Abstract
An industrial waste, 1,2,3-trichloropropane (TCP), is toxic and extremely recalcitrant to biodegradation. To date, no natural TCP degraders able to mineralize TCP aerobically have been isolated. In this work, we engineered a biosafety Pseudomonas putida strain KT2440 for aerobic mineralization of TCP by implantation of a synthetic biodegradation pathway into the chromosome and further improved TCP mineralization using combinatorial engineering strategies. Initially, a synthetic pathway composed of haloalkane dehalogenase, haloalcohol dehalogenase and epoxide hydrolase was functionally assembled for the conversion of TCP into glycerol in P. putida KT2440. Then, the growth lag-phase of using glycerol as a growth precursor was eliminated by deleting the glpR gene, significantly enhancing the flux of carbon through the pathway. Subsequently, we improved the oxygen sequestering capacity of this strain through the heterologous expression of Vitreoscilla hemoglobin, which makes this strain able to mineralize TCP under oxygen-limited conditions. Lastly, we further improved intracellular energy charge (ATP/ADP ratio) and reducing power (NADPH/NADP+ ratio) by deleting flagella-related genes in the genome of P. putida KT2440. The resulting strain (named KTU-TGVF) could efficiently utilize TCP as the sole source of carbon for growth. Degradation studies in a bioreactor highlight the value of this engineered strain for TCP bioremediation.
Introduction
Synthetic biology has become a powerful tool to construct complete heterologous metabolic pathways in a host cell by functional assembly of various enzymes from different organisms1. Currently, most studies have focused on implantation of diverse synthetic pathways into microbial cells to synthesize high value-added products2. Although synthetic biology tools are not widely applied for engineering the organics catabolic pathways in microorganisms so far, they have enormous potential for rational tuning of pathways for efficient degradation of environmental pollutants.
A toxic persistent pollutant, 1,2,3-trichloropropane (TCP), has been recognized as an emerging contaminant in groundwater. The global yield of TCP can reach about 50,000 tons annually, and TCP is widely used as a solvent and as building block for the synthesis of other chemicals3. TCP is frequently detected in groundwater due to improper waste disposal, its moderate water solubility, and its recalcitrance to biodegradation4. TCP contamination of groundwater poses a serious threat to drinking water sources and human health. Therefore, there is an urgent need to develop efficient technologies for the remediation of TCP-contaminated sites.
Disposal of TCP by physical or chemical methods is expensive. Bioremediation, which is a simple, safe and cost-effective technique to fight pollution, utilizes and manipulates the biodegradation abilities of living organisms to transform toxic organic pollutants into harmless products5. To date, there are no reports on the aerobic degradation of TCP by natural microorganisms. Metabolic engineering has opened up new avenues for the evolution of efficient degradation pathways6, which allows the construction of recombinant microorganisms with the capability to degrade TCP under aerobic conditions.
Haloalkane dehalogenases (DhaA) catalyze the dehalogenation of TCP to the (R) and (S) enantiomers of 2,3-dichloropropane-1-ol (DCP)7. To improve the catalytic efficiency and enantioselectivity, two DhaA mutants8, 9, DhaA31 and DhaA90R, were generated by directed evolution. Furthermore, haloalcohol dehalogenases (HheC) and epoxide hydrolases (EchA) are responsible for the conversion of DCP to glycerol (GLY)10, 11. Recently, a synthetic biodegradation pathway capable of aerobic biotransformation of TCP into harmless GLY was assembled by the heterologous expression of DhaA, HheC and EchA in Escherichia coli 12, 13. However, expression of the synthetic pathway is dependent on IPTG induction, which is not suitable for practical application in the large-scale degradation of TCP. Moreover, the plasmid-based expression systems tend to lose the introduced heterologous genes in the absence of selective pressure and the diffusion of antibiotic resistance markers on plasmids poses potential risks to the environment. More importantly, lab-born E. coli strains are not regarded as good candidates for in situ bioremediation due to their strict nutrient demand, poor adaptability and weak competitiveness. In another study, a dhaA31 gene encoding DhaA31 was integrated into the chromosome of the DCP-degrading bacterium Pseudomonas putida MC4 using a transposon delivery system4. However, the engineered strain needs to be intensively researched with regard to its biosafety prior to its application for in situ bioremediation due to legislative barriers on the field release of genetically engineered microorganisms.
The GRAS (generally recognized as safe) strain P. putida KT2440 is considered as a potential agent for environmental bioremediation of industrial waste. The genome of P. putida KT2440 has been sequenced and multiple tools for genome editing have been devised and implemented14–18, which have laid a foundation for the metabolic engineering of P. putida KT2440. Recently, P. putida KT2440 has been highlighted as a robust metabolic chassis for catabolic pathway assembly19.
In this work, we engineered P. putida KT2440 for the efficient degradation of TCP using combinatorial engineering strategies by carrying out the following four tasks: (1) constructed a synthetic pathway for the conversion of TCP into GLY (Fig. 1), in particular, prochiral TCP was converted predominantly into (R)-DCP (ee 90%) by the enantioselective DhaA90R and the accumulation of (S)-DCP in the pathway caused by the poor activity of HheC toward (S)-DCP was avoided; (2) deleted the glpR gene to eliminate the growth lag-phase of using GLY as a carbon source; (3) enhanced aerobic metabolism by promoting oxygen delivery through the heterologous expression of Vitreoscilla hemoglobin (VHb); (4) increased intracellular energy charge (ATP/ADP ratio) and reducing power (NADPH/NADP+ ratio) by deleting the flagellar operon (a ~ 70 kb DNA segment of the genome). Through the engineering of TCP biodegradation pathway, the enhancement of oxygen transport, and the modulation of energy status and reducing power availability, the resulting strain could mineralize TCP aerobically and utilize TCP as the sole source of carbon for growth.
Figure 1.
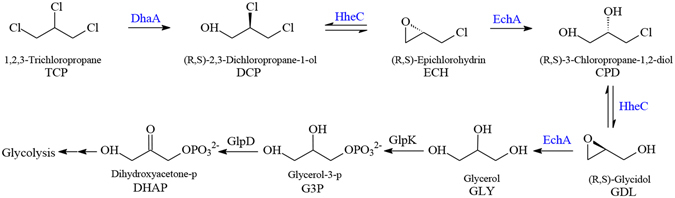
Construction of a synthetic pathway for aerobic mineralization of TCP in P. putida KT2440. Enzyme sources: DhaA from Rhodococcus rhodochrous NCIMB 13064, HheC from Agrobacterium radiobacter AD1, and EchA from Agrobacterium radiobacter AD1.
