Abstract
Objectives:
Approximately 35% of colorectal cancer (CRC) risk is attributable to heritable factors known hereditary syndromes, accounting for 6%. The remainder may be due to lower penetrance polymorphisms particularly of DNA repair genes. DNA repair pathways, including base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), direct reversal repair (DRR), and double-strand break repair are complex, evolutionarily conserved, and critical in carcinogenesis. Germline mutations in these genes are associated with high-penetrance CRC syndromes such as Lynch syndrome. However, the association of low-penetrance polymorphisms of DNA repair genes with CRC risk remains unclear.
Methods:
A systematic literature review of PubMed, Embase, and HuGENet databases was conducted. Pre-specified criteria determined study inclusion/exclusion. Per-allele, pooled odds ratios disclosed the risk attributed to each variant. Heterogeneity was investigated by subgroup analyses for ethnicity and tumor location; funnel plots and Egger’s test assessed publication bias.
Results:
Sixty-one polymorphisms in 26 different DNA repair genes were identified. Meta-analyses for 22 polymorphisms in 17 genes revealed that six polymorphisms were significantly associated with CRC risk within BER (APE1, PARP1), NER (ERCC5, XPC), double-strand break (RAD18), and DRR (MGMT), but none within MMR. Subgroup analyses revealed significant association of OGG1 rs1052133 with rectal cancer risk. Egger’s test revealed no publication bias.
Conclusions:
Low-penetrance polymorphisms in DNA repair genes alter susceptibility to CRC. Future studies should therefore analyze whole-genome polymorphisms and any synergistic effects on CRC risk.
Translational impact:
This knowledge may enhance CRC risk assessment and facilitate a more personalized approach to cancer prevention.
Introduction
The genetic architecture of colorectal cancer (CRC) susceptibility encompasses a broad spectrum of risk, from rare, highly penetrant germline mutations to common low-penetrance polymorphisms, each individually conferring small risks.1 Some models are consistent with a polygenic basis of cancer susceptibility; however, the majority of heritable risk remains unexplained.
Heritable factors account for ~35% of CRC risk.2 The remaining inherited variation is thought to be due to low-penetrance variants.3 The common disease–common variant hypothesis suggests that many common low-penetrance alleles confer very slight increased risk and work synergistically to increase overall risk.4 In contrast, the common disease-rare variant model proposes alleles, which exert a higher risk but have a frequency of less than 5%. Founder effects resulting from genetic drift mean these variants are largely population-specific.4
Complex pathways involving many molecules have evolved to perform DNA repair. These include base excision repair (BER), nucleotide excision repair (NER), double-strand break repair, consisting of non-homologous end joining (NHEJ) and homologous recombination repair (HRR), direct reversal repair (DRR), and mismatch repair (MMR). Thousands of random DNA mutations occur during DNA replication; however, because of these highly conserved DNA repair mechanisms, fewer than one in 1000 persist.5 Given that carcinogenesis requires the acquisition of somatic mutations by susceptible cells, it follows that DNA repair mutations would increase the risk of CRC and other malignancies.6 In fact, recent studies have shown that mutations in DNA repair and replication genes are present in over 58% of cancer cell lines.7 However, the role of low-penetrance polymorphisms of DNA repair genes in CRC risk remains unclear.
High-penetrance germline mutations in BER and MMR result in known hereditary CRC syndromes such as MUTYH-associated polyposis and Lynch Syndrome, respectively.8 Furthermore, mutations in BER are also linked to non-CRCs such as xeroderma pigmentosum, underlining the importance of DNA repair mutations as high-penetrance risk factors for cancer.5
Knowledge of which DNA repair gene low-penetrance polymorphisms have a role in CRC will enable risk assessment and individualized treatment, allowing a more personalized approach to medicine. A better understanding of CRC carcinogenesis is therefore imperative and, consequently, a systematic review and meta-analysis investigating the association of DNA repair gene polymorphisms with CRC risk was conducted.
