Abstract
Gametogenesis is a complicated biological process by which sperm and egg are produced for genetic transmission between generations. In many animals, the germline is segregated from the somatic lineage in early embryonic development through the specification of primordial germ cells (PGCs), the precursors of gametes for reproduction and fertility. In some species, such as fruit fly and zebrafish, PGCs are determined by the maternally provided germ plasm which contains various RNAs and proteins. Here, we identified a germ plasm/PGC-specific microRNA miR-202-5p for the first time in zebrafish. MiR-202-5p was specifically expressed in gonad. In female, it was expressed and accumulated in oocytes during oogenesis. Quantitative reverse transcription PCR and whole mount in situ hybridization results indicated that miR-202-5p exhibited a typical germ plasm /PGC-specific expression pattern throughout embryogenesis, which was consistent with that of the PGC marker vasa, indicating that miR-202-5p was a component of germ plasm and a potential PGC marker in zebrafish. Our present study might be served as a foundation for further investigating the regulative roles of miRNAs in germ plasm formation and PGC development in zebrafish and other teleost.
Introduction
Primordial germ cells (PGCs), the progenitor cells of gametes, play critical roles in genetic information transmission from generation to generation1. In most animals, PGCs segregate from somatic cells at early developmental stages and migrate to genital ridge where PGCs and gonadal somatic precursors together form primordial gonad2. Germ plasm is an amorphous, non-membrane-bound, electron-dense structure comprising of abundant RNAs, proteins and organelles, which is essential for PGC development in most animals3–5. By presence or absence of maternal germ plasm, two modes of PGC specification, preformation and induction, are proposed in animals6. In worms, flies and zebrafish, PGC specification is dependent on the maternal germ plasm accumulated in eggs (preformation)7. In urodele amphibians and mammals, extracellular signals induce PGC specification during gastrulation (induction). Many components of germ plasm have been revealed to play important roles in PGC development. The RNA binding protein Vasa regulates localization of another germ plasm component nanos which mediated transcriptional repression is critical for PGC maintenance8. Daz, Dazl and Boule are three key factors in regulating germ cells entering meiosis9, 10. Dnd protects critical germ plasm mRNA from degradation in germ cells by inhibiting miR-43011, 12.
MicroRNAs (miRNAs) are about 22 nucleotide endogenous non-coding RNAs that play important roles in posttranscriptional regulation by targeting mRNA for degradation or translational repression13. Recently, it has been reported that miRNAs play an important regulatory role in germ cell proliferation, specification, migration and maintenance in flies and mammals14–16. Targeted depletion of the miRNA-processing enzyme Dicer1 in germ cell of worms, flies and mice led to PGC reduction and gametogenesis defects17–20. In chicken, microarray assay revealed some germ cell-enriched miRNAs, suggesting that miRNAs might regulate germ cell development21. In human, it had been found that a miR-372/let-7 axis regulated germ cell maintenance and differentiation22. In zebrafish, little is known about the role of miRNAs in germ cell development, and it has been remained uncertain whether there are germ plasm/PGC-specific miRNAs in zebrafish.
MiR-202–5p is a miRNA abundantly expressed in gonad of some mammals and lower vertebrates. In mouse, miR-202-5p is specifically expressed in Sertoli cells of embryonic gonads23. In human, the Sertoli cell-specific miR-202-5p might play an interaction role between somatic cells and germ cells in spermatogenesis24. In frog, miR-202-5p is a germ cell-enriched miRNA in testis and ovary, indicating miR-202-5p might also be involved in germ cell development in amphibians25, 26. Recently, it has been reported that miR-202-5p and miR-202-3p are expressed throughout gonad development in zebrafish by transcriptomic analysis of miRNA expression in gonads27. Previously we reported that miR-202-5p was highly expressed in spermatozoa and developing male germ cells in zebrafish28. However, the expression patterns of miR-202-5p and -3p are remained unknown in embryos. In this study, the expression patterns of miR-202-5p and -3p were characterized in tissues and embryos, and found that miR-202-5p rather than miR-202-3p was highly and specifically expressed in gonads of zebrafish. Moreover, whole mount in situ hybridization (WISH) and fluorescence in situ hybridization (FISH) revealed that miR-202-5p was a maternal component of germ plasm and was specifically expressed in PGCs throughout embryogenesis. Therefore, for the first time, we identified a germ plasm-specific miRNA miR-202-5p in zebrafish, and it might be a potential marker for PGCs.
