Abstract
Background
Rice blast is the most serious disease afflicting rice and there is an urgent need for the use of disease resistance (R) genes in blast tolerance breeding programs. Pb1 is classified as a quantitative resistance gene and it does not have fungal specificity. Pb1-mediated resistance develops in the latter stages of growth. However, some cultivars, such as Kanto209 (K209), cultivar name Satojiman, despite possessing Pb1, do not exert resistance to rice blast during the reproductive stage.
Results
We found that the expression of WRKY45 gene downstream of Pb1 was weakly induced by rice blast inoculation at the full heading stage in K209. Genetic analysis using the SNP-based Golden Gate assay of K209 crossing with Koshihikari Aichi SBL (KASBL) found at least four regions related to the resistance in the rice genome (Chr8, Chr9, Chr7, Chr11). Mapping of QTL related to Chr7 confirmed the existence of factors that were required for the resistance of Pb1 in the 22 to 23 Mbp region of the rice genome.
Conclusion
We clarified how the K209 cultivar is vulnerable to the blast disease despite possessing Pb1 and found the DNA marker responsible for the quantitative resistance of Pb1. We identified the QTL loci required for Pb1-mediated resistance to rice panicle blast. Pb1 was negatively dependent on at least three QTLs, 7, 9 and 11, and positively dependent on one, QTL 8, in the K209 genome. This finding paves the way for creating a line to select optimal QTLs in order to make use of Pb1-mediated resistance more effectively.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-017-0175-0) contains supplementary material, which is available to authorized users.
Keywords: Oryza sativa, Pyricularia oryzae, Panicle blast, QTL, Salicylic acid, Resistance gene
Background
Rice blast is a pathogenicity that affects rice plants worldwide. In order to minimize the damage of rice blast, resistance (R)-gene cultivars are commonly used. The use of R genes to develop resistant cultivars has become an urgent goal in rice breeding programs. Several molecular approaches to study rice blast resistance genes have been developed during the last decade. So far, hundreds of blast R genes have been mapped in the rice genome. A large group of R genes is clustered in several genomic regions reviewed in (Liu et al. 2014). Most of the genes encode nucleotide-binding site (NBS) leucine-rich repeat (LRR) proteins that interact with pathogen effectors and trigger defense reactions following a gene-for-gene model of recognition (Bryan et al. 2000; Okuyama et al. 2011; Cesari et al. 2013). One of the plant defense mechanisms is the recognition of Microbe-Associated Molecular Patterns (MAMPs) by receptors, leading to generally weak defense responses called MAMP-Triggered Immunity (MTI). Some pathogens overcome the resistance by secreting effector proteins that interfere with host resistance and promote pathogenicity. Plants have evolved to recognize against the effectors through Effector-Triggered Immunity (ETI). True resistance genes are specific and provide strong resistance to the rice blast, but the effect of R genes decline after 2 or 3 years.
In contrast to MTI or ETI, the quantitative resistance genes are durable to the blast, but are less understood (Cook et al. 2015; Fukuoka et al. 2015). Recently, intensive genetic analyses of quantitative “leaf” blast resistance genes including pi21, Pi34, Pi35, Pi39 and Pi63 have been carried out (Nguyen et al. 2006; Zenbayashi-Sawata et al. 2007; Terashima et al. 2008; Fukuoka et al. 2009; Xu et al. 2014). In contrast to these leaf blast resistance genes, the panicle blast (Pb1) resistance gene has been used as the durable form that confers rice plants partial resistance to the rice blast with no fungal race specificity (Fujii et al. 2000). Pb1 gene has been introduced into rice cultivars in Korea (Lee et al. 2015). The cultivars have remained resistant to panicle blast for over 35 years. Cloning of Pb1 has identified that the resistance is weak on the early stage and gradually becomes strong for the reproductive stage of the plant, and is dependent on the Pb1 expression (Hayashi et al. 2010). After heading stage, the resistance of Pb1 cultivar is very strong to rice blast.
The resistance mechanism of Pb1 is attributed to its interaction with WRKY45 (Inoue et al. 2013), which plays a crucial role in the salicylic acid (SA) pathway in rice immunity (Shimono et al. 2007). WRKY45 is regulated by a ubiquitin-proteasome system in the rice nucleus (Matsushita et al. 2012). The Pb1 coiled-coil domain interacts with the nuclear-localized WRKY45 (Inoue et al. 2013), resulting in a Pb1-WKRY45 complex is a weaker target than WRKY45 for protein degradation. WRKY45 overexpressed transgenic rice plants have strong resistance not only to the leaf rice blast but also to the panicle blast (SHIMONO et al. 2011). The genes in the downstream pathway of WRKY45, including OsNAC4, OsHSF1, OsOPR4 (Nakayama et al. 2013), and diterpenoid phytoalexin (DP), a bHLH transcription factor (Yamamura et al. 2015), play a crucial role in disease resistance. Cytokinins (CKs) play a role in mediating the signal of Pyricularia oryzae infection to trigger the induction of DP biosynthetic genes in benzothiadiazole- (BTH) primed plants (AKAGI et al. 2014).
