Abstract
Ralstonia solanacearum is the causal agent of the devastating bacterial wilt disease in many high value Solanaceae crops. R. solanacearum secretes around 70 effectors into host cells in order to promote infection. Plants have, however, evolved specialized immune receptors that recognize corresponding effectors and confer qualitative disease resistance. In the model species Arabidopsis thaliana, the paired immune receptors RRS1 (resistance to Ralstonia solanacearum 1) and RPS4 (resistance to Pseudomonas syringae 4) cooperatively recognize the R. solanacearum effector PopP2 in the nuclei of infected cells. PopP2 is an acetyltransferase that binds to and acetylates the RRS1 WRKY DNA-binding domain resulting in reduced RRS1-DNA association thereby activating plant immunity. Here, we surveyed the naturally occurring variation in PopP2 sequence among the R. solanacearum strains isolated from diseased tomato and pepper fields across the Republic of Korea. Our analysis revealed high conservation of popP2 sequence with only three polymorphic alleles present amongst 17 strains. Only one variation (a premature stop codon) caused the loss of RPS4/RRS1-dependent recognition in Arabidopsis. We also found that PopP2 harbors a putative eukaryotic transcriptional repressor motif (ethylene-responsive element binding factor-associated amphiphilic repression or EAR), which is known to be involved in the recruitment of transcriptional co-repressors. Remarkably, mutation of the EAR motif disabled PopP2 avirulence function as measured by the development of hypersensitive response, electrolyte leakage, defense marker gene expression and bacterial growth in Arabidopsis. This lack of recognition was partially but significantly reverted by the C-terminal addition of a synthetic EAR motif. We show that the EAR motif-dependent gain of avirulence correlated with the stability of the PopP2 protein. Furthermore, we demonstrated the requirement of the PopP2 EAR motif for PTI suppression. A yeast two-hybrid screen indicated that PopP2 does not interact with any well-known Arabidopsis transcriptional co-repressors. Overall, this study reveals high conservation of the PopP2 effector in Korean R. solanacearum strains isolated from commercially cultivated tomato and pepper genotypes. Importantly, our data also indicate that the PopP2 conserved repressor motif could contribute to the effector accumulation in plant cells.
Keywords: effector recognition, natural variation, repressor motif, transcriptional co-repressor, disease resistance
Introduction
The soil-borne pathogen Ralstonia solanacearum is the cause of devastating bacterial wilt in a wide range of host species including agronomically important Solanaceae species. Due to the wide genetic and host range diversity of strains, the concept of an R. solanacearum species complex (RSSC) is now generally accepted (Genin and Denny, 2012). The first RSSC sequenced strain, GMI1000, belongs to phylotype I isolated from tomato plants (Salanoubat et al., 2002). GMI1000 harbors around 70 predicted type 3 effectors (T3Es), also termed Rips (Ralstonia injected proteins), which are secreted into host cells to promote infection and enable bacterial growth (Mukaihara et al., 2010; Peeters et al., 2013). The R. solanacearum T3E repertoire is extensive when compared to other bacterial pathogens such as Xanthomonas spp. and Pseudomonas syringae, which both possess 30–40 T3Es (Alfano and Collmer, 2004; Büttner and Bonas, 2010). Moreover, further analysis of all sequenced R. solanacearum strains revealed that there is a large number of conserved core effectors (>30) (Genin and Denny, 2012). This suggests that the common ancestor already possessed a large arsenal of T3Es.
One of the primary roles for the T3Es is to dampen or suppress host defense responses. These responses are initially induced by the recognition of conserved microbial features termed pathogen/microbe-associated molecular patterns (PAMPs/MAMPs), such as flagellin, the building block of the bacterial flagellum or peptidoglycan from the bacterial envelope (Felix et al., 1999; Gust et al., 2007). Activation of the host pattern-recognition receptors (PRRs) by these molecules leads to pattern-triggered immunity (PTI), an efficient defense response impeding pathogen growth (Zipfel et al., 2004; Jones and Dangl, 2006). Consequently, numerous T3Es have been shown to target and inhibit components of the PTI signaling pathway, restoring susceptibility in the host plant (Macho and Zipfel, 2015). In turn, plants have evolved an intracellular set of immune receptors belonging to the nucleotide-binding leucine-rich repeat resistance (NLR) protein family that can detect corresponding T3Es and activate effector-triggered immunity (ETI) (Jones et al., 2016). The T3Es that activate ETI are termed avirulence proteins. ETI is often associated with a strong programmed cell death (PCD) of the infected cells, the hypersensitive response (HR), which participates in pathogen growth restriction (Dodds and Rathjen, 2010).
At least 7 of the ∼70 Rips trigger HR in R. solanacearum host species. The first identified avirulent Rip, AvrA, triggers HR in tobacco (Nicotiana tabacum) (Carney and Denny, 1990; Robertson et al., 2004). PopP1, Awr2, Awr5 and RipTPS also trigger HR in tobacco leaf cells (Poueymiro et al., 2009, 2014; Solé et al., 2012). Awr2 and Awr5 trigger HR in other Nicotiana species, while PopP1 also acts as an avirulence gene in a Petunia line (Lavie et al., 2002; Solé et al., 2012). In wild eggplant (Solanum torvum), the putative Zn-dependent protease, RipAX2 (formerly Rip36), induces a strong HR (Peeters et al., 2013; Nahar et al., 2014). Finally, the well-characterized acetyltransferase effector, PopP2, is one of two sequence-unrelated effectors that are recognized in the model plant species Arabidopsis by the paired NLRs RPS4 and RRS1-R (Deslandes et al., 2002, 2003; Narusaka et al., 2009). Besides PopP2 being the sole R. solanacearum effector recognized in Arabidopsis so far, the system allowing for its recognition is of particular interest due to the unusual structure of the RRS1-R receptor that harbors a WRKY-DNA binding domain (Deslandes et al., 2002). Multiple WRKY transcription factors (TFs) are involved notably in wound or defense response (Eulgem and Somssich, 2007). Of note, in the Arabidopsis ecotype Col-0, a shorter form of the RRS1 protein caused by a premature stop codon after the WRKY domain loses the ability to recognize PopP2 and is therefore denoted as RRS1-S (Deslandes et al., 2002). For the clarity of this report, we refer to RRS1-R (Ws-2), the allele conferring PopP2 recognition, as RRS1. Importantly, as a result of the conservation of immune signaling in plants, transgenic tomato expressing RPS4 and RRS1 from Arabidopsis are resistant to infection by R. solanacearum RS1002 strain that carries popP2 (Narusaka et al., 2013). Considering the large plasticity of the R. solanacearum effector repertoire across the species complex and geographic regions, investigation into PopP2 contribution to virulence and the structural requirement for its recognition by the RPS4/RRS1 complex is essential to envisage the deployment of these R genes in crop species (Dangl et al., 2013; Peeters et al., 2013).
PopP2 belongs to the YopJ-like family of cysteine proteases, which share conserved catalytic triad residues [histidine (H), aspartate (D)/glutamate (E), cysteine (C)]. However, several YopJ-like effectors from mammals and plant pathogens can modify their host target by trans-acetylation rather than proteolytic activity (Ma and Ma, 2016). Indeed, PopP2 exhibits acetyltransferase activity, which is fully dependent on the catalytic cysteine residue, C321. Auto-acetylation of a lysine residue (K383) is required for the trans-acetylation activity of PopP2 and RPS4/RRS1-mediated recognition (Tasset et al., 2010). PopP2 co-localizes with RRS1 to the plant cell nucleus; however, the N-terminal 148 amino acids of PopP2 that include a putative nuclear localization signal (NLS) are dispensable for nuclear localization and avirulence (Deslandes et al., 2003; Sohn et al., 2014). Two recent studies brought evidence that PopP2 specifically targets the WRKY domain of RRS1. Acetylation of a key lysine residue in the RRS1 WRKY domain results in dissociation from the DNA and RPS4-dependent ETI. The RRS1 WRKY domain hence acts as a decoy to trap PopP2 activity, which may otherwise target WRKY TFs to disable plant defense (Le Roux et al., 2015; Sarris et al., 2015).