Results and Discussion
Construction of a synthetic TCP mineralization pathway in P. putida KT2440
Currently, synthetic biology approaches can be applied for implantation of the synthetic biodegradation pathways into an ideal host strain for bioremediation20. In this work, a synthetic pathway for aerobic mineralization of TCP comprising DhaA from Rhodococcus rhodochrous NCIMB 13064, HheC from Agrobacterium radiobacter AD1, and EchA from Agrobacterium radiobacter AD1 was assembled in P. putida KT2440 (Fig. 1). All these genes (dhaA90R, hheC and echA) were integrated into the chromosome of P. putida KT2440 using a previously developed genome editing method with upp as a counter-selectable marker16. To improve the efficiency of the synthetic biodegradation pathway, the vgb gene was inserted into the P. putida chromosome, and both the glpR gene and the flagellar operon were knocked out. The resulting mutant strain was designated as P. putida KTU-TGVF. The successful construction of strain KTU-TGVF was verified by PCR and DNA sequencing. The correct amplicons were obtained by PCR with chromosomal DNA of strain KTU-TGVF as the template (Fig. S1). The nucleotide sequences of the inserted DNA fragments on chromosome were in accordance with those of four synthetic gene cassettes (Fig. S2).
The poor activity of DhaA toward TCP and cellular toxicity of TCP and intermediates represent primary bottlenecks of the TCP mineralization pathway, limiting the flux of carbon through the pathway3, 12. To overcome the bottlenecks of aerobic TCP mineralization, previous studies focused on engineering of DhaA, the first enzyme of the TCP mineralization pathway. Two DhaA mutants, DhaA31 and DhaA90R, were obtained using computer-assisted directed evolution. Mutant DhaA318 showed 26-fold higher catalytic efficiency toward TCP than the wild-type enzyme (DhaAwt: k cat = 0.08 s−1, K m = 2.2 mM, k cat/K m = 36 M−1 s−1; DhaA31: k cat = 1.26 s−1, K m = 1.2 mM, k cat/K m = 1050 M−1 s−1), while the (S)-DCP was accumulated in the pathway because of equimolar production of (R)-DCP and (S)-DCP by the non-enantioselective DhaA31 and the poor activity of HheC toward (S)-DCP. Mutant DhaA90R9 possessed similar catalytic efficiency toward TCP as the wild-type enzyme (DhaA90R: k cat = 0.16 s−1, K m = 6.5 mM, k cat/K m = 25 M−1 s−1), while DhaA90R converted prochiral TCP predominantly into (R)-DCP (ee 90%), which is the preferred substrate for the enantioselective HheC. In a previous study, the synthetic pathway with DhaA90R reconstructed in E. coli BL21 (DE3) failed to provide sufficient carbon flux to support bacterial growth in minimal medium with TCP12. In another study, a novel TCP biodegradation pathway including a chromosome-borne dhaA31 gene and an intrinsic nonselective DCP degradation pathway was reconstructed in a natural DCP-degrading bacterium P. putida MC4. Because of cellular toxicity of TCP and intermediates, the engineered P. putida MC4 showed a prolonged growth lag-phase when cultured in minimal medium with TCP4. To construct a practically applicable TCP-degrading bacterium, in this study, a robust environmental bacterium P. putida KT2440, which is resistant to toxic organic solvents, was chosen as a host strain for reconstructing the TCP mineralization pathway. To avoid the accumulation of (S)-DCP in the pathway, in this study, the enantioselective DhaA90R was selected for the dehalogenation of TCP to DCP. As discussed later in detail, the insufficient carbon flux through the pathway caused by the less active DhaA90R was further improved by eliminating the growth lag-phase of using GLY as a carbon source, enhancing aerobic metabolism, and increasing intracellular energy charge and reducing power.
Heterologous expression of the synthetic TCP mineralization pathway in P. putida KT2440
To achieve the optimal expression levels, the optimized gene expression regulatory elements21, including a strong constitutive promoter, two Shine-Dalgarno sequences, and a terminator, were designed for controlling the transcription and translation of exogenous genes (Fig. S3). In order to verify whether these exogenous genes (dhaA90R, hheC, echA and vgb) are transcribed to mRNA in P. putida KTU-TGVF, RT-PCR assays were performed. As expected, these target products were obtained by PCR with cDNA or genomic DNA as template. In contrast, no PCR products were obtained with mRNA or ddH2O as template (Fig. S4). These results indicated that transcription of these exogenous genes had occurred in P. putida KTU-TGVF.
Production of DhaA, HheC and EchA in KTU-TGVF cells was demonstrated by Western blot analysis. Immunoreactive bands, which matched well with the theoretical molecular weights of DhaA (34 kDa), HheC (29 kDa) and EchA (35 kDa), were detected in whole-cell lysates using antibodies against DhaA, HheC and EchA (Fig. S5).
Function validation for the synthetic TCP mineralization pathway
The functionality of DhaA, HheC and EchA in KTU-TGVF cells was validated by GC-MS analyses of resting-cell transformation products. Resting cells of KTU-T1 containing dhaA90R produced a new chromatographic peak with a retention time (RT) of 11.7 min when incubated with TCP, and this peak was identified as DCP by comparing its MS pattern with that of DCP standard (Fig. 2A). Transformation of DCP by resting cells of KTU-T2 containing hheC produced a new chromatographic peak with a RT of 5.1 min, which had the same RT and MS pattern as ECH standard (Fig. 2B). After the transformation of ECH by resting cells of KTU-T3 containing echA, a new chromatographic peak appeared at a RT of 12.3 min, the RT and MS pattern of this peak were the same as those of CPD standard (Fig. 2C). Therefore, KTU-T1, KTU-T2 and KTU-T3 cells expressed functional DhaA, HheC and EchA, respectively.
Figure 2.

(A) GC-MS analysis of transformation of TCP by resting cells of P. putida KTU-T1. Two chromatographic peaks representing TCP and DCP had a RT of 11.3 min and 11.7 min, respectively. (B) GC-MS analysis of transformation of DCP by resting cells of P. putida KTU-T2. Two chromatographic peaks corresponding to ECH and DCP appeared at a RT of 5.1 min and 11.7 min, respectively. (C) GC-MS analysis of transformation of ECH by resting cells of P. putida KTU-T3. Two chromatographic peaks representing ECH and CPD had a RT of 5.1 min and 12.3 min, respectively.