Results
Gonad-specific expression of miR-202-5p in zebrafish
In zebrafish, the pri-miR-202 encodes a pair of mature miRNAs, miR-202-5p and miR-202-3p (Fig. 1A). The tissue distribution patterns of miR-202-3p and -5p were examined by quantitative reverse transcription polymerase chain reaction (RT-PCR) in eight tissues including heart, liver, spleen, kidney, brain, muscle, testis and ovary. As shown in Fig. 1B, miR-202-3p was universally expressed in all examined tissues, and its expression level was comparatively high in heart, moderate in ovary, muscle and spleen, low in liver, testis, brain and kidney. However, miR-202-5p was specifically expressed in gonad, with a higher expression level in testis than in ovary. The dynamic expression patterns of miR-202-3p and -5p in oocytes at different developmental stages were further examined. As shown in Fig. 1C, miR-202-3p was almost undetectable in oocytes at stages I-III until stage IV, and the highest expression level was observed in mature oocytes. Whereas, the expression level of miR-202-5p in stage I and II oocytes was low, but the expression level gradually increased from stage III, and peaked in mature oocytes. In comparison with miR-202-3p, the expression of miR-202-5p was much higher and exclusive in mature oocytes.
Figure 1.
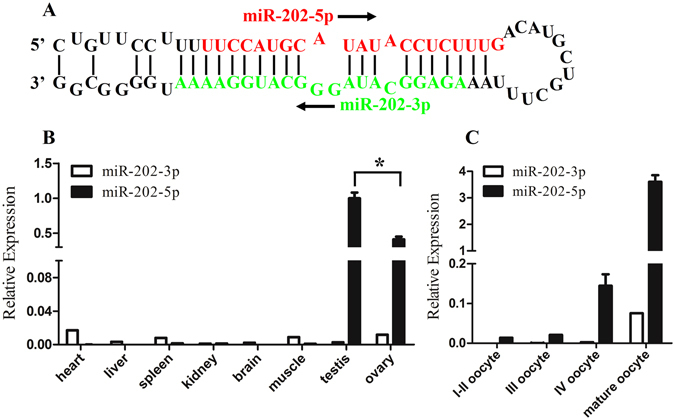
Tissue distribution pattern of miR-202 by RT-PCR. (A) Sequence of miR-202 including stem loop, miR-202-5p (in red) and miR-202-3p (in green). The arrow indicated the direction of miRNA sequences from 5′-3′. (B) Tissue distribution of miR-202-3p/5p. (C) Dynamic expression of miR-202-3p/5p in oocytes at different developmental stages. U6 was used as an internal control. Asterisk indicated significant difference (p < 0.05).
MiR-202-5p is a maternal miRNA in zebrafish
Subsequently, the embryonic expression pattern of miR-202-5p was examined by quantitative RT-PCR. As shown in Fig. 2, miR-202-5p was highly expressed in mature oocytes, and there was a slight up-regulation in 1-cell embryos after fertilization. During blastulation, the expression of miR-202-5p gradually decreased, and only very low expression was detected from gastrulation to long pec stage. These results indicated that miR-202-5p was a maternal miRNA.
Figure 2.