Kanto 209 (K209) cultivar is one of the cultivars harboring Pb1; however, the resistance of the cultivar is weak in comparison with other Pb1 cultivars. In the K209 cultivar of rice, the expression of WRKY45 is lower than that of Koshihikari Aichi SBL (KASBL). The lower expression of the gene might be due to the genotype of the cultivar. To identify the mechanism of this rice blast resistance, we crossed the K209 and KASBL cultivars. Genomic DNA of their F3progeny was subjected to SNP-based Golden Gate assay and we found at least four quantitative trail loci (QTL) to the panicle rice blast resistance. Among the four QTLs, we focused on QTL7, which was located on chromosome 7 and showed Pb1-dependent defense against panicle blast in field tests, and performed a map-based cloning.
Results
Field tests were performed to examine the resistance to rice blast on a large number of Pb1 cultivars available in Japan. To test the resistance, we evaluated the index of panicle blast severity of the cultivars at the Aichi Agricultural Research Center, Mountainous Resin Agricultural Research Institute. A lower index is indicative of higher resistance to panicle blast. Figure 1 shows the gradual increase in severity index to panicle blast with time, and it is due to the high pressure of the rice blast. The resistance of Tsukinohikari and its sister cultivar Aichi67 were stronger than that of K209 and its progeny Hosijirushi (Fig. 1). These cultivars had similar full heading date within plus or minus 2 days from August 20th 2014. The resistance of Kanto HD2, which does not harbor Pb1, was similar to the other cultivars close to the heading date (Fig. 1).
Fig. 1.
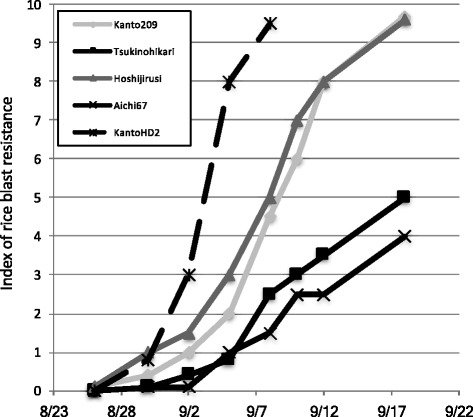
Development of panicle blast on Pb1-mediated resistance gene cultivars. Rice blast index of cultivars grown in an experimental paddy field at the Aichi Agricultural Research Center field in 2014. The cultivars, Aichi67, Tsukinohikari, Hoshijirushi, K209 and Kanto HD2 were evaluated in the field. The resistances of Tsukinohikari and Aichi 67 were stronger than that of Kanto 209 and Hoshijirushi in the full heading stage. The heading date of Aichi67, Tsukinohikari, Hoshijirushi, K209 and Kanto HD2 were 21st, 20th, 20th, 19th and 22nd of August, 2013, respectively. The Kanto HD2 cultivar was the negative control for the rice panicle blast resistance
K209 has other potential mutations for blast resistance
The only information available on Pb1-mediated resistance is that it is dependent on WRKY45 (Inoue et al. 2013), so we checked the sequence of Pb1 and WRKY45 in the K209 cultivar at the DNA and RNA level. At the DNA level, K209 and KASBL were sequenced for the Pb1 and WRKY45 regions. DNA from plants was extracted using CTAB method and the Pb1 and WRKY45 regions were amplified using PCR. The sequences of PCR products were then determined using specific primers on an ABI sequencer. As a result, there were no sequence differences in the Pb1 open reading frame (ORF) or the 2 kb upstream sequence between the KASBL and K209 cultivars (data not shown). Similarly, no differences were observed in the sequence of WRKY45, including the promoter region of 1.7 kbp with ORF and 3′ region site (data not shown). Previously, the expression of WRKY45 in rice was shown to be markedly enhanced by BTH treatment and resulted in resistance to rice blast fungus, whereas RNA interference–mediated knockdown of WRKY45 compromised BTH-inducible resistance to blast disease (Shimono et al. 2007). So we tested if the WRKY45 sequence still remains intact in the K209 cultivars and found that K209 cultivar treated with BTH exhibited resistance for rice blast while nontreated plants showed susceptibility (Fig. 2a).
Fig. 2.