TCP degradation experiment was performed with KTU-TGVF in M9 minimal medium supplemented with 0.5 mM TCP. As shown in Fig. 3A, 0.5 mM TCP was almost completely converted to GLY and the vast majority of GLY were further metabolized within 32 h, as revealed by quantification of DCP, ECH, CPD and GLY throughout the experiment. Accompanying with TCP degradation, cell growth was observed (Fig. 3B), which indicated that this strain could utilize TCP as the sole source of carbon for growth. In contrast, the concentration of TCP did not change and no growth was observed when inoculated with KTU. These results indicated that TCP could be converted to GLY by the heterologous pathway assembled in P. putida KT2440 (Fig. 1). Furthermore, GLY could be utilized as the carbon source for cell growth via the intrinsic pathway of KT2440. Taken together, we concluded that the pathway for aerobic mineralization of TCP was functionally assembled in P. putida KT2440.
Figure 3.
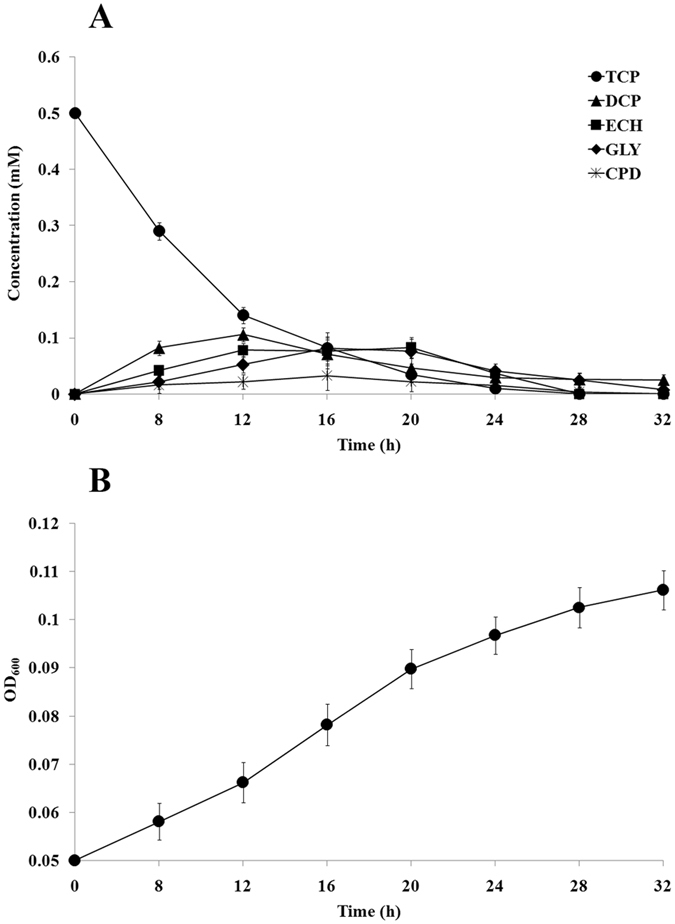
(A) Aerobic mineralization of TCP by P. putida KTU-TGVF. P. putida KTU-TGVF was incubated in M9 minimal medium supplemented with 0.5 mM TCP in a shaking incubator at 200 rpm and 30 °C. The initial inoculum density was OD600 = 0.05. TCP, DCP, ECH and CPD were quantified by GC-MS. GLY was quantified by colorimetric analysis. (B) Growth curve of P. putida KTU-TGVF. The OD600 was measured to estimate cell growth. Bars represent the mean values ± standard deviation of triplicate measurements from three independent experiments.
Elimination of the growth lag-phase of using GLY as a carbon source
GlpR functions as a negative regulator for controlling the utilization of GLY as a growth precursor in P. putida KT2440 and the regulatory mechanisms have been elucidated22. Deletion of glpR may eliminate the growth lag-phase of using GLY as a carbon source. In this work, the strain KTU-TGVF (ΔglpR) could grow aerobically without the lag-phase in minimal medium with TCP as the sole carbon source. In contrast, no growth was observed with the strain KTU-T123 (glpR +) when using TCP as the sole carbon source. These results suggest that the efficient utilization of GLY may support bacterial growth, alleviate cellular toxicity of TCP and intermediate metabolites and improve the flux of carbon through the TCP mineralization pathway.
VHb enhances TCP mineralization under oxygen-limited conditions
CO-difference spectrum assays were used to detect VHb activity. The cell extracts from the strain KTU-TGVF showed a characteristic absorption peak at 420 nm when fed with CO, while the peak was not observed with the cell extracts from the strain KTU fed with CO (Fig. S6). These results indicated that the strain KTU-TGVF expressed functional VHb.
VHb delivers the O2 to the respiratory chain, enhancing ATP generation and NADH consumption at low O2 concentrations. This process could improve the efficiency of bacterial aerobic respiration, increase the carbon flux through the tricarboxylic acid cycle and improve the aerobic growth of bacteria. Heterologous expression of VHb in a variety of hosts has been shown to improve cell growth, protein synthesis, metabolite productivity, and bioremediation under oxygen-limited conditions23, 24. In a previous study, integrating vgb into the chromosomes of P. aeruginosa and Burkholderia sp. strain DNT could improve growth and degradation of 2,4-dinitrotoluene or benzoic acid under hypoxic conditions25. In another study, P. putida KT2440 was engineered for the anoxic biodegradation of 1,3-dichloroprop-1-ene by introducing various genes retrieved from facultative anaerobe and aerotolerant bacteria26.
Sufficient oxygen is the key factor for complete oxidative mineralization of TCP. The success of aerobic mineralization of TCP in oxygen-restricted environments will largely depend on the oxygen sequestering capability of TCP-degrading bacteria. In this work, significant growth was observed for the strain KTU-TGVF expressing VHb under oxygen-limited conditions, while no growth was observed for the strain KTU-TGF without VHb expression under oxygen-limited conditions (Fig. 4). These results suggest that VHb may improve the ability of P. putida KT2440 to compete for limited oxygen in hypoxic environments, thus making this bacterium more competitive in actual environments such as a packed-bed bioreactor. This study not only underscores the value of P. putida KT2440 as a versatile biocatalyst for biotransformation under oxygen-limited conditions but also highlights the value of metabolic engineering for expanding the catalytic repertoire of P. putida KT2440.
Figure 4.