PopP2, though not belonging per se to the core-effector repertoire, contributes significantly to R. solanacearum virulence when present (Macho et al., 2010). This contribution to virulence could now be attributed to the ability of PopP2 to acetylate multiple host WRKY TFs probably resulting in their dissociation from DNA (Le Roux et al., 2015). WRKY TFs are integral for the regulation of plant innate immunity and are implicated in PTI, ETI and systemic acquired resistance (SAR) responses (Eulgem and Somssich, 2007; Rushton et al., 2010). PopP2-mediated WRKY TF acetylation has been shown to abrogate PAMP-triggered immunity (PTI) and contribute to R. solanacearum virulence (Le Roux et al., 2015). Thus, one can infer that the virulence function of the nuclear localized PopP2 is to manipulate host defense gene transcription via inhibition of WRKY TF DNA binding.
WRKY TFs play roles as both activators and repressors in plant immune signaling (Xu et al., 2006). Transcriptional repression is mainly achieved by chromatin modification at different levels (Berger, 2007). Transcriptional repressors can interact with co-repressors that recruit histone deacetylase for epigenetic silencing of gene expression (Long et al., 2006; Krogan et al., 2012). Direct interaction between repressor and co-repressor is mediated by the ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif (Song and Galbraith, 2006; Szemenyei et al., 2008). For instance, the co-repressor TOPLESS (TPL) is involved in the regulation of jasmonic acid (JA) signaling. Jasmonate ZIM-domain (JAZ) proteins function as transcriptional repressors of JA-regulated genes (Santner and Estelle, 2007). The majority of JAZ proteins directly bind to an adapter protein, Novel Interactor of JAZ (NINJA), which possesses an EAR motif and recruits TPL to epigenetically silence gene transcription via the histone deacetylase HDA19 (Pauwels et al., 2010; Pauwels and Goossens, 2011). TPL and TOPLESS-RELATED (TPRs) belong to the Groucho (Gro)/Tup1-like family of co-repressors encompassing 13 members in Arabidopsis (Liu and Karmarkar, 2008). Similar to TPL involvement in JA regulation of gene expression, TPRs play a role in the repression of negative regulators of defense during infection (Zhu et al., 2010). Although the recruitment of co-repressors for the repressor activity of some WRKY TFs has not been demonstrated yet, it is interesting to note that three members of the WRKY family contain an EAR motif (Kagale et al., 2010).
To gain further insights into the surveillance system that monitors virulence activity in the plant cell, we analyzed the natural sequence variation of PopP2 in R. solanacearum strains isolated from diseased tomato and pepper fields across Republic of Korea. We found that the sequence is highly conserved and identified that, among several, only one PopP2 allele lacks avirulence activity. This indicates that RPS4/RRS1-mediated recognition can tolerate multiple natural polymorphisms in PopP2. Furthermore, we identified a conserved EAR motif in PopP2, which we show to be required for in planta recognition, PTI suppression and protein accumulation. Besides providing valuable insight into the natural variation of PopP2 in virulent R. solanacearum strains, our study also unveils a novel mechanism by which a pathogenic effector could maintain its stability in the host cell.
Results
PopP2 Is Highly Conserved among Korean R. solanacearum Isolates and Harbors a Putative Transcriptional Repressor Motif
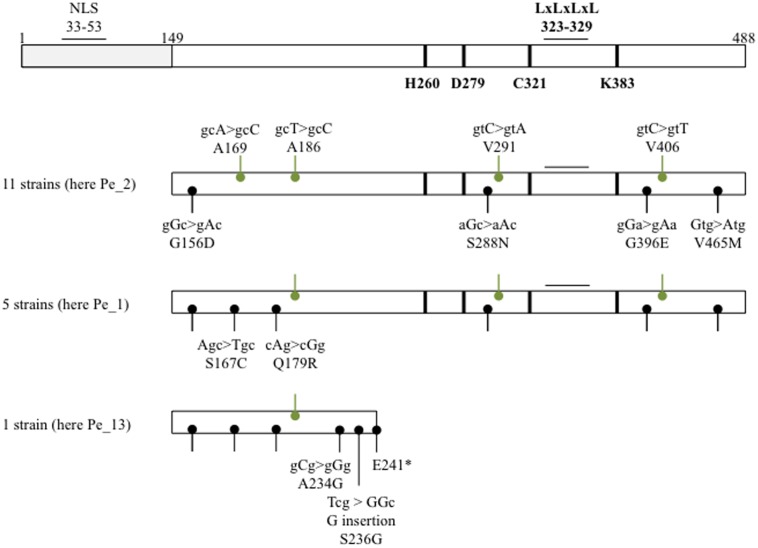
In order to survey naturally occurring sequence variation in PopP2, we first selected 20 R. solanacearum strains isolated from commercially grown pepper or tomato plants showing wilting symptoms in the Republic of Korea, on the basis of their geographic location, the host plant they were collected from (Pepper, strains ‘Pe_’ and Tomato, strains ‘To_’) and the year of collection (Table 1 and Supplementary Figure 1). Using gene-specific primers, we could amplify and confirm the presence of popP2 in 17 of the 20 R. solanacearum strains (Table 1). The genomic sequence encoding the C-terminal region of PopP2 that is necessary and sufficient for avirulence in Arabidopsis, amino acids 149–488, was analyzed in the 17 popP2-harboring strains and compared to the GMI1000 reference (Salanoubat et al., 2002; Sohn et al., 2014). 11 strains (Pe_2, Pe_3, Pe_18, Pe_24, Pe_27, Pe_42, Pe_45, Pe_56, To_1, To_7, and To_42) harbored four SNPs resulting in the following amino acid residue changes: G156D, S288N, G396E, and V465M. These 11 strains also harbored four synonymous mutations at A169, A186, V291, and V406. Five other strains (Pe_1, Pe_26, Pe_28, Pe_40, and To_63) harbored the four aforementioned non-synonymous SNPs as well as two additional SNPs resulting in S167C and Q179R. These five isolates all harbored the same aforementioned synonymous SNPs except for the mutation at A169. Finally, the Pe_13 strain harbored G156D, S167C, and Q179R as well as the synonymous mutation at A186. In addition to this, Pe_13 harbored a SNP resulting in A234G and a single nucleotide insertion, which resulted in a premature stop codon, E241∗ (Figure 1). Therefore, our survey identified three novel polymorphic PopP2 variant groups, which we termed PopP2Pe_2 (four non-synonymous SNPs, present in 11 strains), PopP2Pe_1 (six non-synonymous SNPs, present in five strains) and PopP2Pe_13 (four non-synonymous SNPs and a frameshift insertion, present in only one of the selected strains) (Figure 1). The majority of strains analyzed here (11 of 17) harboring the “Pe_2” popP2 allele were isolated from regions spanning the length of the Republic of Korea from Hwacheon in the north down to the southern coastal region, Haenam (Supplementary Figure 1). Among the five strains harboring the “Pe_1” allele, four were isolated from pepper fields in western regions; the other was isolated from tomato in eastern Bongwha. The Pe_13 strain carrying the truncated PopP2 variant was isolated from Imsil in the south–west (Supplementary Figure 1). The specific host cultivar genotypes are unknown. Thus, no obvious correlation between the host or the location of the isolated strains and the presence of a specific popP2 allele could be revealed by this survey.
Table 1.