Methods
The systematic review and meta-analysis was conducted according to the principles endorsed in the PRISMA-P statement.9
Literature search
An extensive literature search of the PubMed and Embase databases from inception was conducted. Initially the following terms were searched: “DNA Repair Polymorphism” odds ratio (OR) “DNA repair risk” AND “colorectal cancer”. Within Embase “DNA Repair” AND “Colorectal Cancer” were also searched.
For each specific gene or polymorphism identified, a subsequent search was performed using the terms: “rs number” or “polymorphism (e.g., T656G)” AND “colorectal cancer” or “colorectal carcinoma” or “polymorphism” or “variant”.
A further literature search was conducted using the Human Genome Epidemiology Network (HuGENet) database: “[Gene name]” and “colorectal neoplasia”.
Eligibility criteria
Two independent reviewers (N.A. and S.S.) assessed the search strategy results. The study titles were examined for potential relevance and the abstract was then reviewed. Subsequently, the full text was retrieved to ensure eligibility.
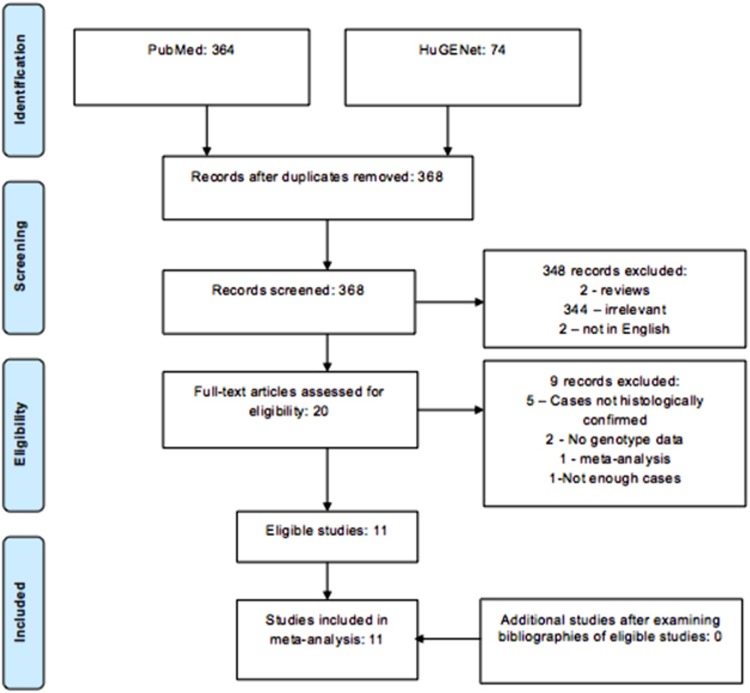
The bibliographies of relevant articles were inspected for further eligible studies. Previous meta-analyses studying polymorphisms of interest were also reviewed (Figure 1).
Figure 1.
Flowchart showing selection process of studies for the polymorphism rs1799782 within gene XRCC1.
If multiple studies included the same cohort of cases or controls, the study with the largest sample size was used. In the case of any uncertainty regarding study inclusion, another investigator (K.M.) was consulted to assess eligibility (Table 1).
Table 1. Table indicating the eligibility criteria used to select relevant studies.
| Inclusion criteria | Exclusion criteria |
|---|---|
| Investigated the association between a polymorphism of DNA repair genes in CRC | Control population is not in Hardy–Weinburg equilibrium |
| Case–control study utilizing at least 100 cases and 100 controls | Case only studies or review articles |
| Sufficient data available for analyses to be conducted | Insufficient information within the article for inclusion/exclusion to be established |
| Histologically confirmed cases of colon or rectal cancer at the time of the study | Study contained subjects with known hereditary syndromes, e.g., FAP or HNPCC |
| Unselected population | Controls with known adenomas or polyps |
| Full articles published in English within a peer-reviewed journal | Insufficient information within the article for inclusion/exclusion to be established |
CRC, colorectal cancer; FAP, familial adenomatous polyposis; HNPCC, hereditary non-polyposis colorectal cancer.
Data extraction
Extracted data includes: title, first author, publication year, polymorphism(s) investigated, number of cases and controls, ethnicity of subjects, country from which subjects were ascertained, number of cases and controls genotyped, genotype frequencies for cases and controls, genotype frequencies within male and female cases, and controls and genotype frequencies stratified according to colon or rectum cancer.