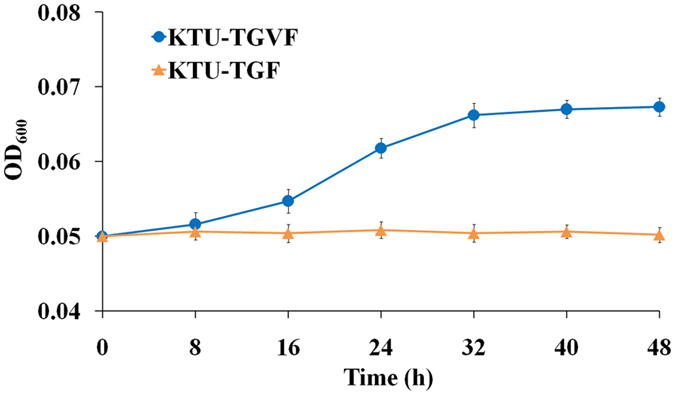
Growth curves of P. putida KTU-TGF and KTU-TGVF under oxygen-limited conditions. P. putida strains were incubated in 25 ml glass vials with a screw cap mininert valve containing 20 ml of M9 minimal medium supplemented with 0.2 mM TCP in a shaking incubator at 80 rpm and 30 °C. Cell growth was estimated by measuring the OD600 of the culture broth. Bars represent the mean values ± standard deviation of triplicate measurements from three independent experiments.
Increased intracellular ATP and NADPH levels improve aerobic mineralization of TCP
Both the production and the motion of flagella are energy-consuming processes for the cell. In E. coli, flagellar production consumes about 2% of the biosynthetic energy expenditure of the cell, while flagellar motion demands about 0.1% of the total energy cost27. In this work, a non-flagellated strain KTU-TGVF was generated from the original strain KTU-TGV by deleting the flagellar operon in the genome. Accordingly, the flagellar operon knockout mutant strain showed the complete absence of the flagellum structure when observed using TEM after negative staining. In contrast, the flagella were observed in the wild-type strain KT2440 (Fig. S7). These results indicated that the flagellar synthesis was completely blocked in the mutant strain.
It has been reported that the non-flagellated strain KT2440 showed increased intracellular ATP and NADPH levels28. In our study, the ATP/ADP ratio in the strain KTU-TGVF was 1.3-fold higher than that in the strain KTU-TGV (Fig. 5A). Since the ATP levels in bacterial cells are tightly regulated, a difference of 30% could significantly influence cellular functions. These results indicated that eliminating the consumption required for flagella synthesis and rotation greatly changed the energy status of the cells.
Figure 5.

Determination of the ATP/ADP (A) and NADPH/NADP+ (B) molar ratios in P. putida. The intracellular levels of ADP, ATP, NADP+ and NADPH were determined as described in Materials and methods. Bars represent the mean values ± standard deviation of triplicate measurements from three independent experiments.
Except for the energy consumption, a considerable amount of reducing power, mainly in the form of NADPH, is indispensable for basic anabolic processes, such as the biosynthesis of building blocks for biomass29. In this work, the strain KTU-TGVF had a 1.2-fold higher NADPH/NADP+ ratio than did the strain KTU-TGV (Fig. 5B). Furthermore, the growth kinetics of the strain KTU-TGVF and KTU-TGV were compared. The strain KTU-TGVF showed a higher growth rate in M9 minimal medium containing 0.5 mM TCP than did the strain KTU-TGV. The KTU-TGVF and KTU-TGV cultures reached a maximum OD600 of 0.104 and 0.088 at 32 h, respectively (Fig. S8). Obviously, the strain KTU-TGVF could utilize TCP more efficiently as the sole carbon source for growth, which may be a reflection of increased resistance to cellular toxicity of TCP and intermediate metabolites. Increased resistance to toxic substrates may be due in part to enhanced anabolic capability resulting from the increase of ATP and NADPH levels within the cell. Taken together, the efficient mineralization of TCP by the strain KTU-TGVF may be attributed to the efficient utilization of GLY as a growth precursor and the increased intracellular ATP and NADPH levels.
Treatment of TCP-contaminated water in a lab-scale bioreactor
The flagella-deficient strain KTU-TGVF showed a distinct change in biofilm formation. The quantities of biofilm formed by the strain KTU-TGVF were 4.3- and 9.5-fold higher at 24 and 96 h, respectively, than those formed by the strain KTU-TGV, as judged by the CV staining assay (Fig. S9), when these strains were grown in M9 minimal medium containing 0.4% (w/v) glucose. The observation is in agreement with a previous report28, in which the non-flagellated P. putida KT2440 cells form more biofilm than the wild-type KT2440 cells. Recently, the biofilm formation capability of P. putida KT2440 was improved for boosting biodegradation of haloalkanes by the modulation of the intracellular c-di-GMP level30. Biofilm offers protection against a hostile environment and helps bacteria persist within the environment. Therefore, the strain KTU-TGVF has potential for use in a packed-bed bioreactor treating TCP-contaminated water because its high biofilm formation ability is favorable for the immobilization of cells on the support material in wastewater treatment.
To explore the feasibility of using strain KTU-TGVF for TCP bioremediation, we designed a packed-bed bioreactor and tested the removal of TCP from wastewater streams in a consecutive process (Fig. S10). KTU-TGVF cells were grown in M9 minimal medium containing 0.5 mM TCP plus 0.4% glucose to OD600 = 0.4 and were subsequently inoculated into the reactor for immobilization on ceramic rings. Previous studies have shown that ceramic rings as the support material are suitable for the immobilization of microbial cells4, 31. After inoculation, the reactor was operated under fed-batch conditions for 4 days to promote attachment of the cells to the support material. Within a period of 4 to 18 days, TCP (0.05 to 0.2 mM) was supplied continuously at a rate of 0.1 ml/min. During the operating period, the maintenance of a high hydraulic retention time (HRT, 133 h) may be favorable for cell growth and biofilm formation. HRT is considered as an important operating parameter directly influencing the bioreactor performance4, 31. After 18 days, the influent concentration of TCP was maintained at 0.2 mM, and the reactor was continuously operated for another 30 days. The reactor performance was evaluated by continuous monitoring of the effluent concentrations of TCP, DCP and chloride (Fig. 6). Under these operating conditions, the removal efficiencies of TCP were 95 to 97% and quantitative release of chloride (TCP/chloride, molar ratio 1:3) was observed. We detected minute quantities of DCP in the reactor effluent. When inoculated with the strain KTU, the TCP effluent concentration remained unchanged relative to the influent concentration.
Figure 6.