Origin of the isolated Ralstonia solanacearum strains.
| Name | Number | Host | Year of isolation | popP2 presence |
|---|---|---|---|---|
| Pe_1 | YKB3030 | Pepper | 2000 | + |
| Pe_2 | YKB3033 | Pepper | 2000 | + |
| Pe_3 | YKB3078 | Pepper | 2001 | + |
| Pe_4 | YKB4598 | Pepper | 2001 | - |
| Pe_13 | YKB5438 | Pepper | 2002 | + |
| Pe_18 | YKB5445 | Pepper | 2002 | + |
| Pe_24 | YKB5458 | Pepper | 2002 | + |
| Pe_26 | YKB5774 | Pepper | 2003 | + |
| Pe_27 | YKB5778 | Pepper | 2003 | + |
| Pe_28 | YKB5861 | Pepper | 1999 | + |
| Pe_40 | YKB6924 | Pepper | 2005 | + |
| Pe_42 | YKB6953 | Pepper | 2005 | + |
| Pe_45 | YKB7024 | Pepper | 2005 | + |
| Pe_56 | YKB7141 | Pepper | 2005 | + |
| Pe_57 | YKB7171 | Pepper | 2005 | - |
| To_1 | YKB9153 | Tomato | 2008 | + |
| To_7 | YKB9174 | Tomato | 2008 | + |
| To_42 | YKB9246 | Tomato | 2008 | + |
| To_52 | YKB9258 | Tomato | 2008 | - |
| To_63 | YKB9274 | Tomato | 2008 | + |
List of the strains used in this study, including host and year of isolation. popP2 presence as assessed by PCR amplification with specific primers is indicated by “+” and popP2 absence is indicated by “-”.
FIGURE 1.
Natural variants and conserved residues of the PopP2 effector. A schematic of PopP2 sequence displaying natural variation and known or putative functional residues/motifs. GMI1000 PopP2 was used as the reference sequence. Labels in black show non-synonymous mutations and labels in green show synonymous mutations. See Table 1 for further details about the strains. The asterisk indicates a stop codon.
Nonetheless, closer analysis of the PopP2 coding sequence highlighted the presence of a putative LxLxL ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif; a motif involved in transcriptional repression via recruitment of transcriptional co-repressors (Hiratsu et al., 2003; Kagale and Rozwadowski, 2011). The reported motif comprises three leucine residues with amino acid spacers; however, PopP2 possesses an additional fourth leucine residue (LxLxLxL) at amino acids 323–329, which is almost adjacent to C321, one of the three conserved catalytic residues, H260, D279, and C321 (Tasset et al., 2010). Similarly to the catalytic residues, the putative EAR motif is fully conserved in all the sequenced PopP2 variants (Figure 1).
Only One of the Newly Identified PopP2 Variants Loses Avirulence Function In Planta
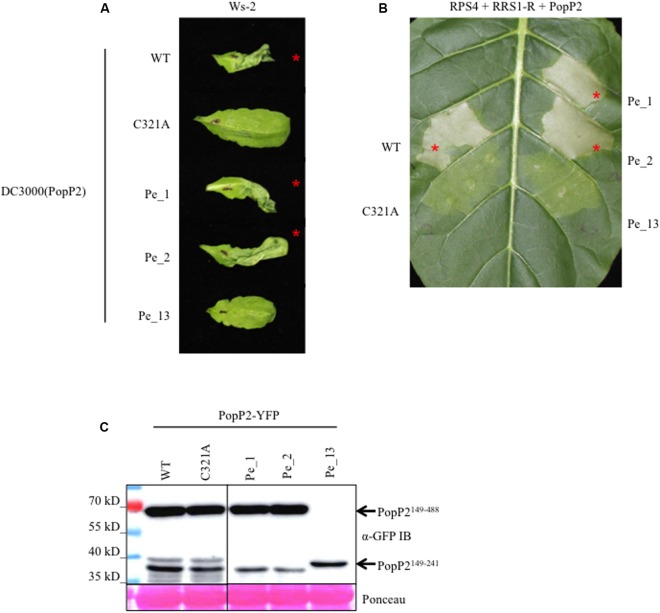
To investigate the effect of natural polymorphism on the avirulence activity of PopP2, the three newly identified PopP2 variants lacking N-terminal region were translationally fused with AvrRps4 N-terminal domain (AvrRps4N) and delivered by the P. syringae pv. tomato (Pto) DC3000 type 3 secretion system into the resistant Ws-2 Arabidopsis accession, carrying functional RPS4 and RRS1-R, to assay for HR (Sohn et al., 2014). At 1 day post-infiltration (dpi) Pto DC3000-delivered AvrRps4N:PopP2Pe_1 and AvrRps4N:PopP2Pe_2 variants triggered a strong HR (Figure 2A). Conversely, the truncated AvrRps4N:PopP2Pe_13 could not trigger HR in Arabidopsis. This lack of avirulence activity was expected as the catalytic residues required for acetyltransferase activity and RPS4/RRS1-mediated recognition are absent in this variant due to a premature stop codon (Tasset et al., 2010; Sohn et al., 2014) (Figure 1). Similar recognition events were observed when PopP2 natural variants were co-expressed with RRS1 and RPS4 in tobacco leaf cells after Agrobacterium-mediated transient transformation (hereafter, agroinfiltration) (Figure 2B). Expression of all three protein variants was confirmed by immuno-detection in Nicotiana benthamiana leaf extracts (Figure 2C). Notably, none of the six identified SNPs present in the avirulent PopP2Pe_1 allele affected recognition by RPS4/RRS1 despite the close proximity of two polymorphisms to catalytic residues (S288N and G396E). This suggests that these polymorphisms do not impair PopP2 acetyltransferase activity and that RPS4/RRS1-mediated recognition can accommodate significant variation in the PopP2 sequence.
FIGURE 2.
PopP2Pe_1 and PopP2Pe_2 are avirulent in Arabidopsis, PopP2Pe_13 is not. (A) PopP2 natural variants, PopP2Pe_1 and PopP2Pe_2, elicit an HR in resistant Arabidopsis accession Ws-2. The truncated natural variant, PopP2Pe_13 is not recognized. PopP2 variants were delivered by Pseudomonas syringae pv. tomato (Pto) DC3000 and photographs were taken 1 day post-infiltration (1 dpi). Red asterisks indicate HR. This experiment was conducted three times with similar results. (B) Agrobacterium-mediated co-expression of PopP2 variants with RPS4 and RRS1-R in tobacco leaf cells. Red asterisks indicate strong programmed cell death (PCD) at 3 dpi. This experiment was conducted three times with similar results. (C) The PopP2 natural variant proteins accumulate to a similar amount in Nicotiana benthamiana after agro-infiltration. Immuno-detection of PopP2 variants C-terminally fused to YFP epitope tag was conducted using anti-GFP antibodies. Ponceau S staining of total protein demonstrates equal loading of the samples.
The Conserved EAR Motif Is Required for PopP2 Avirulence Activity in Arabidopsis
To test if PopP2149-488 (hereafter referred to as PopP2) requires an LxLxL motif (hereafter; EAR motif) for avirulence function, we performed site-directed mutagenesis on the PopP2 LxLxLxL sequence to generate the LxAxAxL variant (hereafter, PopP2LAAL). Mutation of the two central leucine residues ensured that both LxLxL sequences were disrupted. Additionally, we fused a synthetic EAR motif, SRDX (LDLDLELRLGFA, derived from the SUPERMAN repressor domain) or a mutated version of the artificial EAR motif, srdx (FDFDFEFRLGFA) to the C-terminus of PopP2C321A and PopP2LAAL to test the specificity of EAR motif function (Hiratsu et al., 2003).