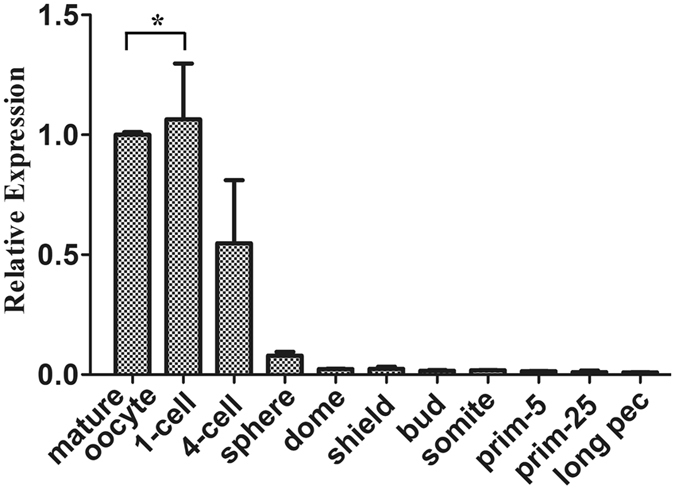
Temporal expression of miR-202-5p in mature oocytes and embryos at different development stages. U6 was used as an internal control. Asterisk indicated significant difference (p < 0.05).
MiR-202-5p is a germ plasm-specific miRNA
Furthermore, the spatial and temporal localization of miR-202-5p was revealed by WISH. As shown in Fig. 3A, miR-202-5p signals were present in a wide cortical band around the animal pole in mature oocytes. After fertilization, miR-202-5p signals were present in numerous granules throughout the cortex of 1-cell embryos (Fig. 3B). Subsequently, miR-202-5p signals were dominantly located at the cleavage furrows in embryos at 2 and 4 cells stages (Fig. 3C and D). At 128 cells stage, some granular miR-202-5p signals were detected at the animal pole (Fig. 3E). At dome stage, miR-202-5p-expressing cells were present at the four corners of the animal pole (Fig. 3F). At shield stage, miR-202-5p-expressing cells were distributed at the marginal region of the blastoderm (Fig. 3G). At 3 somites stage, miR-202-5p-expressing cells aggregated into two lines along the yolk syncytial layer (Fig. 3H). At prim-5 stage, miR-202-5p-expressing cells located in the genital ridge (Fig. 3I). This expression pattern of miR-202-5p was consistent to that of the known PGC marker vasa throughout early embryonic development (Fig. 3J–R). Meanwhile, we observed that miR-202-3p was maternally and universally expressed in almost all cells but not in PGCs during embryogenesis (Supplementary Fig. S1).
Figure 3.
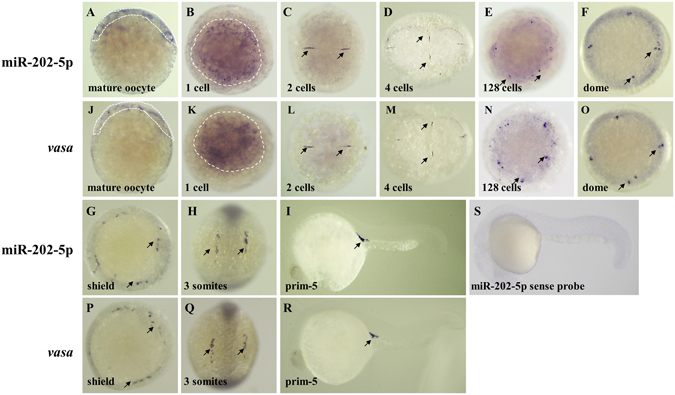
Spatial and temporal localization of miR-202-5p in mature oocytes and embryos at different development stages by WISH. A-I, localization of miR-202-5p; J-R, localization of vasa mRNA; S, negative control. The black arrows indicated the positive signals of miR-202-5p and vasa mRNA.
Co-localization of miR-202-5p and vasa mRNA in germ plasm and PGCs
To further demonstrate the specific distribution of miR-202-5p in germ plasm and PGCs, dual-color FISH was performed to examine the co-localization of miR-202-5p and vasa mRNA during embryogenesis (Fig. 4). At 4-cell stage, miR-202-5p and vasa mRNA co-localized at the cleavage furrows where the maternal germ plasm distributed (Fig. 4A–C). At prim-5 stage, miR-202-5p and vasa mRNA co-localized at the perinuclear germ granules of PGCs (Fig. 4D–F’). Therefore, similar to vasa mRNA, miR-202-5p was also a maternal germ plasm component, and specifically localized in zebrafish PGCs throughout embryogenesis.