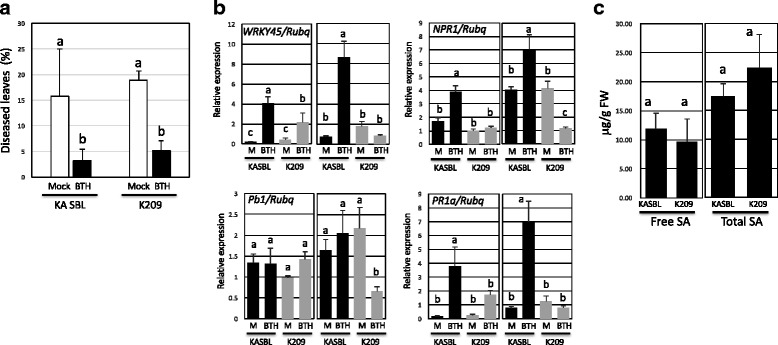
BTH-induced rice blast resistance of 5th leaves, expression analysis by q-RT-PCR, and the concentration of SA in KASBL and K209 cultivars. a. The 5th leaves of KASBL and K209 were sprayed with 50 μΜ BTH or water. After 2 days, the leaves were spray-inoculated with blast fungal conidia suspension (isolate Kyu89–246, 4.0 × 104 spores ml−1). Disease symptoms were evaluated by percentage of diseased leaves 10 days after inoculation. The experiments were performed three times with similar results. b. The expression of Pb1, WRKY45, NH1 and PR1a in the 5th leaves were analyzed by q-RT-PCR. Leaves after mock (M) or BTH treatment (BTH) for 2 days (left column) or 4 days (right column) were used for the expression analyses. The Y-axis indicates the mRNA expression relative to that of UBQ. The averages with SD are shown. Different letters indicate significant differences among the different lines at p < 0.05 (one-way ANOVA). Experiments were repeated twice with similar results. c. Free (left) and total (right) SA content of 25th leaves of KASBL and K209 measured by high performance liquid chromatography (HPLC). Letters indicate significant differences among the different lines at p < 0.05 (one-way ANOVA). Experiments were repeated three times with similar results
To study gene expression by quantitative PCR, total RNAs were extracted from the 25 days old leaf of KASBL and K209 after treatment with water (mock) or BTH for 2 or 4 days. Surprisingly, WRKY45, NH1 and PR1a expression was reduced by BTH treatment on both days (Fig. 2b). The transcript abundance for Pb1 was significantly reduced at 4 days after treatment with BTH (Fig. 2b). The concentrations of SA and total SA were not statistically different between K209 and KASBL plants (Fig. 2c).
To study gene expression, RNA was extracted from K209 and KASBL during the reproductive growth phase. The flag leaves with or without induced rice blast after 1 and 3 days were removed from K209 and KASBL cultivars, RNAs were extracted and quantitative PCR was performed. The Pb1 transcript of K209 exhibited reduced expression compared to KASBL after inoculation (Fig. 3). The WRKY45 transcript of KASBL was induced 1 day and 3 days after the rice blast (Fig. 3). The transcript abundance of WRKY45 was reduced and the expression of NH1 in K209 still remained at the basal level after rice blast inoculation (Fig. 3), suggesting that the transport or the perception of SA was defective. These indicates that the mutation(s) was (were) not in the Pb1 genomic region in the genome of K209.
Fig. 3.
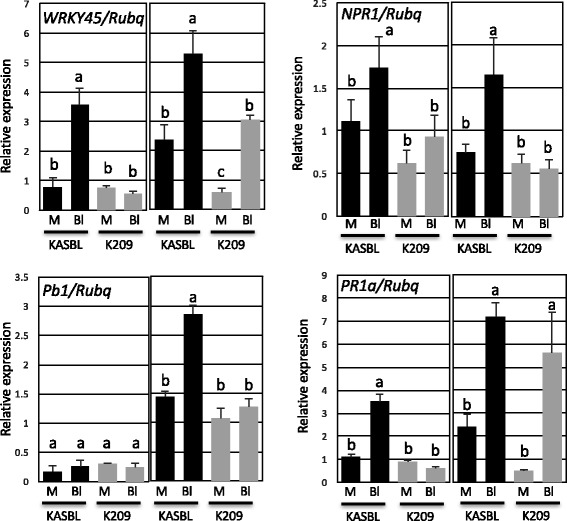
Expression analyses in flag leaves in response to rice blast. The flag leaves of KASBL and K209 were spray-inoculated with water (mock) or blast fungal conidia suspension (Bl) (2.0–1.5 × 105 spores ml−1). The expression of NH1, WRKY45 Pb1 and PR1a in flag leaves 1 day (left column) and 3 days (right column) after inoculation was analyzed by q-RT-PCR. The Y-axis shows relative mRNA expression to UBQ. The average of 12 plants and SD are shown. Different letters indicate significant differences among the different lines at p < 0.05 (one-way ANOVA). Experiments were repeated twice with similar results
K209 has at least four DNA mutations for blast resistance on its genome
It was apparent that K209 had a genomic alteration in the molecular mechanism responsible for Pb1-mediated resistance gene activation. To identify this mechanism, K209 line (+Pb1) was crossed with KASBL (+Pb1). Genomic DNA was extracted from 82 F3 progeny from each line and subjected to the SNP-based Golden Gate assay. Out of 768 SNPs spanning the entire genome, 199 SNPs distinguished K209 from KASBL. The ratio of heterozygosity in the samples was 26.9%. To evaluate rice panicle blast resistance, the F4 plants were tested in an experimental field at the Aichi Agricultural Research Center in 2010. QTL analysis detected four QTLs for Pb1-mediated resistance each one on chromosomes 7, 8, 9 and 11 (Table 1). The alleles from K209 at the QTL 7, 9 and 11 had negative effect on rice blast resistance while that on QTL 8 had positive effect (Fig. 4). QTL7 was located on the long arm, the end of chromosome 7 while QTL9 was on the middle of the long arm. QTL11 and QLT8 were on the short arm, with the former present near the centromere and the latter near the telomere. The effects of resistance QTL alleles of K209 and KASBL on panicle blast resistance to different haplotypes are shown in Table 2.