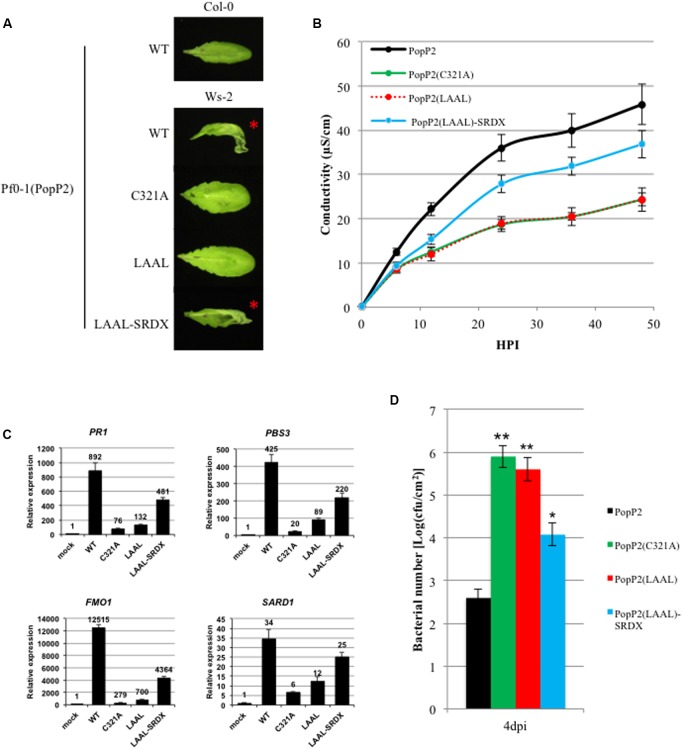
To assay for HR in Arabidopsis, we used a modified Pseudomonas fluorescens strain, Pf0-1, which carries a functional type 3 secretion system [hereafter Pf0-1(T3S)] for delivery of the PopP2 variants (Thomas et al., 2009). As was previously reported, AvrRps4N:PopP2 triggered HR at 24 hours post-infection (hpi) in the resistant Ws-2 accession, but not in the susceptible Col-0 accession when delivered by Pf0-1(T3S) (Sohn et al., 2014). The catalytic cysteine mutant, PopP2C321A, was unable to trigger HR in Ws-2 due to the loss of acetyltransferase activity required for RPS4/RRS1-mediated recognition, and this was unaffected by fusion of C-terminal SRDX or srdx (Supplementary Figure 3) (Tasset et al., 2010; Sohn et al., 2014). Interestingly, the PopP2LAAL variant with a disrupted EAR motif also failed to elicit HR in Ws-2. This suggests that PopP2 requires a functional EAR motif to trigger RPS4/RRS1-mediated HR. Indeed, fusion of the SRDX motif to the C-terminus of PopP2LAAL partially restored the ability of PopP2 to trigger HR in Ws-2, suggesting that PopP2-triggered HR is dependent on a functional EAR motif. Fusion of the mutated artificial EAR motif, srdx, to PopP2LAAL had no effect as expected (Figure 3A). PopP2 variants (WT, C321A, and LAAL) were delivered in planta as demonstrated by a secretion assay in N. benthamiana leaves (Supplementary Figure 5). In addition, we measured ion leakage to quantify the macroscopic HR symptoms and found that PopP2LAAL induced ion leakage to the same extent as the negative control, PopP2C321A. In agreement with the HR data, PopP2LAAL-SRDX induced more ion leakage than PopP2LAAL to a level intermediate between PopP2LAAL and PopP2WT (Figure 3B).
FIGURE 3.
The EAR motif is required for PopP2 avirulence activity in Arabidopsis. (A) PopP2 EAR motif is required for HR elicitation in a resistant Arabidopsis accession (Ws-2). PopP2 variants were delivered by Pseudomonas fluorescens Pf0-1(T3S) and photographs were taken 1 day after infiltration. This experiment was conducted three times with similar results. (B) PopP2 EAR motif is required for ion leakage in Ws-2. Data are means ± SE (n = 6) of one representative experiment. This experiment was conducted three times with similar results. (C) PopP2 EAR motif is required for the upregulation of defense marker genes PR1, PBS3, FMO1, and SARD1. Values shown are the average of values obtained in three independent experiments ± SE. (D) PopP2 EAR motif is required for Pto DC3000 growth restriction. Data are shown as mean colony-forming units (cfu).cm-2 ± SE (n = 5). Asterisks indicate statistically significant differences from Pto DC3000(PopP2) growth (∗P < 0.01, ∗∗P < 0.001). This experiment was conducted three times with similar results.
It became apparent that the PopP2 EAR motif was required to trigger HR in Arabidopsis. However, HR does not always correlate with the defense response leading to immune transcriptional reprogramming (Yu et al., 1998; Cawly et al., 2005; Gassmann, 2005). Therefore, we also investigated the requirement of this motif for the induction of defense-related genes and disease resistance. To this end, we analyzed expression of multiple defense marker genes known to be upregulated by bacterial T3S-delivered PopP2: PR1, FMO1, PBS3, and SARD1 (Sohn et al., 2014). The Arabidopsis accession Ws-2 was infiltrated with Pf0-1(T3S) carrying PopP2 variants and samples were taken 8 hpi for RNA extraction and qRT-PCR analysis. Consistent with the loss of HR induction by PopP2LAAL, the upregulation of all four defense genes was significantly impaired by mutation of the EAR motif. Additionally, fusion of the SRDX motif partially restored defense gene upregulation to a level more similar to PopP2WT (Figure 3C). To further test the requirement of the EAR motif for PopP2 avirulence function, we assayed bacterial growth of Pto DC3000 carrying PopP2 variants in Arabidopsis (Sohn et al., 2014). As expected from our previous findings, Pto DC3000(PopP2LAAL) did not exhibit the growth restriction that PopP2WT did, and grew to a number comparable to Pto DC3000 (PopP2C321A) (Figure 3D). Furthermore, Pto DC3000 (PopP2LAAL-SRDX) growth was partially restricted, corroborating our other evidence that PopP2 recognition in planta is dependent on a functional EAR motif. Overall, we have demonstrated that the PopP2 EAR motif is required for HR elicitation, upregulation of defense marker genes and bacterial growth restriction. The specificity of these effects of PopP2 EAR motif was further confirmed by the gain of avirulence observed with PopP2LAAL fused to a C-terminal synthetic EAR motif.
The Conserved EAR Motif Is Required for PopP2-Mediated PTI Suppression
Delivery of PopP2 but not PopP2C321Avia P. fluorescens Pf0-1(T3S) has been demonstrated to inhibit PTI as indicated by cell death induction by subsequent Pto DC3000 infiltration (Crabill et al., 2010; Badel et al., 2013; Le Roux et al., 2015). This suggested that PopP2 acetyltransferase activity is required for virulence activity to suppress host PTI (Le Roux et al., 2015). Since discovering that the conserved EAR motif was required for avirulence, we sought to investigate its requirement for PopP2 virulence activity as measured by PTI suppression in N. benthamiana leaves. Infiltration of Pf0-1(T3S) carrying PopP2 variants alone [empty vector (EV), PopP2, PopP2C321A, PopP2LAAL, PopP2LAAL-SRDX, and PopP2LAAL-SRDX] induced no cell death while infiltration of Pto DC3000 carrying each aforementioned variant induced a cell death. As previously shown, infiltration of Pf0-1(T3S)(PopP2) followed by infiltration of Pto DC3000 resulted in a cell death response due to PopP2 PTI suppression; however, infiltration of Pf0-1(T3S)(EV) or Pf0-1(T3S) (PopP2C321A) followed by infiltration of Pto DC3000 resulted in no cell death induction due to N. benthamiana PTI-induced inhibition of Pto DC3000-induced cell death. Interestingly, we discovered that Pf0-1(T3S) (PopP2LAAL) infiltration into N. benthamiana leaves was also unable to suppress host PTI, as demonstrated by the lack of cell death induction by subsequent Pto DC3000 infiltration (Supplementary Figure 4). This suggests that the EAR motif is not only required for avirulence activity but also for PopP2 virulence function. Fusion of the artificial EAR motif, SRDX, partially restored the PTI suppression ability of PopP2, whereas fusion of the mutated version, srdx, had no effect (Supplementary Figure 4).