If the required data were unavailable within the full text or any accompanying Supplementary Material online, it was sought via correspondence with the relevant authors.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) was calculated for control groups and is defined as p2+2pq+q2=1, whereby p and q denote the frequencies of dominant and minor alleles, respectively, at a single gene locus. Pearson’s χ2-test was used to assess any statistically significant difference between expected and observed genotype frequencies. An χ2-value greater than 3.84 (P<0.05) signified deviation from HWE.
Quantitative synthesis involved calculating allele-specific pooled OR for three or more studies.
Assuming that AA is the genotype frequency of the single nucleotide polymorphism (SNP), AB the heterozygote genotype frequency and BB the wild-type (Wt) genotype frequency, the frequencies were calculated as follows:
If (A) cases and (A) controls correspond to the allele frequencies of the “at-risk” groups with and without CRC, respectively, and (B) cases and (B) controls are the allele frequencies of the unexposed groups, the following equation was used to determine the pooled ORs:
Corresponding 95% confidence intervals (CIs) were deemed statistically significant if they did not cross 1.
Heterogeneity was determined by Cochrane’s Q test and the I2 statistic. If the I2 value was between 50 and 100%, the DerSimonian and Laird random effects method was used to generate pooled ORs.10 If the I2 value was between 0 and 50, then the Mantel–Haenszel fixed effects method was used.10
Sensitivity analysis was achieved by removing each study individually and repeating the analysis.
Publication bias was assessed using funnel plots and was quantitatively assessed using Egger’s test in meta-analyses containing 10 or more studies.11
Subgroup analyses were conducted for ethnicity and cancer location, provided there were at least three studies. χ2-test was used to assess a significant difference between the subgroups for ethnicity. Colon and rectum cancer cases were compared to controls because it is not possible to segregate controls into colon or rectum cancers.
Statistical analyses were performed using the Metafor package in R (Version 3.2.4).12
Results
The comprehensive literature search identified 61 polymorphisms in 26 different DNA repair genes with case–control studies eligible for inclusion.
Meta-analyses were conducted for 22 of these polymorphisms in 17 genes, including between 1,706 and 9,682 CRC cases per polymorphism. Study numbers ranged from 3 to 13 (Supplementary Table 1 online). For two polymorphisms (RAD18 rs272572 and MSH2 rs2303425) the meta-analysis was constituted by two studies with three different population groups reported. Minor allele frequencies ranged from between 0.0513 and 0.4571, as gathered by the 1000 genomes project.13
Five polymorphisms were identified to have a significantly increased risk of CRC: APE1 rs1130409, PARP1 rs11136410, ERCC5 rs17655, XPC rs2228001, and RAD18 rs373572. However, MGMT rs12917 had a significantly decreased risk of CRC. The remaining polymorphisms did not demonstrate a significant association in meta-analysis (Table 2).
Table 2. Polymorphisms included within the meta-analysis and their association with CRC risk.
