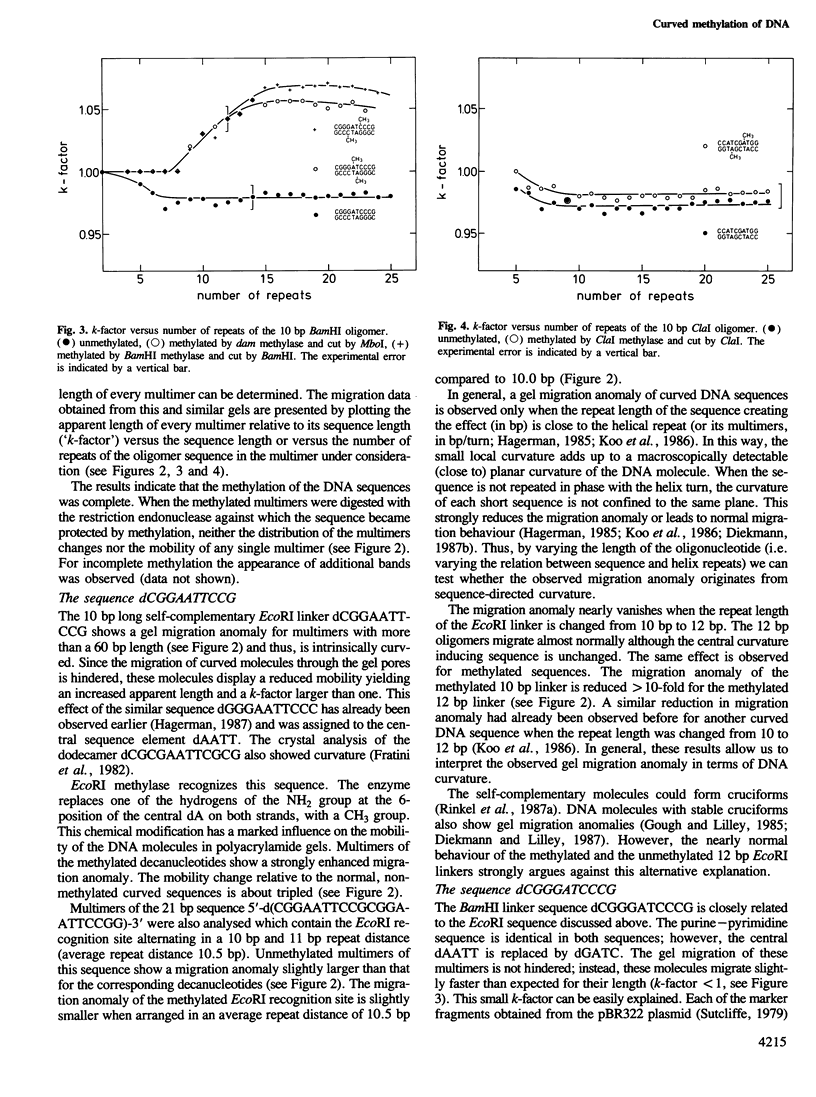
Abstract
Oligomers of different palindromic sequences (EcoRI, BamHI, and ClaI linkers) were ligated to form distributions of multimers. These long ligation ladders were methylated using corresponding methylases. The migration of the unmethylated as well as the methylated multimer distributions was analysed in 10% polyacrylamide gels. The migration anomaly of these sequences is interpreted in terms of the curvature of the DNA helix axis. The double-stranded oligomer dCGGAATTCCG is considerably curved in its unmodified form. Its curvature is strongly enhanced when the central dAs are methylated. This result is predicted by a model for DNA curvature. Multimers of the closely related sequence dCGGGATCCCG are straight. When methylated at the central dAs or at the most central dCs, a small curvature of the helix axis is induced. The double-stranded oligomer dCCATCGATGG is straight in its unmodified form as well as when it is methylated. Thus, DNA curvature can be induced or enhanced by methylation. However, DNA methylation at palindromic sequences seems not always to influence the linear path of the DNA helix axis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexeev D. G., Lipanov A. A., Skuratovskii IYa Poly(dA).poly(dT) is a B-type double helix with a distinctively narrow minor groove. 1987 Feb 26-Mar 4Nature. 325(6107):821–823. doi: 10.1038/325821a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Herman G., Modrich P. Extent of equilibrium perturbation of the DNA helix upon enzymatic methylation of adenine residues. J Biol Chem. 1985 Jan 10;260(1):191–194. [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., Lilley D. M. The anomalous gel migration of a stable cruciform: temperature and salt dependence, and some comparisons with curved DNA. Nucleic Acids Res. 1987 Jul 24;15(14):5765–5774. doi: 10.1093/nar/15.14.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S. Sequence specificity of curved DNA. FEBS Lett. 1986 Jan 20;195(1-2):53–56. doi: 10.1016/0014-5793(86)80128-4. [DOI] [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Zarling D. A. Unique poly(dA).poly(dT) B'-conformation in cellular and synthetic DNAs. Nucleic Acids Res. 1987 Aug 11;15(15):6063–6074. doi: 10.1093/nar/15.15.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson S. P., Johnson W. C., Jr Base tilt of poly[d(A)]-poly[d(T)] and poly[d(AT)]-poly[d(AT)] in solution determined by linear dichroism. Biopolymers. 1985 May;24(5):825–841. doi: 10.1002/bip.360240508. [DOI] [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J Biol Chem. 1978 Feb 10;253(3):927–934. [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Griffith J., Bleyman M., Rauch C. A., Kitchin P. A., Englund P. T. Visualization of the bent helix in kinetoplast DNA by electron microscopy. Cell. 1986 Aug 29;46(5):717–724. doi: 10.1016/0092-8674(86)90347-8. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry. 1985 Dec 3;24(25):7033–7037. doi: 10.1021/bi00346a001. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Nature. 1986 May 22;321(6068):449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- Hattman S. DNA methyltransferase-dependent transcription of the phage Mu mom gene. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5518–5521. doi: 10.1073/pnas.79.18.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P., Squali-Houssaini F., Forterre P., Kohiyama M. In vitro replication of a dam methylated and non-methylated ori-C plasmid. J Mol Biol. 1984 Jun 15;176(1):155–159. doi: 10.1016/0022-2836(84)90386-3. [DOI] [PubMed] [Google Scholar]
- Jernigan R. L., Sarai A., Shapiro B., Nussinov R. Relationship between curved DNA conformations and slow gel migration. J Biomol Struct Dyn. 1987 Feb;4(4):561–567. doi: 10.1080/07391102.1987.10507660. [DOI] [PubMed] [Google Scholar]