PopP2 Does Not Interact with Known Arabidopsis Transcriptional Co-repressors in Yeast
The EAR motif is known to confer transcriptional repression activity via the recruitment of a co-repressor (Kagale and Rozwadowski, 2011). This led us to hypothesize that PopP2 recognition in Arabidopsis requires the recruitment of a transcriptional co-repressor. Therefore, we screened a library of transcriptional co-repressors for interaction with PopP2 using a LexA-based yeast-two-hybrid (Y2H) assay (Gyuris et al., 1993). The library we built comprises several members of the Groucho/Tup1-like family of co-repressors with known LisH domains and WD repeats (Supplementary Table 1) (Liu and Karmarkar, 2008). LEUNIG (LUG) and the closely related LEUNIG_HOMOLOG (LUH) as well as TPL and its close homologs TPR1–4 are the best characterized of the Gro/Tup1-like co-repressors. SEUSS (SEU) and its close homologs, SEUSS-LIKE (SLK1–2), can interact with LUG or LUH to form a functional repressor complex (Franks et al., 2002; Sitaraman et al., 2008; Grigorova et al., 2011; Shrestha et al., 2014). Finally, high expression of osmotically responsive genes 15 (HOS15) and SIN3-associated polypeptide of 18 kDa (SAP18) were included, as they are known to mediate transcriptional repression in Arabidopsis via chromatin modification (Song and Galbraith, 2006; Hill et al., 2008; Zhu et al., 2008).
Protein fusions were assayed for interaction in yeast with LEU2 and lacZ reporter genes under the control of upstream LexA operators. Yeast cells expressing empty vector controls with fusion proteins were assayed for growth and the development of blue color on the induction medium [-His(H)/-Trp(T)/-Ura(U)/-Leu(L)] + X-Gal, to test for auto-activity. TPL, TPR4, and SAP18 in the pB42-AD vector alone activated LEU2 but not lacZ allowing growth on medium lacking leucine; all other yeast cells did not activate reporter genes (Supplementary Figure 2). As expected, LUG interaction with SLK2, used as a positive control, showed strong interaction (Stahle et al., 2009). However, none of the yeast cells co-expressing PopP2-DBD and a transcriptional co-repressor-AD fusion protein showed clear protein-protein interaction in yeast cells (Supplementary Figure 2).
The EAR Motif Is Required for PopP2 Stability In Planta
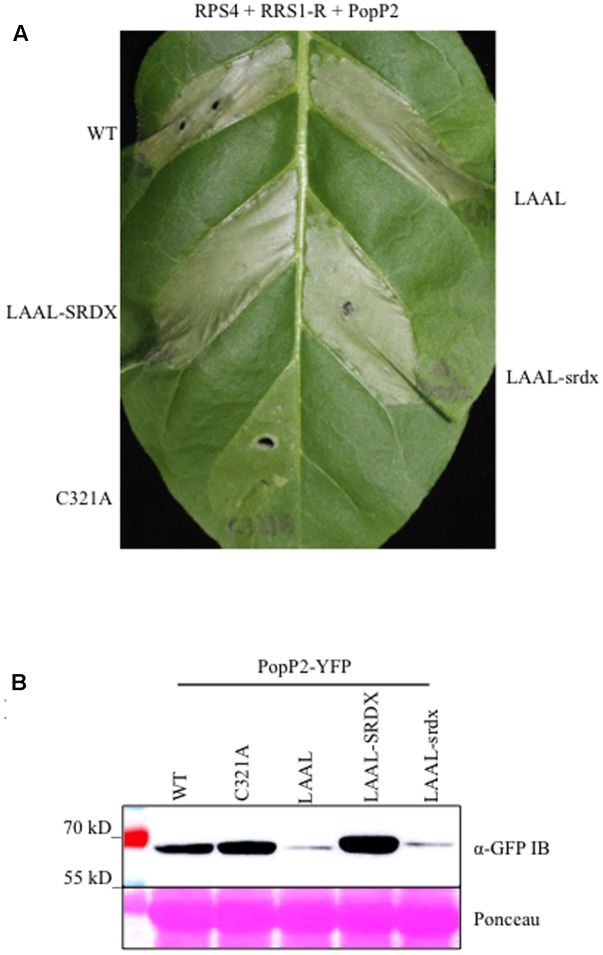
We have assayed the HR activation in response to PopP2 variants in the native Arabidopsis system; we also used a heterologous tobacco overexpression system to assay for RPS4/RRS1 mediated recognition of PopP2 variants (Figure 4A). Agroinfiltration of PopP2WT, RPS4, and RRS1 resulted in a robust PCD response at 3 dpi in tobacco. Conversely, co-expression of the inactive PopP2C321A mutant with RPS4 and RRS1 resulted in significantly reduced PCD (Sohn et al., 2014). Intriguingly, the PopP2LAAL, PopP2LAAL-SRDX, and PopP2LAAL-srdx variants all elicited a strong PCD response when co-expressed with RPS4 and RRS1 (Figure 4A). Thus, loss of PopP2 avirulence activity as a result of EAR motif disruption in Arabidopsis could not be reconstituted in the tobacco overexpression system.
FIGURE 4.
The EAR motif is required for PopP2 stability in planta. (A) Agro-mediated co-expression of the PopP2 variants with RPS4 and RRS1-R in tobacco. PopP2LAAL triggers a cell death response when co-expressed with RPS4 and RRS1-R independent of SRDX fusion. This experiment was conducted 3 times with similar results. (B) Mutation of the PopP2 EAR motif affects PopP2 protein accumulation after transient expression following agro-infiltration in N. benthamiana. The wild-type level of protein accumulation could be restored by fusion of a functional synthetic EAR motif (SRDX), but not by the mutated version (srdx). Immuno-detection of PopP2 variants C-terminally fused to YFP tag was conducted using anti-GFP antibodies. Ponceau S staining of total protein demonstrates equal loading of the samples.
To confirm the expression of PopP2 variants, sequences were fused with a C-terminal YFP epitope tag and transiently expressed in N. benthamiana. Total proteins were extracted and subjected to immuno-detection with an anti-GFP antibody. PopP2WT and PopP2C321A accumulated to similar amounts. Surprisingly, PopP2LAAL protein accumulation was significantly lower as indicated by the low intensity band on the blot (Figure 4B). Furthermore, fusion of the synthetic EAR motif restored the protein level to the WT level while fusion of the non-functional srdx motif, had no effect on protein accumulation. This evidence suggests that the stability of PopP2 inside plant cells is dependent on an EAR motif (Figure 4B).
Discussion
The R. solanacearum acetyltransferase effector, PopP2, is known to activate RPS4/RRS1-mediated resistance in certain Arabidopsis accessions (Deslandes et al., 2003; Sohn et al., 2014). PopP2 acetyltransferase activity is required for auto-acetylation and trans-acetylation of key lysine residues in the RRS1 WRKY domain and activation of the RPS4/RRS1 immune complex (Tasset et al., 2010; Le Roux et al., 2015; Sarris et al., 2015). Here, our results indicate that while the RPS4/RRS1 immune complex can recognize the naturally occurring alleles of PopP2 in Korean R. solanacearum isolates, a conserved EAR motif is necessary for its avirulence activity by regulating the protein stability in the plant cell.
PopP2 Natural Variation in Virulent Ralstonia solanacearum Strains
We surveyed the natural variation of PopP2 sequence in R. solanacearum strains isolated from diseased tomato and pepper fields at different geographic locations across the Republic of Korea. Gene-specific sequencing and subsequent analysis revealed that popP2 is highly conserved across South Korean isolates, as we identified only three polymorphic alleles among the 17 popP2-harboring strains. In fact, all but two of the polymorphisms are conservative; they result in a change to an amino acid with similar properties. However, G156D and G396E result in a change from a single hydrogen radical to a negatively charged side chain. We have shown that, despite these polymorphisms, two of the natural variants, PopP2Pe_1 and PopP2Pe_2, retained avirulence activity in Arabidopsis Ws-2. The significantly truncated variant, PopP2Pe_13, did not trigger HR. This is consistent with the previous report showing that the catalytic triad is required for PopP2 acetyltransferase activity (Tasset et al., 2010).