| Pathway | Gene | rs number | Number of studies | Cases/controls | Wt allele | Minor allele | MAF | Pooled odds ratio | 95% CI | I2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Base excision repair | APE1 | rs1130409 | 5 | 4,606/5,000 | G | G | 0.3756 | 1.15 | 1.06–1.24 | 36.38 |
| MUTYH | rs3219489 | 3 | 5,230/2,756 | G | G | 0.3135 | 1.14 | 0.82–1.58 | 88.23 | |
| OGG1 | rs1052133 | 13 | 9,682/1,2938 | C | G | 0.3021 | 1.05 | 0.98–1.11 | 0.00 | |
| PARP1 | rs1136410 | 3 | 2,132/4,320 | T | G | 0.1969 | 1.16 | 1.04–1.30 | 15.01 | |
| XRCC1 | rs1799782 | 11 | 6,190/10,454 | C | A | 0.1238 | 1.30 | 0.96–1.75 | 86.98 | |
| rs25487 | 13 | 7,981/12,226 | G | T | 0.2604 | 1.08 | 0.92–1.26 | 80.55 | ||
| rs25489 | 3 | 2,402/3,000 | G | T | 0.0671 | 1.03 | 0.83–1.27 | 0.00 | ||
| Homologous recombination repair | RAD18 | rs373572 | 2 | 2,074/2,298 | A | C | 0.3524 | 1.32 | 1.16–1.49 | 0.00 |
| RAD51 | rs1801320 | 5 | 1,706/1,240 | G | C | 0.1432 | 0.83 | 0.50–1.38 | 85.96 | |
| XRCC3 | rs861539 | 8 | 3,850/4,566 | C | A | 0.2169 | 1.15 | 0.85–1.55 | 82.90 | |
| Mismatch repair | MLH1 | rs1799977 | 4 | 8,252/8,802 | A | G | 0.1296 | 1.15 | 0.96–1.37 | 82.88 |
| MSH2 | rs2303425 | 2 | 4,598/5,602 | T | C | 0.1008 | 1.06 | 0.94–1.19 | 0.00 | |
| MSH6 | rs1042821 | 3 | 6,952/8,174 | C | A | 0.2009 | 0.98 | 0.80–1.20 | 80.04 | |
| Nucleotide excision repair | ERCC1 | rs11615 | 4 | 1,890/1,650 | C | A | 0.3311 | 1.03 | 0.90–1.18 | 11.22 |
| ERCC5 | rs17655 | 6 | 7,912/8,772 | C | C | 0.3614 | 1.12 | 1.06–1.20 | 0.04 | |
| TP53 | rs1042522 | 12 | 8,164/10,176 | C | G | 0.4571 | 1.18 | 0.97–1.45 | 89.23 | |
| XPC | rs2228000 | 3 | 3,680/5,212 | C | A | 0.2330 | 1.06 | 0.85–1.32 | 80.80 | |
| rs2228001 | 7 | 6,584/8,930 | A | G | 0.3153 | 1.08 | 1.01–1.15 | 49.46 | ||
| XPD | rs13181 | 10 | 4,932/6,210 | G | G | 0.2366 | 1.21 | 0.95–1.54 | 85.46 | |
| rs1799793 | 3 | 1,520/2,244 | G | T | 0.1954 | 1.13 | 0.87–1.45 | 62.01 | ||
| Direct reversal repair | MGMT | rs12917 | 3 | 1,528/3,332 | C | T | 0.1484 | 0.81 | 0.68–0.98 | 0.00 |
| rs2308321 | 3 | 1,664/7,016 | A | G | 0.0513 | 0.99 | 0.82–1.19 | 0.00 |
CI, confidence interval; CRC, colorectal cancer; MAF, minor allele frequency; Wt, wild type.
Significant results are in bold.
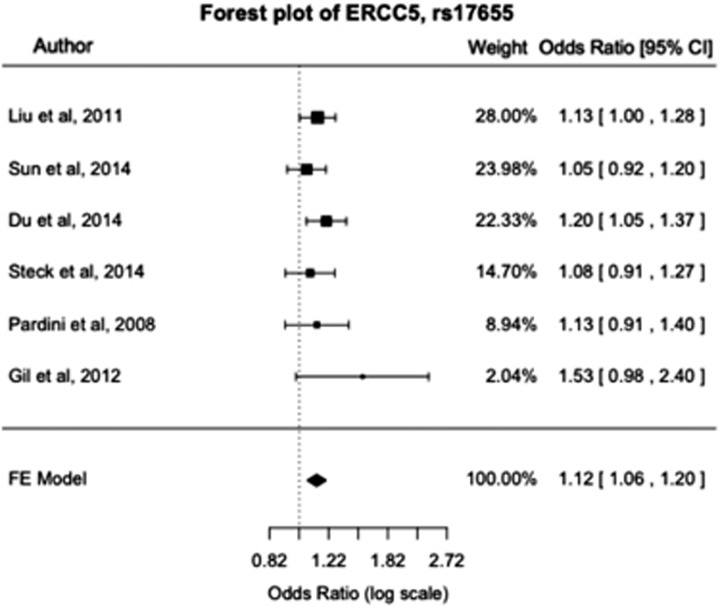
Forest plots for polymorphisms that were meta-analyzed displayed pooled ORs with 95% CI as well as study weightings. The forest plot for the polymorphism ERCC5 rs17655 within the NER pathway is shown in Figure 2.