Table 1.
Putative QTLs associated with panicle blast resistance in K209/KASBL F4 lines in 2010
| Nearest markers | chromosome | Logarithm of odds | Additive effecta | Phenotype variationb
explained (%) |
|---|---|---|---|---|
| b297 = C7–14 | 7 | 4.24 | 0.74 | 12.1 |
| b063 = C8–4 | 8 | 7.06 | −0.94 | 22.0 |
| b586 = C9–7 | 9 | 3.30 | 0.86 | 14.6 |
| b285 = C8–4 | 11 | 4.52 | 0.58 | 12.0 |
aAdditive effects of K209 allele
bPhenotypic variation explained by QTL
Fig. 4.
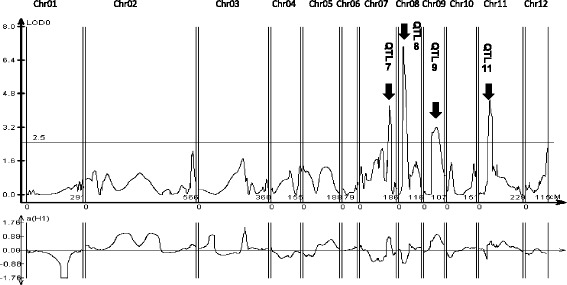
LOD curve plot from QTL analysis for panicle blast resistance tested by using 82 F4 lines from the K209 and KASBL cross evaluated in 2010. Horizontal line at 2.5 indicates LOD thresholds obtained by using 1000 permutations
Table 2.
Eight genotypes defined by four QTLs for panicle blast resistance and their phenotypes
| Lines | QTLs | Leaf disease index | Panicle disease index | Days until heading | |||
|---|---|---|---|---|---|---|---|
| QTL7 | QTL8 | QTL9 | QTL11 | ||||
| Ha21 | + | + | + | + | 3 | 1.8 | 76 |
| Ha55 | + | + | + | + | 5 | 2.8 | 81 |
| Ha06 | + | + | + | 5 | 2.6 | 73 | |
| Ha64 | + | + | + | 5.5 | 2.8 | 76 | |
| Ha13 | + | + | + | 6 | 3.6 | 76 | |
| Ha30 | + | + | + | 6 | 4.1 | 76 | |
| Ha38 | + | + | + | 6.5 | 4.5 | 83 | |
| Ha27 | + | + | + | 5 | 5.0 | 65 | |
| Ha12 | + | 6 | 5.6 | 76 | |||
| Ha22 | 5 | 7.2 | 73 | ||||
| Ha29 | 5 | 8.8 | 70 | ||||
| K209 | + | 5 | 7.6 | 80 | |||
| KASBL | + | + | + | 5 | 4.0 | 70 | |
+: homozygous resistance QTL allele
QTL7 was located within a 1.8 Mbp region on chromosome 7
To delimit the chromosomal region for QTL7 through a map-based procedure, we screened recombinants within and around this QTL in a mapping population and subjected them to a field test. We compared the degree of rice blast resistance between recombinants and their counterpart non-recombinant controls. We evaluated 14 lines in 2014, 122 lines in 2015, and 86 lines in 2016. The positions of QTLs identified by the 3-year field test are shown in Fig. 5a. When disease index differs by 2 or more, we judged that the resistance of two lines is different. We found that NT642/641, NT397/398, NT413/414, NT595/596, NT432/431, NT588/587 and NT618/617 exhibited differences in the panicle rice blast. Two genotypes, NT518/517 and NT525/526 showed no difference in panicle resistance index (Fig 5b). The results led to the conclusion that QTL7 lies in the 1.777 Mb region within the marker loci between RM336 and RM5847 (Fig. 5b).