It must be considered that the R. solanacearum strains from which the popP2 alleles were isolated were highly virulent on pepper or tomato plants. Genome sequence analysis does not reveal any strong homology of Solanaceae disease resistance (R) genes with RPS4 and RRS1 (Consortium, 2012; Kim et al., 2014). However, the existence of PopP2Pe_13 (a truncated variant that lacks avirulence) suggests that there might be a selective pressure in natural host plants of R. solanacearum. This hypothesis is further supported by the three R. solanacearum strains that were shown to lack popP2 in our study. In this regard, it would be interesting to survey the presence/absence and sequence polymorphism of popP2 in R. solanacearum strains from other host plants or geographic regions. Furthermore, in addition to RPS4/RRS1, it is plausible that PopP2 may be recognized by other R protein(s) and that natural variants found in our study may show altered avirulence activity.
EAR Motif-Dependent Protein Stability Control
Our study illustrates the first example of a plant pathogenic effector that is dependent on an EAR motif for avirulence activity. Of note, the Xanthomonas campestris pv. vesicatoria (Xcv) effector XopD possesses two EAR motifs [sequence (L/F)DLN(L/F)(X)P] that are both required for virulence. XopD represses defense gene transcription via the two EAR motifs to enable Xcv growth in tomato (Kim et al., 2008). Likewise, we demonstrated that the PopP2 EAR motif is required for PTI suppression, which may contribute to R. solanacearum virulence. In addition, we showed that disruption of the LxLxLxL amino acid sequence rendered PopP2 unstable, but could be stabilized by addition of the synthetic EAR motif SRDX at the C-terminus. Therefore, PopP2 stability appears to depend specifically on the presence of the LxLxLxL sequence. In the tobacco heterologous system, despite being clearly reduced, accumulation of PopP2LAAL might nonetheless reach a threshold required for detection by the over-expressed RPS4 and RRS1 receptors. Conversely, in the native Arabidopsis system, we can infer that PopP2 EAR motif could play an important role in protein stability, allowing its accumulation above the necessary amount to trigger RRS1 activation.
A novel mechanism of controlling protein stability via an EAR motif has recently been unveiled. Proteasomal and non-proteasomal degradation of the ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) TF is controlled by an LxLxL EAR motif (Le et al., 2016). Similarly to the reduced accumulation of PopP2LAAL, the abundance of the ZAT12 variant carrying a mutation in the EAR motif was lower than the wild-type. Le et al. (2016) hypothesized that the ZAT12 EAR motif is involved in mediating interactions with different partners; at least one of which is H2O2 responsive and another that is a factor of proteasomal degradation targeting, such as an E3-ubiquitin ligase. Conversely, an earlier study investigating a poplar (Populus spp.) ortholog of ZAT12, Pti Cys2/His2 zinc-finger protein 1 (PtiZFP1), reported that the PtiZFP1 EAR motif promotes its degradation by the 26S proteasome through MAPK binding (Hamel et al., 2011). This is in contrast to the ZAT12 EAR motif-mediated protein stability model, but it provides additional clues to investigate the mechanism regulating PopP2 accumulation in planta.
PopP2 contributes to R. solanacearum virulence in tomato, eggplant, bean, and Arabidopsis (Macho et al., 2010; Le Roux et al., 2015). It is conceivable that successful R. solanacearum strains have acquired PopP2 variants carrying an EAR motif to stabilize the secreted effector by circumventing in planta degradation. This mechanism could have been selected to enhance the virulence of R. solanacearum on host plants. Indeed, we have shown that PopP2 stability is dependent on a functional EAR motif and that this is associated with both PopP2-mediated PTI suppression and host recognition ability.
Possible Mechanisms of PopP2 EAR Motif Function
The requirement of PopP2 for an EAR motif to trigger an avirulence response coupled with the numerous reports of EAR motif-mediated recruitment of co-repressors led us to generate yeast two-hybrid constructs of known Arabidopsis transcriptional co-repressors to screen for interaction with PopP2 in yeast cells. The Groucho (Gro)/Tup1-like family of co-repressors make up the largest and best characterized family of co-repressors in Arabidopsis with at least 13 members, including LUG and LUH, TPL and TPRs and HOS15 (Liu and Karmarkar, 2008). LUG and LUH are partially redundant transcriptional co-repressors and together with SEU/SLKs are involved in embryo and floral development and abiotic stress responses (Sitaraman et al., 2008; Shrestha et al., 2014). Similarly, HOS15 and SAP18 have not been implicated in plant defense so far, but mediate gene repression in response to cold or salt stress (Song and Galbraith, 2006; Hill et al., 2008; Zhu et al., 2008). On the other hand, the recruitment of the functionally redundant TPL and TPR1–4 by EAR motif-containing proteins in defense signaling is well-known. TPL is involved in the regulation of JA signaling for disease resistance to necrotrophic pathogens and stomatal defense (Pauwels et al., 2010; Pauwels and Goossens, 2011). Similarly, TPR1 and other TPRs associate with the R gene SNC1 to participate in transcriptional repression of negative regulators of defense (Zhu et al., 2010).
Our yeast-two-hybrid (Y2H) screen data indicate that PopP2 does not interact directly with any of the tested co-repressors in our experimental conditions. Further confirmation of this result could be obtained by testing in planta interaction between PopP2 and transcriptional co-repressors in the future. Nonetheless, based on our findings, we hypothesize that in planta PopP2 stability may be dependent on EAR motif-mediated recruitment of an as yet untested co-repressor or another host component. Identification of the host factor(s) controlling PopP2 stability shall bring new insights into the systems used to monitor pathogen virulence in the plant cell.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis accessions Col-0 and Ws-2 were grown in short day conditions (11 h light/13 h dark) at 22°C. N. benthamiana and N. tabacum W38 plants were grown in long day conditions (14 h light/10 h dark) at 25°C.
Plasmid Constructions
For Pseudomonas-mediated delivery, R. solanacearum GMI1000 PopP2149-488 variants (WT, C321A, LAAL, LAAL-SRDX, and LAAL-srdx) were cloned in the pEDV6 gateway destination vector, which is described in detail in Sohn et al. (2014). The SRDX synthetic EAR motif, LDLDLELRLGFA, is described in Hiratsu et al. (2003). The mutated SRDX, termed srdx, encodes the peptide sequence FDFDFEFRLGFA. SRDX and srdx were fused to the popP2 C-terminus with the cloning primers.
The natural polymorphic popP2 variants (Pe_1, Pe_2, and Pe_13) were amplified from R. solanacearum strains isolated from pepper in Korean fields using gene-specific primers. These were cloned into a modified Golden Gate-compatible pBBR1MCS-5 broad host range vector containing avrRps4 promoter (128 bp) and N-terminus (408 bp), as was used for the pEDV vectors (Sohn et al., 2007). popP2 variants were fused to a C-terminal 6xHA tag.
Golden Gate-compatible pLexA-DBD and pB42-AD vectors were used for the Y2H assay and coding sequences were fused to C-terminal 3xFLAG and 6xHA, respectively. popP2(149–488) was amplified from a previously reported plasmid, pCR8:popP2 (Sohn et al., 2014). Transcriptional co-repressors were amplified from Arabidopsis cDNA (accession Col-0).
The binary vector pICH86988 (provided by Sylvestre Marillonnet) was used for Agrobacterium-mediated delivery into N. tabacum and N. benthamiana. This is a Golden Gate-compatible vector in which popP2 variants, RPS4 (Arabidopsis accession No-0) and RRS1-R (Arabidopsis accession Ws-2) were assembled in fusion with C-terminal YFP, 3xHA and 3xFLAG, respectively.
For assembly into all Golden Gate-compatible vectors, genes were PCR-amplified using gene-specific primers, which also introduced BsaI recognition sequences and specific 4 bp overhangs flanking the sequences. These were cloned into the pICH41021 shuttle vector (modified pUC19 with a mutated BsaI recognition sequence). Golden Gate assembly of the resulting pICH41021 constructs into Golden Gate-compatible destination vectors was carried out to generate final constructs (Engler et al., 2008).