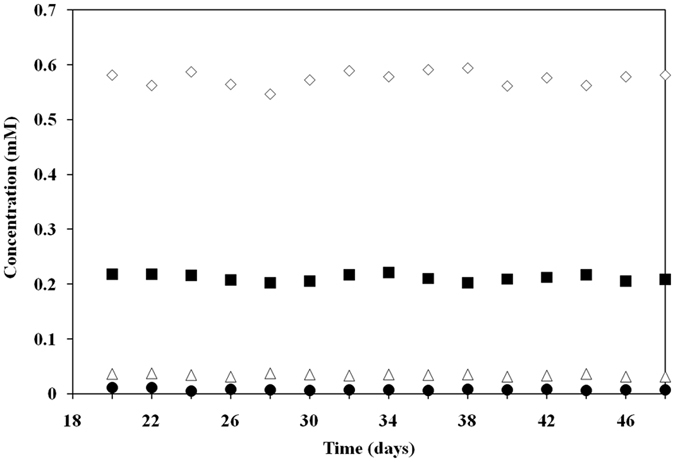
Degradation of TCP by P. putida KTU-TGVF in a lab-scale bioreactor. Reactor performance was assessed by continuously monitoring the effluent concentrations of TCP, DCP and chloride during a 30-day operating period. Symbols: ◾, TCP influent; ⚫, TCP effluent; ▵, DCP effluent; ◊, chloride effluent.
Cells from the reactor effluent were spread on LB agar plates and colonies formed were checked for their identity. All 20 colonies tested were identified as P. putida KT2440 by the 16 S rRNA gene sequencing. Furthermore, both the four gene insertions (dhaA90R +, hheC +, echA +, vgb +) and the two gene deletions (∆glpR, ∆flagellar operon) were detected by PCR from all 20 colonies tested, and the detection results of one colony are shown in Fig. S11. These results demonstrated that the inoculated KTU-TGVF cells thrived and accounted for the observed TCP degradation during a 30-day operating period in the bioreactor. In the future, the efficient TCP-mineralizing strain KTU-TGVF coupled with a field-scale reactor has enormous potential to be applied for the treatment of industrial wastewater containing TCP.
Conclusions
In this work, an efficient TCP-mineralizing strain was created using combinatorial engineering strategies. In brief, we first integrated a synthetic pathway for aerobic TCP mineralization into the chromosome of P. putida KT2440 by functional assembly of various TCP-degrading enzymes from different bacteria. We then enhanced the utilization of TCP as a carbon source by eliminating the growth lag-phase of using GLY as a growth precursor. We additionally engineered the strain with the ability to mineralize TCP under oxygen-limited conditions through the heterologous expression of VHb. Lastly, we further improved the utilization of TCP by increasing intracellular ATP and NADPH levels resulting from deletion of the flagellar operon. We envision that these combinatorial engineering approaches could provide insights into devising engineering strategies to improve the degradation of anthropogenic chemicals by P. putida.
Materials and Methods
Reagents, strains, and culture conditions
TCP, DCP, epichlorohydrin (ECH), 3-chloropropane-1,2-diol (CPD) and GLY (99% pure analytical grade) were purchased from Alta Scientific Co., Ltd., Tianjin, China. All the other chemical reagents were of analytical grade and purchased from Dingguo Biotechnology Co. Ltd., Tianjin, China.
E. coli strains were grown at 37 °C in LB medium supplemented with 50 μg/ml kanamycin (Kan) if necessary. P. putida strains were grown at 30 °C in M9 minimal medium16 containing 0.5 mM TCP as well as in LB medium32 supplemented with 50 μg/ml Kan or 20 μg/ml 5-fluorouracil (5-FU) when required.
DNA manipulation and strain constructions
Nine P. putida mutant strains with multiple gene insertions and/or deletions were constructed as described previously using a chromosomal scarless modification strategy19 and a suicide plasmid pK18mobsacB33. The detailed procedures for the construction of these nine P. putida mutant strains are described in Supplementary methods. The detailed information on the inserted exogenous genes (dhaA90R, hheC, echA and vgb) and deleted target genes (glpR and flagellar operon) is shown in Table 1. These four gene cassettes (dhaA90R, hheC, echA and vgb) were chemically synthesized by Genscript Co. Ltd., Nanjing, China and their nucleotide sequences are shown in Fig. S2. The strains, plasmids, and primers used in this study are listed in Table 2.
Table 1.
Information on the heterologous genes and their chromosomal insertion sites in P. putida KT2440.
| Gene | Length (bp) | Amino acid residues | Function | Strains (GenBank accession no.) | Insertion site/targeted deletion |
|---|---|---|---|---|---|
| dhaA90R | 882 | 294 | haloalkane dehalogenase | Rhodococcus rhodochrous NCIMB13064 (AF060871) | PP_1277 (algA) |
| hheC | 765 | 255 | haloalcohol dehalogenase | Agrobacterium radiobacter AD1 (AF397296) | PP_1278/PP_1279 (algF/algJ) |
| echA | 885 | 295 | epoxide hydrolase | Agrobacterium radiobacter AD1 (Y12804) | PP_1280 (algI) |
| glpR | 756 | 252 | glycerol-3-phosphate regulon repressor | Pseudomonas putida KT2440 (AE015451) | PP_1074 |
| vgb | 441 | 147 | hemoglobin | Vitreoscilla sp. HG1 (AF292694) | PP_3356 (fcs) |
| flagellar operon | 68139 | 69 genes | flagella synthesis | Pseudomonas putida KT2440 (AE015451) | PP_4329-PP_4397 |
Table 2.
Strains, plasmids, and primers used in this study.