Fig. 5.

a QTL region for panicle blast resistance on chromosome 7 estimated by a 3 year field test using recombinant inbred lines with recombination within and around the QTL. b Graphical genotypes of recombinant inbred lines used to determine the QTL7 locus. Positions are based on the International Rice Genome Sequencing Project 1.0 of the Nipponbare genome. Box 1 indicates chromosomes derived from the resistance cultivar, KASBL; box 2, from the susceptible cultivar K209. Box 0 indicates missing data. The location of the QTL7 locus, indicated at the bottom, is based on phenotypic data obtained from field tests obtained in 2016
Discussion
Rice with enhanced Pb1-mediated resistance can be produced by using QTL Analyses
Pb1 is known as a durable blast-resistance gene consisting of a single dominant locus. Introduction of Pb1 gene into rice plants by marker selection breeding has been actively carried out. As a result, there are several Pb1 cultivars to panicle blast resistance of rice in Japan. Among them, K209 and its progeny cultivar Hoshijirushi have been found to exhibit weak resistance to rice panicle blast despite harboring Pb1. The loss of Pb1-mediated resistance over time has generated a negative impact for rice breeders for a long time. To this end, we attempted to identify the mechanism of the loss of Pb1-mediated blast resistance of Pb1 in K209. Traditionally, a functional gene region is introduced into a genomic fragment, so the gene can have a large additive effect in breeding. However, interactions with the genes in the background have been known to cause differential effects with respect to rice bacterial blight resistance (Sun et al. 2004; Cao et al. 2007; Iyer-Pascuzzi et al. 2008). In this study, we attempted to solve the conundrum of the absence of resistance to rice blast despite the presence of the resistance gene Pb1in the K209 line. Our studies revealed the exact locus in the gene responsible for rice blast resistance, underscoring the importance of this research in blast resistance breeding. Genetic analysis of KASBL and K209 revealed that the regions Chr8, Chr9, Chr7, and Chr11 were involved in rice blast resistance in at least four rice genomes (Fig. 4). Among them, three QTLs (Chr9, Chr7 and Chr11) negatively affected the resistance despite the presence of Pb1. Chr8, on the other hand, had a positive influence on signal transduction, so it is expected that these QTLs will specifically change Pb1-mediated resistance single handedly or in combination with others (Fig. 4). This paves the way for creating a line in future containing just the positively acting QTL and excluding the negatively affecting QTLs in order to make a more effective Pb1-mediated resistance. Therefore, this result is important in developing highly persistent blast resistance disease varieties by selection of markers.
Investigations into finding the factors responsible for resistance reveal the signal transduction of several factors interacting with each other to produce enhanced resistance to the blast. Although several causative genes have been identified as factors involved in highly quantitative resistance, detailed studies on downstream factors are lacking. Analyses of factors responsible for highly effective, true resistance to rice blast have, however, been progressing. For example, Pit resistance is a true blast resistance gene located downstream of OsRAC1 (Os01g0229400) (Kawasaki et al. 1999; Kawano et al. 2010), OsRAR1 (Os02g0535400) and OsSGT1 (Os01g0624500) (Wang et al. 2008). They are located outside of all the four QTLs loci. Other downstream pathways involved in basic resistance of the blast such as Rice Chitin Receptor CEBiP (Os03g0133400) (Kaku et al. 2006), Rice LysM receptor-like kinase, OsCERK1 (Os08g0538300) (Shimizu et al. 2010), cytoplasmic kinase, OsRLCK176 (Os05g0110900), OsRLCK185 (Os05g0372100), lysin motif-containing proteins, OsLYP6 (Os06g0208800) (Ao et al. 2014) have been shown. They all exist in different QTLs in rice genome. They all exist in different loci in rice genome, except OsLYP4 (Os09g0452200) that may be one of candidates for the QTL on Chr9 (Ao et al. 2014). Altogether, we found that the QTLs of Chr7 and Chr11 are new genes exhibiting resistance to blast disease and this will contribute toward the efforts to elucidate the network of genes responsible for rice blast resistance.
K209 exhibits lower induction of SA-responsive genes with blast infection compared to KASBL cultivar
When K209 and KASBL cultivars were inoculated with rice blast, we found that WRKY45 and NH1 were weakly induced in K209 (Figs. 2b and 3) and the SA concentration was similar to that of KASBL (Fig. 2c). This result indicates that the transport or perception of SA was defective in the K209 cultivar. In Arabidopsis, there there is evidence for SA reception by AtNPR proteins (Wu et al. 2012; Fu et al. 2012). Rice has orthologous NPR proteins in the genome, OsNPR1 (Os01g0194300), OsNPR2 (Os01g0767966) and OsNPR3 (Os03g0667100), but these genes did not correspond to the QTLs derived from qualitative reasoning in these experiments.