Figure 4.
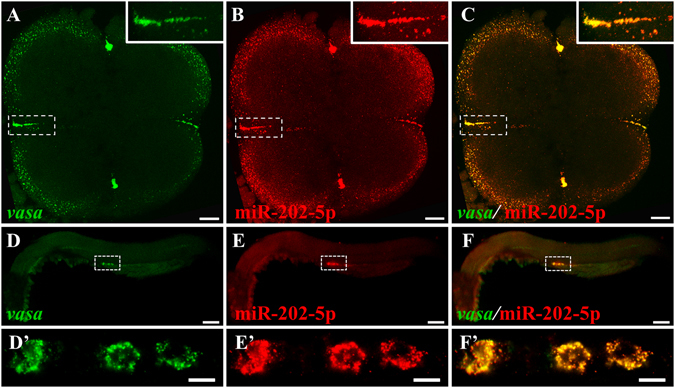
Co-localization of miR-202-5p and vasa mRNA by FISH. (A and D), vasa mRNA (green), (B and E) miR-202-5P (red); (C and F), merged. (A–C), embryo at 4-cell stage; D–F’, embryo at prim-5 stage. The smaller panels on top right of (A–C) indicated the high magnification of the white dotted areas, and D’–F’ are the high magnification of the white dotted areas in (D–F). Scale bars: 50 μm in (A–C), and 100 μm in (D–F).
Discussion
In this study, we characterized the expression patterns of miR-202-5p and -3p. The expression patterns of these two miRNAs were quite different in tissues and embryos. Comparison to the universe expression of miR-202-3p, miR-202-5p was specifically expressed in gonads and was accumulated during oocyte maturation in female. Importantly, miR-202-5p was a maternal miRNA that was specifically expressed in germ plasm and subsequently in PGCs throughout embryogenesis. MiR-202-5p is the first miRNA identified in germ plasm and PGCs in zebrafish.
In vertebrates, pri-miR-202 encoded two opposing miRNAs, miR-202-3p and -5p. In mammals, miR-202-3p was not only involved in gonad development, but also identified as an important tumor repressor in inhibiting tumor progression29–31. It has been reported that miR-202-5p was highly expressed in Sertoli cells in embryonic mouse gonads23. In human, miR-202-5p was specifically localized in Sertoli cells and its expression significantly differed between fertile and sterile men, suggesting a novel interaction for regulating miRNA expression between the somatic and germ cell components of the seminiferous epithelium24. Therefore, the somatic cells expressing miR-202 regulated spermatogenesis in mammals. Differently, in lower vertebrates such as frog and zebrafish, miR-202 was expressed in germ cells. In frog, miR-202-5p was highly expressed in testis and was dramatically down-regulated in sterile males, suggesting a role of miR-202-5p in spermatogenesis26. Meanwhile, frog miR-202-5p was a gonad-specific miRNA and enriched in oocytes, indicating miR-202-5p also regulated oogenesis25. In zebrafish, dynamics of miRNA transcriptome showed that the expression of miR-202-3p and -5p were increased during oogenesis and spermatogenesis27. Our previous study revealed that miR-202-5p was highly expressed in spermatozoa and developing male germ cells at various stages in zebrafish28. Our present study demonstrated the different expression patterns between miR-202-5p and miR-202-3p in zebrafish, indicating the different functions of miR-202-3p and miR-202-5p. The low and universe expression of miR-202-3p in many tissues suggested a broad function. However, the gonad-specific expression pattern of miR-202-5p and its dynamic expression during oocyte maturation suggested its potential role in germ cell development. The high expression level of miR-202-5p in early embryos inherited from mature eggs and spermatozoa by fertilization strongly suggested that miR-202-5p might be an important regulator in PGC development.