| Strain, plasmid or primer | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| Trans1 T1 | F−, φ80 (lacZ), ΔM15, ΔlacX74, hsdR (rK −, mK +), ΔrecA1398, endA1, tonA | Transgen |
| P. putida | ||
| KT2440 | Wild type | ATCC 47054 |
| KTU | upp-deficient KT2440 | This study |
| KTU-T1 | KT2440 mutant (Δupp, ΔalgA, dhaA90R +) | This study |
| KTU-T2 | KT2440 mutant (Δupp, ΔalgF&algJ, hheC +) | This study |
| KTU-T3 | KT2440 mutant (Δupp, ΔalgI, echA +) | This study |
| KTU-T12 | KT2440 mutant (Δupp, ΔalgA, ΔalgF&algJ, dhaA90R +, hheC +) | This study |
| KTU-T123 | KT2440 mutant (Δupp, ΔalgA, ΔalgF&algJ, ΔalgI, dhaA90R +, hheC +, echA +) | This study |
| KTU-TG | KT2440 mutant (Δupp, ΔalgA, ΔalgF&algJ, ΔalgI, ΔglpR, dhaA90R +, hheC +, echA +) | This study |
| KTU-TGV | KT2440 mutant (Δupp, ΔalgA, ΔalgF&algJ, ΔalgI, ΔglpR, Δfcs, dhaA90R +, hheC +, echA +, vgb +) | This study |
| KTU-TGF | KT2440 mutant (Δupp, ΔalgA, ΔalgF&algJ, ΔalgI, ΔglpR, Δflagellar operon, dhaA90R + , hheC + , echA +) | This study |
| KTU-TGVF | KT2440 mutant (Δupp, ΔalgA, ΔalgF&algJ, ΔalgI, ΔglpR, Δfcs, Δflagellar operon, dhaA90R + , hheC + , echA + , vgb +) | This study |
| Plasmids | ||
| pK18mobsacB | Kanr, suicide plasmid for gene knockout | 33 |
| pKU | Kanr, pK18mobsacB containing upp gene | This study |
| pKU-T1 | Kanr, pK18mobsacB containing upp and dhaA90R | This study |
| pKU-T2 | Kanr, pK18mobsacB containing upp and hheC | This study |
| pKU-T3 | Kanr, pK18mobsacB containing upp and echA | This study |
| pKU-G | Kanr, pK18mobsacB containing upp and ΔglpR | This study |
| pKU-V | Kanr, pK18mobsacB containing upp and vgb | This study |
| pKU-F | Kanr, pK18mobsacB containing upp and Δflagellar operon | This study |
| Primers | ||
| T1-1 | 5′-TTCTGGTGGTAGCGGTTCCCGTCTG-3' | This study |
| T1-2 | 5′-CTGCGCGATGGTCTTCACCGAAACG-3' | This study |
| T2-1 | 5′-GATGACCCTGCTCAACGGCAAGCTG-3' | This study |
| T2-2 | 5′-GGTCAGTTGCTGGTCGCTCAGGTTC-3' | This study |
| T3-1 | 5′-CCTGTTCCTGTTCTTGCCGATCTTC-3' | This study |
| T3-2 | 5′-CTGGAAGTAAAGGAACGGCGAGTAG-3' | This study |
| G-1 | 5′-CCCGAATTCCGAGTATTTCGCCAGCAAGG-3' | This study |
| G-2 | 5′-GCGGCATGCATGCCGGACAAATTCTGCAAT-3' | This study |
| G-3 | 5′-TTGTCCGGCATGCATGCCGCCAGACTTTTG-3' | This study |
| G-4 | 5′-CCCGAATTCAGGTGGTCGTAGAGGAACAG-3' | This study |
| V-1 | 5′-GCCCGGACACCACTTTCATC-3' | This study |
| V-2 | 5′-CTTGGGCATCGCGGTTCAAG-3' | This study |
| F-1 | 5′-AGACTTCCATTGCCAAAGCCCTCAC-3' | This study |
| F-2 | 5′-ACTGCGCGATGGTCTTCACCGAAAC-3' | This study |
| F-3 | 5′-AGACTTCCATTGCCAAAGCCCTCAC-3' | This study |
| F-4 | 5′-ACTGCGCGATGGTCTTCACCGAAAC-3' | This study |
| T1-F | 5′-CAGCTACCTGTGGCGCAACATCATC-3' | This study |
| T1-R | 5′-GTGCAGCCAGTTCATGTAGGCTTCC-3' | This study |
| T2-F | 5′-CACGACGAGAGCTTCAAGCAGAAGG-3' | This study |
| T2-R | 5′-AGAAGTACGGGCTGTCTTCGCTGTG-3' | This study |
| T3-F | 5′-CCTGAGCAAGTACAGCCTGGACAAG-3' | This study |
| T3-R | 5′-GTACTTCGGCACGAACTCGATCAGC-3' | This study |
| V-F | 5′-ATGTTAGACCAGCAAACCATTAACA-3' | This study |
| V-R | 5′-TTATTCAACCGCTTGAGCGTACAAA-3' | This study |
Transformation of plasmid into P. putida was carried out using the electroporation method34. Screening of P. putida mutant strains was performed as described previously19. In brief, the single-crossover recombinants were screened by incubating at 30 °C for 24 h on LB agar plates supplemented with 50 μg/ml Kan. The selected recombinants were then incubated at 30 °C for 24 h in LB medium. To further screen the double-crossover recombinants, the culture broths that had been diluted to 10−5 were spread on LB agar plates supplemented with 20 μg/ml 5-FU. The selected recombinants showing 5-FUr and Kans were further checked by PCR using primers listed in Table 2. All the constructed mutant strains were validated by DNA sequencing.
RT-PCR assays
Total RNA was extracted from KTU-TGVF cells using a RNApure Bacteria Kit (CWBIO, China) and treated with DNase I at 37 °C for 30 min. RNA quality was checked by an Agilent 2100 Bioanalyzer. cDNA was synthesized with 0.5 μg total RNA as template using a PrimeScript RT Master Mix Kit (TaKaRa, Japan). PCR was carried out with cDNA as the template using PrimeSTAR HS DNA polymerase (TaKaRa) and primers listed in Table 2 on an ABI 2720 thermal cycler (Applied Biosystems). Genomic DNA, mRNA or ddH2O was used as the template in the control reactions. PCR products were detected by agarose gel electrophoresis.
Resting-cell transformation assays
To verify the functionality of DhaA, HheC and EchA in P. putida, transformation assays were carried out with resting cells of KTU-T1, KTU-T2 or KTU-T3. Cells were pre-grown in M9 minimal medium containing 0.4% glucose at 200 rpm and 30 °C for 24 h, harvested by centrifugation, washed twice with 50 mM potassium phosphate buffer (pH 7.2), and resuspended (OD600 = 1.0) in the same buffer. Subsequently, resting-cell suspension was incubated with 0.2 mM TCP, DCP or ECH at 200 rpm and 30 °C. Samples were removed at 6 h for the analysis of transformation products by gas chromatography-mass spectrometer (GC-MS).
TCP degradation studies
KTU-TGVF cells were inoculated at OD600 = 0.05 into 50 ml Erlenmeyer flasks containing 10 ml of M9 minimal medium supplemented with 0.5 mM TCP and samples were incubated in a shaking incubator at 200 rpm and 30 °C. Aliquots (1 ml) of samples were withdrawn at regular time intervals and extracted with equal volume of acetone. The mixture was centrifuged for 10 min at 4,000 rpm and 4 °C and the organic layer was recovered carefully. The extraction process was repeated three times. The organic layers were pooled and then dried over anhydrous Na2SO4. Then, the concentrations of TCP, DCP, ECH and CPD were measured by GC-MS. GLY was quantified using a free GLY assay kit (Sigma, USA).