Bacterial Strains, Culture Conditions, and Manipulations
Escherichia coli DH5α was used to maintain and replicate plasmids, as well as for bacterial conjugation. Modified P. fluorescens Pf0-1(T3S) was used for HR assays and ion leakage assays (Thomas et al., 2009). P. syringae pv. tomato (Pto) DC3000 was used for HR assays and in planta bacterial growth assays. To transform Pf0-1(T3S) and Pto DC3000 with the appropriate construct, we used triparental mating with E. coli HB101 (pRK2013) as a helper strain (Sohn et al., 2007) or Pseudomonas electroporation (Choi et al., 2006). Agrobacterium tumefaciens strain AGL1 was transformed by electroporation with appropriate binary constructs for the transient expression assays.
Plant Pathology Experiments
For HR assays, bacteria were grown on King’s B (KB) agar containing appropriate antibiotics and harvested in 10 mM MgCl2. The final concentration of Pf0-1(T3S) suspensions was adjusted to OD600 = 0.2; the final concentration of Pto DC3000 suspensions was adjusted to OD600 = 0.1. Leaves of 5-week old Arabidopsis plants were hand-infiltrated on the abaxial surface using a blunt-end syringe, and macroscopic symptoms were observed and photographed at 20–24 hours post-infiltration (hpi). For ion leakage assays with Pf0-1(T3S), leaf disks were sampled at 0.5 hpi, washed in water for 30 min (with gentle shaking at room temperature) and transferred to fresh water (0 hpi sample). Ion leakage measurements were taken at 6, 12, 24, 36, and 48 hpi using a conductivity meter (Horiba B-173). For in planta bacterial growth assays, Pto DC3000 strains were grown and harvested as for HR assays; however, bacterial suspensions were adjusted to OD600 = 0.001. Leaves of 5-week old Arabidopsis plants were hand-infiltrated on the abaxial surface using a blunt-end syringe, and sampled at 4 days post-infiltration (dpi). Samples were ground in 10 mM MgCl2, serially diluted and spotted on KB agar containing appropriate antibiotics. These were incubated at 28°C for 2 days prior to counting colonies in order to calculate the number of colony forming units (cfu)/cm2 of infected leaf.
Yeast-Two-Hybrid (Y2H) Assays
The Saccharomyces cerevisiae strains used for the Y2H assays were EGY48 Mat(α) and RFY206 Mat(a). RFY206 carries the pSH18–34 vector, which encodes the lacZ reporter gene under the control of 8 upstream LexA operators. Additionally, pSH18–34 encodes the URA3 selectable marker, allowing growth on media lacking uracil. EGY48 and RFY206(pSH18–34) were transformed with pB42-AD and pLexA-DBD constructs, respectively, using the ‘Frozen-EZ Yeast Transformation II Kit’ according to the manufacturer’s recommendations (Zymo Research). pB42-AD encodes the TRP1 selectable marker, which allows yeast growth on media lacking tryptophan (Trp), pLexA encodes the HIS3 selectable marker, allowing growth on media lacking histidine (His). After transformation of yeast with the appropriate constructs, mating and interaction assays were performed as described in the Yeast Protocols Handbook (Clontech).
Agrobacterium-Mediated Transient Transformation of Nicotiana benthamiana and Nicotiana tabacum
Agrobacterium tumefaciens AGL1 carrying the binary constructs were grown in liquid L-medium supplemented with the appropriate antibiotics for 24 h. Cells were harvested by centrifugation and re-suspended in infiltration medium (10 mM MgCl2 and 10 mM MES-KOH pH 5.6). Suspensions were then adjusted to OD600 = 0.1. Bacterial suspensions were mixed in a 1:1 ratio and infiltrated into the abaxial surface of 5-week old N. benthamiana or N. tabacum leaves using a blunt-end syringe. Cell death was observed and photographed at 2–3 dpi.
Protein Extraction and Immunoblotting
Plant protein samples were prepared from N. benthamiana 48 h after Agrobacterium-mediated transformation. One full infiltrated leaf was ground in liquid nitrogen and total proteins were extracted in GTEN buffer (10% glycerol, 150 mM Tris-HCl pH 7.5, 1 mM EDTA, 150 mM NaCl) supplemented with 5 mM DTT, plant protease inhibitor tablet (1 tablet/50 ml extraction solution) (Roche) and 0.2% (vol/vol) Nonidet P-40. Lysates were centrifuged for 15 min at 5000 rpm at 4°C. The supernatants were filtered through miracloth mesh (Millipore) and used as input samples. Proteins were separated by SDS-PAGE and immunoblotted using anti-GFP-HRP conjugated antibodies (Santa Cruz). Proteins were detected with a mix of SuperSignal West Pico and SuperSignal West Femto chemiluminescent substrates (Pierce). Membranes were stained with Ponceau S to visualize protein loading.
Author Contributions
CS, TN, and KS conceived and designed the study. CS, TN, SC, JJ, DC, GJ, and KS carried out the experiments. HC and YL provided the R. solanacearum isolates and related information. CS, TN, and KS analyzed and interpreted the data. CS, TN, and KS prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer LD and handling Editor declared their shared affiliation, and the handling Editor states that the process met the standards of a fair and objective review.
Acknowledgments
This work was carried out with the support of Next-Generation BioGreen 21 Program (SSAC) of Rural Development Administration (PJ01188701) and the TJ Park Science Fellowship of POSCO TJ Park Foundation, South Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01330/full#supplementary-material
References
- Alfano J. R., Collmer A. (2004). Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42 385–414. 10.1146/annurev.phyto.42.040103.110731 [DOI] [PubMed] [Google Scholar]
- Badel J. L., Piquerez S. J., Greenshields D., Rallapalli G., Fabro G., Ishaque N., et al. (2013). In planta effector competition assays detect Hyaloperonospora arabidopsidis effectors that contribute to virulence and localize to different plant subcellular compartments. Mol. Plant Microbe Interact. 26 745–757. 10.1094/MPMI-06-12-0154-R [DOI] [PubMed] [Google Scholar]
- Berger S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447 407–412. 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- Büttner D., Bonas U. (2010). Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34 107–133. 10.1111/j.1574-6976.2009.00192.x [DOI] [PubMed] [Google Scholar]
- Carney B. F., Denny T. P. (1990). A cloned avirulence gene from Pseudomonas solanacearum determines incompatibility on Nicotiana tabacum at the host species level. J. Bacteriol. 172 4836–4843. 10.1128/jb.172.9.4836-4843.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawly J., Cole A. B., Király L., Qiu W., Schoelz J. E. (2005). The plant gene CCD1 selectively blocks cell death during the hypersensitive response to Cauliflower mosaic virus infection. Mol. Plant Microbe Interact. 18 212–219. 10.1094/MPMI-18-0212 [DOI] [PubMed] [Google Scholar]
- Choi K.-H., Kumar A., Schweizer H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64 391–397. 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Consortium T. G. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabill E., Joe A., Block A., Van Rooyen J. M., Alfano J. R. (2010). Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 154 233–244. 10.1104/pp.110.159723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Horvath D. M., Staskawicz B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Peeters N., Feng D. X., Khounlotham M., Boucher C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100 8024–8029. 10.1073/pnas.1230660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Theulières F., Hirsch J., Feng D. X., Bittner-Eddy P., et al. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. U.S.A. 99 2404–2409. 10.1073/pnas.032485099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11 539–548. 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Engler C., Kandzia R., Marillonnet S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3:e3647 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Somssich I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol 10 366–371. 10.1016/j.pbi.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Felix G., Duran J. D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18 265–276. 10.1046/j.1365-313X.1999.00265.x [DOI] [PubMed] [Google Scholar]
- Franks R. G., Wang C., Levin J. Z., Liu Z. (2002). SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129 253–263. [DOI] [PubMed] [Google Scholar]
- Gassmann W. (2005). Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol. Plant Microbe Interact. 18 1054–1060. 10.1094/MPMI-18-1054 [DOI] [PubMed] [Google Scholar]
- Genin S., Denny T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50 67–89. 10.1146/annurev-phyto-081211-173000 [DOI] [PubMed] [Google Scholar]
- Grigorova B., Mara C., Hollender C., Sijacic P., Chen X., Liu Z. (2011). LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138 2451–2456. 10.1242/dev.058362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust A. A., Biswas R., Lenz H. D., Rauhut T., Ranf S., Kemmerling B., et al. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282 32338–32348. 10.1074/jbc.M704886200 [DOI] [PubMed] [Google Scholar]