So far, little is known about the embryonic expression pattern of miR-202 in lower vertebrates. In this study, RT-PCR examination revealed that miR-202-5p was maternally exist in unfertilized eggs, and there was a little up-regulation of miR-202-5p in 1-cell embryos, which might be due to an additional amount of miR-202-5p from spermatozoa upon fertilization. WISH and FISH detection further showed that miR-202-5p was localized in cortex around the animal pole in mature oocyte and was exclusively expressed in germ plasm and PGCs post fertilization. The decreased expression of miR-202-5p detected by RT-PCR was inconsistent with the increased number of miR-202-5p positive cells because of a lower proliferation rate of PGC than that of somatic cells. In zebrafish, miR-34 was the only reported maternal miRNA, which was involved in embryonic neurogenesis32. This study showed that miR-202-5p was the second maternal miRNA and it was the first germ plasm-specific miRNA in zebrafish. In animals with preformation mode, the specification of PGCs depends on the germ plasm, which comprises of various maternal proteins and RNAs. It had been found that germ plasm-specific long non-coding RNA pgc was important for transcriptional silence and chromatin structure33, 34. Since miRNA-mediated posttranscriptional regulation always led to gene silence, much attention was paid to miRNAs that might be involved in germ cell development. In animals with inductive mode, many miRNAs related to germ cell development had been identified and investigated. In mouse, the LIN28/let-7/BLIMP1 axis, in which RNA binding protein LIN28 negatively regulated let-7 to protect the expression of BLIMP1, was essential for PGC induction in vitro 35, 36. In human, the miR-372/let-7 axis regulated germ cell formation from embryonic stem cells18. In chicken, several PGC-enriched miRNAs were identified by microarray, and these miRNAs were highly expressed in male germ cells and even PGCs17. In preformation mode, although it had been demonstrated that miRNA pathways were important for germ cell development, no germ plasm-specific miRNA was identified18. Our present study showed that miR-202-5p was a novel germ plasm-specific miRNA in zebrafish. These results indicated the importance to further investigate the functions of miRNAs in germ cell development in animals with PGC preformation mode.
In present study we characterized the expression pattern of miR-202, and found that miR-202-5p was a novel germ plasm-specific miRNA in zebrafish. The unique expression pattern of miR-202-5p suggested that it might play an important role in PGC development in zebrafish, and further work was needed to investigate the function of miR-202-5p and the molecular mechanism by which miR-202-5p works.
Methods
Fish
Zebrafish, AB line wild type, were raised at 28 °C with 10 hours (h) darkness and 14 h light. All embryos were collected after natural spawning and staged as previous reported37. All procedures with zebrafish were approved by the Ethics Committee of Sun Yat-Sen University and the methods were carried out in accordance with the approved guidelines.
Sample collection
Embryo samples were collected at different developmental stages including mature oocytes, and embryos at 1-cell stage, 4-cell stage, sphere, dome, shield, bud, somite stage, prim-5 (24 hours post fertilization, 24 hpf), prim-25 (36 hpf) and long pec (48 hpf) stages into Trizol reagent (Invitrogen) for RNA isolation. Embryos at above stages were also collected and fixed in 4% paraformaldehyde (PFA) in PBS at 4 °C overnight and stored in 100% methanol at −20 °C for WISH. Tissues including heart, liver, spleen, kidney, brain, muscle, ovary and testis, were collected from adult zebrafish into Trizol reagent for RNA isolation. Oocytes at different developmental stages were separated under a Leica stereomicroscope as previously described38.
RNA isolation and RT-PCR
Total RNA was isolated by Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The total RNA was reverse-transcripted using Mir-X™ miRNA Frist-Strand Synthesis Kit (Takara). The quantitative RT-PCR was performed in a Roche LightCycle 480 II (Roche) and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) as previously described39. The RT-PCR cycling conditions were: 94 °C for 2 min, followed by 40 cycles of 94 °C for 12 s, 60 °C for 12 s, 72 °C for 15 s. U6 was used as the internal control (The U6 primers were provide by Mir-X™ miRNA Frist-Strand Synthesis Kit). The relative gene expression was calculated with the 2−ΔΔCT methods. Data were shown as mean ± SD from three independent experiments in triplicates. The primer sequences were listed in Table S1.