Samples of 1 μl (diluted if necessary) were injected directly for GC-MS analysis, which was obtained on an Agilent Technologies 7890A-5975C (Agilent Technologies, Palo Alto, CA, USA). The GC was equipped with a HP-5 capillary column (30 m × 0.25 mm × 0.25 µm) and operated in split ratio of 20:1. Helium (>99.999%) was used as carrier gas with a constant flow rate of 0.6 ml/min. The GC conditions were as follows: injector temperature of 250 °C, oven temperature maintained at 35 °C for 6 min and then increased to 250 °C at a rate of 10 °C/min, and interface temperature of 230 °C. The mass spectrum conditions were as follows: ionization energy of 70 eV, scan range of 30–300 amu, and ion chamber temperature of 250 °C with tungsten filament used for ionization of molecules. The concentration of metabolites was determined using a standard curve of peak area versus concentration. For identification of metabolites, the mass spectra of metabolites were compared with those of their authentic standards or the same compound available in the National Institute of Standard Technology library, USA.
VHb detection
VHb activity was detected by carbon monoxide (CO)-difference spectra. The complex consisting of CO and reduced hemoglobin has a characteristic peak at 420 nm35. KTU-TGVF or KTU cells were suspended in 50 mM potassium phosphate buffer (pH 7.2) and broken using an ultrasonic disruptor on ice (200 W for 15 min with cycles of sonication of 10 s each and 15 s pause). The crude extracts were centrifuged at 10,000 rpm and 4 °C for 15 min to remove cell debris. The soluble cellular fraction was exposed to CO for 2 min. Then, hemoglobin levels were determined by CO-difference spectra using a SHIMADZU UV-1800 spectrophotometer (SHIMADZU, Japan).
Flagellum observation
For electron microscope, KTU-TGVF or KT2440 cells were negatively stained with 0.05% (w/v) uranyl acetate for 20 s on 200-mesh carbon-coated copper grids. Electron microscope images were captured using a JEOL JEM-1400 transmission electron microscope (TEM) (JEOL, Japan) operated at 80 kV and equipped with a GATAN Orius 832 CCD (GATAN, USA).
Determination of the ATP/ADP and NADPH/NADP+ molar ratios
Cells were grown in M9 minimal medium containing 0.2% (w/v) glucose at 30 °C for 12 or 20 h. The intracellular levels of ADP and ATP were measured using an ADP/ATP ratio bioluminescence assay kit (BioVision, USA). The intracellular concentrations of NADP+ and NADPH were measured using an EnzyChrom™ NADP/NADPH assay kit (BioAssay Systems, USA).
Biofilm formation assays
Biofilm formation was quantified using the crystal violet (CV) staining assay30. Cells were grown overnight at 30 °C and 170 rpm in M9 minimal medium supplemented with 0.4% (w/v) glucose. The OD600 of the cultures was adjusted to 0.05 before inoculating 200 μl into 96-well microtiter plates. The plates were incubated at 30 °C without shaking for 24 or 96 h, after which the culture broth was removed from the plates and the OD600 was measured to estimate the planktonic cells in the culture. The plates were washed three times with H2O to remove all non-adhered cells, and stained with 0.1% (w/v) CV for 30 min at room temperature. After discarding the dye, the plates were washed with H2O, dried and the remaining stain was dissolved in 33% (v/v) acetic acid. The plates were agitated gently for 1 h. Biofilm was quantified by measuring the absorbance at 590 nm (A590) using a BIOLOG microplate reader. The biofilm index was calculated as the ratio of biofilm formation to planktonic cell density (A590/OD600).
Degradation of TCP by KTU-TGVF in a lab-scale bioreactor
A lab-scale reactor was designed for treatment of TCP-contaminated water (Fig. S10), which was mainly composed of a cylindrical vessel of polymethyl methacrylate with a volumetric capacity of 2 liters and a working volume of 0.8 liter. The reactor was filled with 500 g ceramic Raschig rings (outer diameter, 8 mm; internal diameter, 4 mm; length, 8 mm; Pingxiang Tower Packing Co.) as packing material. TCP-contaminated water was introduced into the reactor using a peristaltic pump from a glass vessel (0.5 liter) containing TCP in water. The airflow was regulated with a gas flow controller. The pH was controlled by adding 0.1 M NaOH when necessary.
For startup, the reactor was inoculated with a 800 ml suspension of cells grown on 0.5 mM TCP plus 0.4% glucose (OD600 = 0.4), which was left for 4 days without a supply of TCP-contaminated water. During this period, air was introduced into the reactor at a rate of 0.1 ml/min. After 4 days of inoculation, a wastewater stream containing TCP was introduced continuously, and the influent concentration of TCP was increased gradually from 0.05 to 0.2 mM. From day 18 onward, about 0.2 mM TCP was introduced into the reactor at a rate of 0.1 ml/min, and the flow rate was maintained during the operating period. The airflow was maintained at 1 ml/min to provide sufficient oxygen for aerobic mineralization of TCP. The effluent concentrations of TCP, DCP, and chloride ions were measured by GC-MS and a chloride assay kit (Sigma, USA) during a 30-day operating period.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31570035), the National Key Technology Support Program of China (No. 2015BAD16B04), Open Fund of State Key Laboratory of Microbial Technology, Shandong University (No. M2016-03), and Open Fund of State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University (No. MMLKF16-02).
Author Contributions
T.G., R.H.L., C.J.S. and C.Y. designed the research. T.G., X.Q.X., Y.C., W.X.G., F.J.Z. and J.N.L. performed the research. T.G., X.Q.X., Y.C., R.H.L., H.L.Y., P.X., C.J.S. and C.Y. analyzed the data. T.G., R.H.L., C.J.S. and C.Y. wrote the paper. All of the authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07435-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ruihua Liu, Email: yangyangliu@nankai.edu.cn.
Cunjiang Song, Email: songcj@nankai.edu.cn.
Chao Yang, Email: yang_chao2008@hotmail.com.