The mechanism of the induced resistance of WRKY45 depended on the resistance of Pb1, and could not be induced by inoculation with blast disease in this cultivar. The expression of WRKY45 in rice is induced by BTH by activating the SA pathway (Shimono et al. 2007). Therefore, we investigated whether the induction of WRKY45 by BTH still remained in these cultivars. In this experiment, the expression of WRKY45 in both genotypes was maintained normally, and the resistance induced by BTH was conserved (Fig. 2b). However, when treated with rice blast in KASBL and K209 in reproductive growth, the induction of WRKY45 expression was not observed only in the K209 varieties (Fig. 3). Therefore, it is highly probable that the upstream signaling pathway up to WRKY45 is compromised by some mutations in this cultivar for infection of rice blast. Little is known on the upstream signaling activation mechanism of WRKY45 in rice plants. WRKY45 is regulated by several enzymes, including mitogen-activated protein kinase kinase OsMKK10–2 and mitogen-activated protein kinase, some phosphorylation events such as OsMPK4 and OsMPK6 phosphorylation, and also dephosphorylation events such as the tyrosine dephosphorylation of MAPKs by PTPases (OsPTP1/2) in the SA pathway (Ueno et al. 2015). OsMKK10–2 is located on chromosome 3 (Hamel et al. 2006), and the loci of OsMPK4 (Chromosome 10), OsMPK6 (Chromosome 10), OsPTP1 (Chromosome 12) and OsPTP2 (Chromosome 11), were all different from the QTLs derived from qualitative reasoning in these experiments. The mechanism of WRKY45 activation against the rice panicle blast of Pb1-mediated resistance is still unknown.
Conclusion
In this report, we have identified the QTL loci of Pb1 based on the resistance to rice panicle blast with map-based screening. We found that Pb1 was negatively dependent at least on three QTLs, 7, 9 and 11, and positively on one, QTL 8. This understanding of the molecular mechanism of blast resistance paves the way for creating a line in future containing just the positively acting QTL in order to make a more effective Pb1-mediated resistance.
Methods
Plant materials
Kanto 209 and Koshihikari Aichi SBL are cultivars that are susceptible and resistant to panicle blast, respectively, despite possessing Pb1. 82 F3 plants derived from a cross between these cultivars developed by single seed descent method were used for DNA genotyping. The F4 lines derived from respective F3 plants were used for assessing resistance to panicle blast to detect putative QTLs. To confirm and delimit putative QTLs on chromosome 7, a total of 320 F4 or F5 lines whose QTL regions were fixed (except for that on chromosome 7) were used to select recombinant inbred plants on and around the putative QTL on chromosome 7. Selected progeny lines (F5 or F6) were subjected to field tests in 2014, 2015 and 2016.
DNA sequencing
To sequence Pb1 and WRKY45, we amplified the following regions from the K209 cultivar: for Pb1, we amplified both the Pb1 upstream sequence (−2200 to −1) and ORF region using the primers 5′-AAGGTGAGGTGAGTCATTAGTG-3′ and 5′-TCATGGTTCATTACATTTAA-3′. For WRKY45, we amplified both the upstream sequence (−1700 to −1) and ORF genomic region using the primers 5′-GCCCAATCGGCTGTAATAC-3′ and 5′- AGGCACGTGAAGCTATATGT-3′. The DNA fragments were purified by Nucleo Spin Gel and PCR clean-up kit (Machery-Nagel, Germany). Then, we performed DNA sequencing using BigDye terminator v3.1 Cycle Sequencing kit (ThermoFisher, Japan) with 3130 genetic Analyzer. The obtained sequences were aligned with Nipponbare genome by a plasmid editor software, APE version 2.04 (http://biologylabs.utah.edu/jorgensen/wayned/ape/).
Evaluation of panicle blast resistance
Panicle blast resistance was evaluated in an experimental paddy field at the Aichi Agricultural Research Center, Mountainous Resin Agricultural Research Institute, where the high pressure of blast disease and its progress are well controlled. Thirteen plants were transplanted for each line in early June. Disease severity was visually evaluated 2–3 weeks after the heading date, and scored from 0 (no diseased grain) to 10 (100% diseased grain) according to the report (Asaga 1981). Some varieties developed for the evaluation of partial resistance to panicle blast (Hayashi et al., 1993) were used for comparison.
RNA analysis
Total RNA was isolated from rice tissue using Trizol (Invitrogen). For quantitative RT-PCR, total RNA was treated with DNA remover to remove contaminating genomic DNA. cDNA was synthesized using ReverTra Ace reverse transcriptase (Toyobo, Japan). To determine Pb1 expression, quantitative RT-PCRs were run on a Thermal Cycler Dice TP800 system (Takara Bio, Japan) as shown previously (Hayashi et al. 2010) using primers Pb1sp4Fw/Pb1s4Rv. Rice ubiquitin 1 (Rubq1; AK121590) was used as an internal standard. The primers used in quantitative RT-PCR were listed in Additional file 1: Table S1.