Whole mount in situ hybridization (WISH)
The vasa cDNA sequence for synthesis of digoxin (DIG) or fluorescein isothiocyanate (FITC) labelled vasa sense and anti-sense RNA probes was as previously described40. The 1.1-kb vasa cDNA were cloned and sequenced by using a pair of vasa primers which containing a T7 core promoter at the 5′ end of the reverse primers. The primer sequences were listed in Table S1. The probes were synthesized by using DIG- or FITC-RNA labeling kit according to the manufacturer’s instructions (Roche). The sense and anti-sense DIG labelled RNA probes of miR-202-5p (Genbank accession number: MIMAT0003406) were synthesized by Ribobio (Guangzhou, China). WISH was performed as previously described41. Embryos were fixed in 4% PFA at 4 °C overnight, dehydrated in methanol at −20 °C overnight, rehydrated by a series of methanol/PBS gradient into 100% PBST, and treated with proteinase K and re-fixed in 4% PFA. Embryos were prehybridized with hybridization mix at 59 °C for 2-4 h, and hybridized with DIG-labelled miR-202-5p or vasa anti-sense probe at 59 °C overnight. After hybridization, embryos were washed in a series of SSCT gradient at 64 °C, and in PBST at room temperature. Subsequently, the embryos were blocked PBST with 2 mg/mL BSA and 2% normal goat serum for 2 hours at room temperature, and then incubated in alkaline phosphatase conjugated anti-DIG antibody (1:5000 diluted in blocking solution, Roche) at 4 °C overnight. Finally, the embryos were washed in PBST for 15 min six times, and the signal was detected using NBT/BCIP as the chromogenic substrate. The images were documented with a Leica stereomicroscope MZ10F equipped with a digital camera.
Fluorescent in situ hybridization (FISH)
The dual-color FISH was performed as previously described42. The procedure before hybridization and washing steps were same to that of WISH. Differently, embryos were hybridized with DIG-labelled miR-202-5p and FITC-labelled vasa probe at 59 °C overnight. Embryos were washed in MABT (0.1 M Maleic acid, 0.15 M NaCl, 0.1% Tween-20, PH 7.5) for 10 min twice at room temperature before blocking, and blocked with 2% blocking reagent (Roche) in MABT for 2 h at room temperature, and then incubated in anti-fluorescein-POD sheep monoclonal antibody (1:2000 diluted in blocking solution, Roche) at 4 °C overnight. The signal was detected by using the TSATM Plus Fluorescence Systems according to the product manual (NEL756, NEN Life Science). FISH images were acquired with a Leica TCS-SP8 confocal microscope (Leica), and confocal Z-series image stacks collected at 1 μm intervals were assembled by LAS X basic software.
Statistics analysis
All statistics were calculated using SPSS version 20. Differences between control and treatment groups were assessed by one-way ANOVA. p < 0.05 was considered to be statistically significant.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31271576), the Zhuhai Scholar Professor Program (2015) and the Guangdong Natural Science Foundation (2015A030308012).
Author Contributions
M.Y., K.J. and W.L. designed the research and wrote the manuscript. J.Z., W.L., Y. J. and P.J. carried out the experiments. Y.J. and P.J. and W.L. analyzed the data. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jing Zhang and Wei Liu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07675-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kuntong Jia, Email: jiakt3@mail.sysu.edu.cn.
Meisheng Yi, Email: jiakt3@mail.sysu.edu.cn.