References
- 1.Keasling JD. Synthetic biology and the development of tools for metabolic engineering. Metab. Eng. 2012;14:189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Cho C, Choi SY, Luo ZW, Lee SY. Recent advances in microbial production of fuels and chemicals using tools and strategies of systems metabolic engineering. Biotechnol. Adv. 2015;33:1455–1466. doi: 10.1016/j.biotechadv.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Dvorak P, Bidmanova S, Damborsky J, Prokop Z. Immobilized synthetic pathway for biodegradation of toxic recalcitrant pollutant 1,2,3-trichloropropane. Environ. Sci. Technol. 2014;48:6859–6866. doi: 10.1021/es500396r. [DOI] [PubMed] [Google Scholar]
- 4.Samin G, et al. A Pseudomonas putida strain genetically engineered for 1,2,3-trichloropropane bioremediation. Appl. Environ. Microbiol. 2014;80:5467–5476. doi: 10.1128/AEM.01620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R. Bioremediation approaches for organic pollutants: a critical perspective. Environ. Int. 2011;37:1362–1375. doi: 10.1016/j.envint.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Copley SD. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol. 2009;5:559–566. doi: 10.1038/nchembio.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulakova AN, Larkin MJ, Kulakov LA. The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB 13064. Microbiology. 1997;143:109–115. doi: 10.1099/00221287-143-1-109. [DOI] [PubMed] [Google Scholar]
- 8.Pavlova M, et al. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat. Chem. Biol. 2009;5:727–733. doi: 10.1038/nchembio.205. [DOI] [PubMed] [Google Scholar]
- 9.van Leeuwen JG, Wijma HJ, Floor RJ, van der Laan JM, Janssen DB. Directed evolution strategies for enantiocomplementary haloalkane dehalogenases: from chemical waste to enantiopure building blocks. Chembiochem. 2012;13:137–148. doi: 10.1002/cbic.201100579. [DOI] [PubMed] [Google Scholar]
- 10.van Hylckama Vlieg JE, et al. Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases. J. Bacteriol. 2001;183:5058–5066. doi: 10.1128/JB.183.17.5058-5066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rink R, Fennema M, Smids M, Dehmel U, Janssen DB. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J. Biol. Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 12.Kurumbang NP, et al. Computer-assisted engineering of the synthetic pathway for biodegradation of a toxic persistent pollutant. ACS Synth. Biol. 2014;3:172–181. doi: 10.1021/sb400147n. [DOI] [PubMed] [Google Scholar]
- 13.Dvorak P, et al. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microb. Cell Fact. 2015;14:201. doi: 10.1186/s12934-015-0393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H. Martins dos Santos, V. A., Fouts, D. E., Gill, S. R., et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 15.Belda E, et al. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 2016;18:3403–3424. doi: 10.1111/1462-2920.13230. [DOI] [PubMed] [Google Scholar]
- 16.Graf N, Altenbuchner J. Development of a method for markerless gene deletion in Pseudomonas putida. Appl. Environ. Microbiol. 2011;77:5549–5552. doi: 10.1128/AEM.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-García E, de Lorenzo V. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 2011;13:2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x. [DOI] [PubMed] [Google Scholar]
- 18.Luo, X. et al. Pseudomonas putida KT2440 markerless gene deletion using a combination of λRed recombineering and Cre/loxP site-specific recombination. FEMS Microbiol. Lett. 363, fnw014 (2016). [DOI] [PubMed]
- 19.Gong T, et al. Metabolic engineering of Pseudomonas putida KT2440 for complete mineralization of methyl parathion and γ-hexachlorocyclohexane. ACS Synth. Biol. 2016;5:434–442. doi: 10.1021/acssynbio.6b00025. [DOI] [PubMed] [Google Scholar]
- 20.Nikel PI, Martínez-García E, de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014;12:368–379. doi: 10.1038/nrmicro3253. [DOI] [PubMed] [Google Scholar]
- 21.Zobel S, et al. Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth. Biol. 2015;4:1341–1351. doi: 10.1021/acssynbio.5b00058. [DOI] [PubMed] [Google Scholar]
- 22.Escapa IF, del Cerro C, García JL, Prieto MA. The role of GlpR repressor in Pseudomonas putida KT2440 growth and PHA production from glycerol. Environ. Microbiol. 2013;15:93–110. doi: 10.1111/j.1462-2920.2012.02790.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Recent developments and future prospects of Vitreoscilla hemoglobin application in metabolic engineering. Biotechnol. Adv. 2007;25:123–136. doi: 10.1016/j.biotechadv.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Stark BC, Pagilla KR, Dikshit KL. Recent applications of Vitreoscilla hemoglobin technology in bioproduct synthesis and bioremediation. Appl. Microbiol. Biotechnol. 2015;99:1627–1636. doi: 10.1007/s00253-014-6350-y. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y, Webster DA, Stark BC. Improvement of bioremediation by Pseudomonas and Burkholderia by mutants of the Vitreoscilla hemoglobin gene (vgb) integrated into their chromosomes. J. Ind. Microbiol. Biotechnol. 2005;32:148–154. doi: 10.1007/s10295-005-0215-4. [DOI] [PubMed] [Google Scholar]
- 26.Nikel PI, de Lorenzo V. Engineering an anaerobic metabolic regime in Pseudomonas putida KT2440 for the anoxic biodegradation of 1,3-dichloroprop-1-ene. Metab. Eng. 2013;15:98–112. doi: 10.1016/j.ymben.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Liu R, Ochman H. Stepwise formation of the bacterial flagellar system. Proc. Natl. Acad. Sci. USA. 2007;104:7116–7121. doi: 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-García E, Nikel PI, Chavarría M, de Lorenzo V. The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ. Microbiol. 2014;16:291–303. doi: 10.1111/1462-2920.12309. [DOI] [PubMed] [Google Scholar]
- 29.Russell JB, Cook GM. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benedetti I, de Lorenzo V, Nikel PI. Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab. Eng. 2016;33:109–118. doi: 10.1016/j.ymben.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Gungormusler M, Gonen C, Azbar N. Use of ceramic-based cell immobilization to produce 1,3-propanediol from biodiesel-derived waste glycerol with Klebsiella pneumoniae. J. Appl. Microbiol. 2011;111:1138–1147. doi: 10.1111/j.1365-2672.2011.05137.x. [DOI] [PubMed] [Google Scholar]
- 32.Green, M. R. & Sambrook, J. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA (2012).
- 33.Schäfer A, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 34.Cho JH, Kim EK, So JS. Improved transformation of Pseudomonas putida KT2440 by electroporation. Biotechnol. Tech. 1995;9:41–44. doi: 10.1007/BF00152998. [DOI] [Google Scholar]
- 35.Zhang W, et al. Chromosome integration of the Vitreoscilla hemoglobin gene (vgb) mediated by temperature-sensitive plasmid enhances γ-PGA production in Bacillus amyloliquefaciens. FEMS Microbiol. Lett. 2013;343:127–134. doi: 10.1111/1574-6968.12139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