- Jernigan R. L., Sarai A., Ting K. L., Nussinov R. Hydrophobic interactions in the major groove can influence DNA local structure. J Biomol Struct Dyn. 1986 Aug;4(1):41–48. doi: 10.1080/07391102.1986.10507645. [DOI] [PubMed] [Google Scholar]
- Jollès B., Laigle A., Chinsky L., Turpin P. Y. The poly dA strand of poly dA.poly dT adopts an A-form in solution: a UV resonance Raman study. Nucleic Acids Res. 1985 Mar 25;13(6):2075–2085. doi: 10.1093/nar/13.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J., Stirdivant S. M., Singleton C. K., Zacharias W., Wells R. D. Effects of 5 cytosine methylation on the B-Z transition in DNA restriction fragments and recombinant plasmids. J Mol Biol. 1983 Jul 25;168(1):51–71. doi: 10.1016/s0022-2836(83)80322-2. [DOI] [PubMed] [Google Scholar]
- Knebel D., Doerfler W. N6-methyldeoxyadenosine residues at specific sites decrease the activity of the E1A promoter of adenovirus type 12 DNA. J Mol Biol. 1986 May 20;189(2):371–375. doi: 10.1016/0022-2836(86)90518-8. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner K. D., Vardimon L., Renz D., Doerfler W. DNA methylation of three 5' C-C-G-G 3' sites in the promoter and 5' region inactivate the E2a gene of adenovirus type 2. Proc Natl Acad Sci U S A. 1984 May;81(10):2950–2954. doi: 10.1073/pnas.81.10.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Ott J., Eckstein F. Filter disc supported oligonucleotide synthesis by the phosphite method. Nucleic Acids Res. 1984 Dec 11;12(23):9137–9142. doi: 10.1093/nar/12.23.9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vrieling H., Van de Putte P. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-coded transactivator. Nature. 1983 Jan 27;301(5898):344–347. doi: 10.1038/301344a0. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Reik W., Collick A., Norris M. L., Barton S. C., Surani M. A. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987 Jul 16;328(6127):248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of N6-methylation of residue A(5) on the conformational behaviour of d(C-C-G-A-A-T-T-C-G-G) in solution studied by 1H-NMR spectroscopy. 1. The duplex form. Eur J Biochem. 1987 Mar 2;163(2):275–286. doi: 10.1111/j.1432-1033.1987.tb10798.x. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of N6-methylation of residue A(5) on the conformational behaviour of d(C-C-G-A-A-T-T-C-G-G) in solution studied by 1H-NMR spectroscopy. 2. The hairpin form. Eur J Biochem. 1987 Mar 2;163(2):287–296. doi: 10.1111/j.1432-1033.1987.tb10799.x. [DOI] [PubMed] [Google Scholar]
- Sapienza C., Peterson A. C., Rossant J., Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987 Jul 16;328(6127):251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Szyf M., Gruenbaum Y., Urieli-Shoval S., Razin A. Studies on the biological role of DNA methylation: V. The pattern of E.coli DNA methylation. Nucleic Acids Res. 1982 Nov 25;10(22):7247–7259. doi: 10.1093/nar/10.22.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillandier E., Ridoux J. P., Liquier J., Leupin W., Denny W. A., Wang Y., Thomas G. A., Peticolas W. L. Infrared and Raman studies show that poly(dA).poly(dT) and d(AAAAATTTTT)2 exhibit a heteronomous conformation in films at 75% relative humidity and a B-type conformation at high humidities and in solution. Biochemistry. 1987 Jun 16;26(12):3361–3368. doi: 10.1021/bi00386a017. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartell R. M., Harrell J. T. Characteristics and variations of B-type DNA conformations in solution: a quantitative analysis of Raman band intensities of eight DNAs. Biochemistry. 1986 May 6;25(9):2664–2671. doi: 10.1021/bi00357a056. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Annu Rev Biochem. 1981;50:285–319. doi: 10.1146/annurev.bi.50.070181.001441. [DOI] [PubMed] [Google Scholar]
- von Kitzing E., Diekmann S. Molecular mechanics calculations of dA12.dT12 and of the curved molecule d(GCTCGAAAAA)4.d(TTTTTCGAGC)4. Eur Biophys J. 1987;15(1):13–26. doi: 10.1007/BF00255031. [DOI] [PubMed] [Google Scholar]