References
- 1.Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 2.Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat. Rev. Mol. Cell. Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Mukhtar KAK, Webb AC. An ultrastructural study of primordial germ cells, oogonia and early oocytes in Xenopus laevis. J. Embryol. Exp. Morphol. 1971;26:195–217. [PubMed] [Google Scholar]
- 4.Toyooka Y, et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 2000;93:139–149. doi: 10.1016/S0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 5.Wylie C. Germ cells. Cell. 1999;96:165–174. doi: 10.1016/S0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- 6.Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- 7.Xu HY, Li MY, Gui JF, Hong YH. Fish germ cells. Sci. China Life Sci. 2010;53:435–446. doi: 10.1007/s11427-010-0058-8. [DOI] [PubMed] [Google Scholar]
- 8.Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Dev. Dynam. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- 9.Kee K, Angeles VT, Flores M, Nguyen HN, Pera RAR. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat. Cell. Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- 11.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 12.Weidinger G, et al. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003;13:1429–1434. doi: 10.1016/S0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- 15.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIver SC, Roman SD, Nixon B, McLaughlin E. A. miRNA and mammalian male germ cells. Human Reproduction Update. 2012;18:44–59. doi: 10.1093/humupd/dmr041. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi K, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PloS One. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 19.Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 20.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhin J, et al. PGC-enriched mirnas control germ cell development. Mol. Cells. 2015;38:895–903. doi: 10.14348/molcells.2015.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran ND, et al. A miR-372/let-7 axis regulates human germ versus somatic cell fates. Stem Cells. 2016;34:1985–1991. doi: 10.1002/stem.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wainwright EN, et al. SOX9 regulates microRNA miR-202-5p/3p expression during mouse testis differentiation. Biol. Reprod. 2013;89:34. doi: 10.1095/biolreprod.113.110155. [DOI] [PubMed] [Google Scholar]
- 24.Dabaja AA, et al. Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific microRNA, miR-202-5p, in human testis. Basic Clin. Androl. 2015;25:2. doi: 10.1186/s12610-015-0018-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res. 2009;19:1766–1775. doi: 10.1101/gr.093054.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michalak P, Malone JH. Testis-derived microRNA profiles of African clawed frogs (Xenopus) and their sterile hybrids. Genomics. 2008;91:158–164. doi: 10.1016/j.ygeno.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Presslauer, C., Bizuayehu, T. T., Kopp, M., Fernandes, J. M. & Babiak, I. Dynamics of miRNA transcriptome during gonadal development of zebrafish. Sci. Rep. 7, doi:10.1038/srep43850 (2017). [DOI] [PMC free article] [PubMed]
- 28.Jia KT, et al. Identification of micrornas in zebrafish spermatozoa. Zebrafish. 2015;12:387–397. doi: 10.1089/zeb.2015.1115. [DOI] [PubMed] [Google Scholar]
- 29.Ma G, Zhang F, Dong X, Wang X, Ren Y. Low expression of microRNA-202 is associated with the metastasis of esophageal squamous cell carcinoma. Exp. Ther. Med. 2016;11:951–956. doi: 10.3892/etm.2016.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng X, et al. MicroRNA-202 inhibits tumor progression by targeting LAMA1 in esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2016;473:821–827. doi: 10.1016/j.bbrc.2016.03.130. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. miR-202 suppresses cell proliferation in human hepatocellular carcinoma by downregulating LRP6 post-transcriptionally. FEBS Lett. 2014;588:1913–1920. doi: 10.1016/j.febslet.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Soni K, et al. miR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res. 2013;41:4470–4480. doi: 10.1093/nar/gkt139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNA polII-dependent transcription in primordial germ cells. Curr. Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- 35.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West JA, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 38.Lubzens, E., Young, G., Bobe, J. & Cerda, J. Oogenesis in teleosts: How fish eggs are formed. Gen. Comp. Endocrinol, doi:S0016-6480(09)00231-7 (2009). [DOI] [PubMed]
- 39.Liu W, et al. Complete depletion of primordial germ cells in an All-female fish leads to sex-biased gene expression alteration and sterile all-male occurrence. BMC Genomics. 2015;16:1. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 41.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 42.Xu H, Li Z, Li M, Wang L, Hong Y. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One. 2009;4:e6097. doi: 10.1371/journal.pone.0006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