The measurement of SA and SAG content
Free SA and SAG were quantified as described previously (Raskin et al. 1989; Malamy et al. 1992).
The SNP genotyping
The genotypes of the F3 plants, (which we characterized by using 199 SNP markers), were determined by using 768-plex SNPs for the Illumina Golden Gate Bead Array technology platform (Illumina, Inc., San Diego, CA, USA) based on previously reported information (Ebana et al. 2010; Nagasaki et al. 2010). DNA was extracted from 50 mg of fresh rice leaves using the DNeasy 96 Plant Kit (Qiagen). The designed OPA and 250 ng of DNA were used for the preparation of bead chips according to the protocol for the Golden Gate Genotyping Assay.
QTL analysis and progeny tests
We constructed a genetic map using MAPMAKER/EXP v. 3.0 software (Lander et al. 1987). QTL analysis for grain length and grain width was performed using version 2.5 of QTL Cartographer software (Basten et al. 2005), and the threshold was obtained by using 1000 permutations. The progeny lines of F4 or F5 plants were grown to select recombinants within the QTL in order to develop inbred lines with recombinations within and around QTL7. Plants with independent recombination events were selected from on the basis of genotypes around the QTL region determined by using simple-sequence repeat (SSR) markers. The primers for SSR markers were listed in Additional file 1: Table S1.
Acknowledgments
The cultivar of K209, Kanto HD2 and Hoshijirushi were kindly gifted by Dr. Sato, NARO Institute of Crop Science, Tsukuba, Ibaraki, Japan. The chemical analysis of SA by HPLC was performed by Dr. Shigemi Seo, NARO Institute of Crop Science, Tsukuba, Ibaraki, Japan.
Funding
This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement, LCT-0013) and by JSPS KAKENHI Grant Number 17 K07678.
Additional file
The primers (OsWRKY45, NH1, PR1a, Pb1 and Rubq1) were used for quantitative RT-PCR analysis. The others (RM336, RM1017, RM3826, RM5623, RM5847, RM6885, RM21773 and RM21784) were used for QTL analysis. (PDF 27 kb)
Authors’ contributions
HI, NH designed research; HI, MN, TM, AT, SS, SF, NH performed research; HI, SF, and NH wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-017-0175-0) contains supplementary material, which is available to authorized users.
Contributor Information
Haruhiko Inoue, Phone: +81(29) 838-8385, Email: haruhiko@affrc.go.jp.
Nagao Hayashi, Phone: +81(29) 838-7020, Email: nhayash@affrc.go.jp.
References
- AKAGI A, Fukushima S, Okada K, et al. WRKY45-dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol Biol. 2014;86:171–183. doi: 10.1007/s11103-014-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Y, Li Z, Feng D, et al. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014;80:1072–1084. doi: 10.1111/tpj.12710. [DOI] [PubMed] [Google Scholar]
- Asaga K. A procedure for evaluating field resistance to blast in rice varieties. J Cent Agric Exp Stn. 1981;35:51–138. [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB (2005) QTL cartographer, 1.17
- Bryan GT, Wu KS, Farrall L, et al. tA single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene pi-ta. Plant Cell. 2000;12:2033–2046. doi: 10.1105/tpc.12.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Ding X, Cai M, et al. The expression pattern of a rice disease resistance gene xa3/xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics. 2007;177:523–533. doi: 10.1534/genetics.107.075176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Thilliez G, Ribot C, et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BPHJ. Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol. 2015;53:541–563. doi: 10.1146/annurev-phyto-080614-120114. [DOI] [PubMed] [Google Scholar]
- Ebana K, Yonemaru J-I, Fukuoka S, et al. Genetic structure revealed by a whole-genome single-nucleotide polymorphism survey of diverse accessions of cultivated Asian rice (Oryza Sativa L.) Breed Sci. 2010;60:390–397. doi: 10.1270/jsbbs.60.390. [DOI] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Hayano-Saito Y, Saito K, et al. Identification of a RFLP marker tightly linked to the panicle blast resistance gene, Pb1, in Rice. Breed Sci. 2000;50:183–188. doi: 10.1270/jsbbs.50.183. [DOI] [Google Scholar]
- Fukuoka S, Saka N, KOGA H, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- Fukuoka S, Saka N, Mizukami Y, et al. Gene pyramiding enhances durable blast disease resistance in rice. Sci Rep. 2015;5:7773. doi: 10.1038/srep07773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L-P, Nicole M-C, Sritubtim S, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Saka N, Koide T, et al. Selection of standard rice [Oryza Sativa] varieties for panicle blast resistance in different heading group. Res Bull Aichi Agric Res Ctr. 1993;25:13–22. [Google Scholar]