- Gyuris J., Golemis E., Chertkov H., Brent R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75 791–803. 10.1016/0092-8674(93)90498-F [DOI] [PubMed] [Google Scholar]
- Hamel L.-P., Benchabane M., Nicole M.-C., Major I. T., Morency M.-J., Pelletier G., et al. (2011). Stress-responsive mitogen-activated protein kinases interact with the EAR motif of a poplar zinc finger protein and mediate its degradation through the 26S proteasome. Plant Physiol. 157 1379–1393. 10.1104/pp.111.178343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S. E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53 172–185. 10.1111/j.1365-313X.2007.03336.x [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34 733–739. 10.1046/j.1365-313X.2003.01759.x [DOI] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Jones J. D., Vance R. E., Dangl J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354 aaf6395. 10.1126/science.aaf6395 [DOI] [PubMed] [Google Scholar]
- Kagale S., Links M. G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152 1109–1134. 10.1104/pp.109.151704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Rozwadowski K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6 141–146. 10.4161/epi.6.2.13627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-G., Taylor K. W., Hotson A., Keegan M., Schmelz E. A., Mudgett M. B. (2008). XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell 20 1915–1929. 10.1105/tpc.108.058529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Park M., Yeom S.-I., Kim Y.-M., Lee J. M., Lee H.-A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46 270–278. 10.1038/ng.2877 [DOI] [PubMed] [Google Scholar]
- Krogan N. T., Hogan K., Long J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139 4180–4190. 10.1242/dev.085407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie M., Shillington E., Eguiluz C., Grimsley N., Boucher C. (2002). PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant Microbe Interact. 15 1058–1068. 10.1094/MPMI.2002.15.10.1058 [DOI] [PubMed] [Google Scholar]
- Le C. T. T., Brumbarova T., Ivanov R., Stoof C., Weber E., Mohrbacher J., et al. (2016). ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) Interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) Linking Iron Deficiency and Oxidative Stress Responses. Plant Physiol. 170 540–557. 10.1104/pp.15.01589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C., Huet G., Jauneau A., Camborde L., Trémousaygue D., Kraut A., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161 1074–1088. 10.1016/j.cell.2015.04.025 [DOI] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13 137–144. 10.1016/j.tplants.2007.12.005 [DOI] [PubMed] [Google Scholar]
- Long J. A., Ohno C., Smith Z. R., Meyerowitz E. M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312 1520–1523. 10.1126/science.1123841 [DOI] [PubMed] [Google Scholar]
- Ma K.-W., Ma W. (2016). YopJ family effectors promote bacterial infection through a unique acetyltransferase activity. Microbiol. Mol. Biol. Rev. 80 1011–1027. 10.1128/MMBR.00032-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A. P., Guidot A., Barberis P., Beuzón C. R., Genin S. (2010). A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol. Plant Microbe Interact. 23 1197–1205. 10.1094/MPMI-23-9-1197 [DOI] [PubMed] [Google Scholar]
- Macho A. P., Zipfel C. (2015). Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 23 14–22. 10.1016/j.mib.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Mukaihara T., Tamura N., Iwabuchi M. (2010). Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23 251–262. 10.1094/MPMI-23-3-0251 [DOI] [PubMed] [Google Scholar]
- Nahar K., Matsumoto I., Taguchi F., Inagaki Y., Yamamoto M., Toyoda K., et al. (2014). Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum. Mol. Plant Pathol. 15 297–303. 10.1111/mpp.12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M., Kubo Y., Hatakeyama K., Imamura J., Ezura H., Nanasato Y., et al. (2013). Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS ONE 8:e55954 10.1371/journal.pone.0055954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., et al. (2009). RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60 218–226. 10.1111/j.1365-313X.2009.03949.x [DOI] [PubMed] [Google Scholar]
- Pauwels L., Barbero G. F., Geerinck J., Tilleman S., Grunewald W., Pérez A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464 788–791. 10.1038/nature08854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23 3089–3100. 10.1105/tpc.111.089300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N., Carrère S., Anisimova M., Plener L., Cazalé A.-C., Genin S. (2013). Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics 14:859 10.1186/1471-2164-14-859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro M., Cazalé A.-C., François J.-M., Parrou J.-L., Peeters N., Genin S. (2014). A Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose-6-phosphate. MBio 5 e02065–14. 10.1128/mBio.02065-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro M., Cunnac S., Barberis P., Deslandes L., Peeters N., Cazale-Noel A.-C., et al. (2009). Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant Microbe Interact. 22 538–550. 10.1094/MPMI-22-5-0538 [DOI] [PubMed] [Google Scholar]
- Robertson A. E., Wechter W. P., Denny T. P., Fortnum B. A., Kluepfel D. A. (2004). Relationship between avirulence gene (avrA) diversity in Ralstonia solanacearum and bacterial wilt incidence. Mol. Plant Microbe Interact. 17 1376–1384. 10.1094/MPMI.2004.17.12.1376 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Salanoubat M., Genin S., Artiguenave F., Gouzy J., Mangenot S., Arlat M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415 497–502. 10.1038/415497a [DOI] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2007). The JAZ proteins link jasmonate perception with transcriptional changes. Plant Cell 19 3839–3842. 10.1105/tpc.107.056960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris P. F., Duxbury Z., Huh S. U., Ma Y., Segonzac C., Sklenar J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161 1089–1100. 10.1016/j.cell.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Shrestha B., Guragain B., Sridhar V. V. (2014). Involvement of co-repressor LUH and the adapter proteins SLK1 and SLK2 in the regulation of abiotic stress response genes in Arabidopsis. BMC Plant Biol. 14:54 10.1186/1471-2229-14-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman J., Bui M., Liu Z. (2008). LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 147 672–681. 10.1104/pp.108.115923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K. H., Lei R., Nemri A., Jones J. D. (2007). The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19 4077–4090. 10.1105/tpc.107.054262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K. H., Segonzac C., Rallapalli G., Sarris P. F., Woo J. Y., Williams S. J., et al. (2014). The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 10:e1004655 10.1371/journal.pgen.1004655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé M., Popa C., Mith O., Sohn K. H., Jones J. D., Deslandes L., et al. (2012). The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant Microbe Interact. 25 941–953. 10.1094/MPMI-12-11-0321 [DOI] [PubMed] [Google Scholar]
- Song C.-P., Galbraith D. W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60 241–257. 10.1007/s11103-005-3880-9 [DOI] [PubMed] [Google Scholar]
- Stahle M. I., Kuehlich J., Staron L., von Arnim A. G., Golz J. F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21 3105–3118. 10.1105/tpc.109.070458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319 1384–1386. 10.1126/science.1151461 [DOI] [PubMed] [Google Scholar]
- Tasset C., Bernoux M., Jauneau A., Pouzet C., Briere C., Kieffer-Jacquinod S., et al. (2010). Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 6:e1001202 10.1371/journal.ppat.1001202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. J., Thireault C. A., Kimbrel J. A., Chang J. H. (2009). Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. 60 919–928. 10.1111/j.1365-313X.2009.03998.x [DOI] [PubMed] [Google Scholar]
- Xu X., Chen C., Fan B., Chen Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326. 10.1105/tpc.105.037523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.-C., Parker J., Bent A. F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. U.S.A. 95 7819–7824. 10.1073/pnas.95.13.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jeong J. C., Zhu Y., Sokolchik I., Miyazaki S., Zhu J.-K., et al. (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. U.S.A. 105 4945–4950. 10.1073/pnas.0801029105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Xu F., Zhang Y., Cheng Y. T., Wiermer M., Li X., et al. (2010). Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. U.S.A. 107 13960–13965. 10.1073/pnas.1002828107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D., Felix G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.