- Hayashi N, Inoue H, Kato T, et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hayashi N, Matsushita A, et al. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction. Proc Natl Acad Sci. 2013;110:9577–9582. doi: 10.1073/pnas.1222155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jiang H, Huang L, McCouch SR. Genetic and functional characterization of the rice bacterial blight disease resistance gene xa5. Phytopathology. 2008;98:289–295. doi: 10.1094/PHYTO-98-3-0289. [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, et al. Activation of a Rac GTPase by the NLR family disease resistance protein pit plays a critical role in Rice innate immunity. Cell Host Microbe. 2010;7:362–375. doi: 10.1016/j.chom.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, et al. The small GTP-binding protein rac is a regulator of cell death in plants. Proc Natl Acad Sci. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E., P. Green, J. Abrahamson, A. Barlow, M. Daly, E. Lincoln, A. Newberg (1987) Mapmaker: an. interactive computer package for constructing primary genetic linkage maps of experimental and natural. populations. Genomics 1:174–181 [DOI] [PubMed]
- Lee J-H, Lee J-Y, Yoon Y-N, et al. Enhancement of panicle blast resistance in Korean Rice cultivar “Saeilmi” by marker assisted backcross breeding. Plant Breed Biotechnol. 2015;3:1–10. doi: 10.9787/PBB.2015.3.1.001. [DOI] [Google Scholar]
- Liu W, Liu J, Triplett L, et al. Novel insights into Rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 2014;52:213–241. doi: 10.1146/annurev-phyto-102313-045926. [DOI] [PubMed] [Google Scholar]
- Malamy J, Hennig J, Klessig DF. Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell. 1992;4:359–366. doi: 10.1105/tpc.4.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Inoue H, GOTO S, et al. Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J. 2012;73:302–313. doi: 10.1111/tpj.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Ebana K, Shibaya T, et al. Core single-nucleotide polymorphisms—a tool for genetic analysis of the Japanese rice population. Breed Sci. 2010;60:648–655. doi: 10.1270/jsbbs.60.648. [DOI] [Google Scholar]
- Nakayama A, Fukushima S, GOTO S, et al. Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 2013;13:150. doi: 10.1186/1471-2229-13-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TTT, Koizumi S, La TN, et al. Pi35(t), a new gene conferring partial resistance to leaf blast in the rice cultivar Hokkai 188. Theor Appl Genet. 2006;113:697–704. doi: 10.1007/s00122-006-0337-8. [DOI] [PubMed] [Google Scholar]
- Okuyama Y, Kanzaki H, Abe A, et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011;66:467–479. doi: 10.1111/j.1365-313X.2011.04502.x. [DOI] [PubMed] [Google Scholar]
- Raskin I, Turner IM, Melander WR. Regulation of heat production in the inflorescences of an arum lily by endogenous salicylic acid. Proc Natl Acad Sci. 1989;86:2214–2218. doi: 10.1073/pnas.86.7.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, et al. Rice WRKY45 plays a crucial role in Benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIMONO M, KOGA H, AKAGI A, et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol Plant Pathol. 2011;13:83–94. doi: 10.1111/j.1364-3703.2011.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. Oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313X.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Terashima T, Fukuoka S, Saka N, Kudo S. Mapping of a blast field resistance gene Pi39(t) of elite rice strain Chubu 111. Plant Breed. 2008;127:485–489. doi: 10.1111/j.1439-0523.2007.01451.x. [DOI] [Google Scholar]
- Ueno Y, Yoshida R, Kishi-Kaboshi M, et al. Abiotic stresses antagonize the Rice Defence pathway through the tyrosine-Dephosphorylation of OsMPK6. PLoS Pathog. 2015;11:e1005231. doi: 10.1371/journal.ppat.1005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao M, Li Q, et al. OsRAR1 and OsSGT1 physically interact and function in Rice basal disease resistance. Mol Plant-Microbe Interact. 2008;21:294–303. doi: 10.1094/MPMI-21-3-0294. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, et al. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1:639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Xu X, Hayashi N, Wang C-T, et al. Rice blast resistance gene Pikahei-1(t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol Breed. 2014;34:691–700. doi: 10.1007/s11032-014-0067-6. [DOI] [Google Scholar]
- Yamamura C, Mizutani E, Okada K, et al. Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 2015;84:1100–1113. doi: 10.1111/tpj.13065. [DOI] [PubMed] [Google Scholar]
- Zenbayashi-Sawata K, Fukuoka S, Katagiri S, et al. Genetic and physical mapping of the partial resistance gene, pi34, to blast in rice. Phytopathology. 2007;97:598–602. doi: 10.1094/PHYTO-97-5-0598. [DOI] [PubMed] [Google Scholar]