Figure 2.
Forest plot of ERCC5 rs17655 within the NER pathway. The author, OR, 95% CI and study weightings are shown. Analysis using the Mantel–Haenszel fixed effects model shows ERCC5 rs17655 is associated with a significantly increased risk of CRC. CI, confidence interval; CRC, colorectal cancer; NER, nucleotide excision repair; OR, odds ratio.
Subgroup analysis
Subgroup analysis was performed by tumor location and ethnicity. There was insufficient data for subgroup analysis by gender.
Tumor location
For OGG1, the polymorphism rs1052133 is associated with a significantly increased risk of rectum cancer 1.18 (1.03–1.34 95% CI) but not of colon cancer 0.93 (0.81–1.06 95% CI). No other significant associations were identified.
Ethnicity
Subgroup analysis by ethnicity was performed for OGG1 rs1052133, XRCC1 rs25487, and ERCC1 rs11615 (Table 3). For all three, neither European nor Chinese patients were associated with risk of CRC. There were also no significant differences between ethnicities and the 95% CI calculated for the ORs overlap between the two subgroups.
Table 3. Subgroup analysis by ethnicity for OGG1 rs1052133, XRCC1 rs25489, and ERCC1 rs11615.
| Pathway | Gene | rs number | Wt allele |
European per allele |
Chinese per allele |
P value | ||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | |||||
| BER | OGG1 | rs1052133 | C | 1.11 | 1.00–1.23 | 1.03 | 0.93–1.14 | 0.32 |
| XRCC1 | rs25489 | G | 1.04 | 0.94–1.15 | 0.92 | 0.82–1.02 | 0.10 | |
| NER | ERCC1 | rs11615 | C | 1.11 | 0.96–1.30 | 1.13 | 0.97–1.32 | 0.90 |
BER, base excision repair; CI, confidence interval; NER, nucleotide excision repair; OR, odds ratio; Wt, wild type.
European patients were compared with controls. Chinese patients were compared with controls. A χ2-test to detect subgroup difference found nonsignificant P values.
Sensitivity analysis
Sensitivity analysis was performed for 12 polymorphisms. For both XRCC1 rs1799782 and XPD rs13181, removal of the studies by Gsur et al.14 and Sliwinski et al.15 yielded ORs indicating significantly increased risk of CRC, although previously the result was insignificant. For XPC rs2228001, removal of the studies by Wu et al.,16 Liu et al.,17 and Aizat et al.18 yielded nonsignificant ORs, indicating no association with CRC, although previously the result was significant. However, a difference in heterogeneity was found after removal of studies by Wu et al.16 and Liu et al.17 (Table 4). Removal of all other studies did not change pooled OR significance for any polymorphism.
Table 4. Polymorphisms for which sensitivity analysis revealed significantly different pooled ORs.
| Pathway | Gene | rs number | Wt allele |
Original |
Removed study |
After removal of study |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | I2 | OR | 95% CI | I2 | |||||
| BER | XRCC1 | rs1799782 | C | 1.30 | 0.96–1.75 | 86.98 | Gsur et al.14 | 1.38 | 1.02–1.85 | 86.27 |
| NER | XPC | rs2228001 | A | 1.08 | 1.01–1.15 | 49.46 | Wu et al.16 | 1.07 | 0.96–1.19 | 50.14 |
| Liu et al.17 | 1.09 | 0.96–1.23 | 56.50 | |||||||
| Aizat et al.18 | 1.06 | 0.99–1.13 | 40.66 | |||||||
| XPD | rs13181 | G | 1.21 | 0.95–1.54 | 85.46 | Sliwinski et al.15 | 1.28 | 1.01–1.62 | 83.15 | |
BER, base excision repair; CI, confidence interval; NER, nucleotide excision repair; OR, odds ratio.
Significant ORs and 95% CI are in bold.
Publication bias
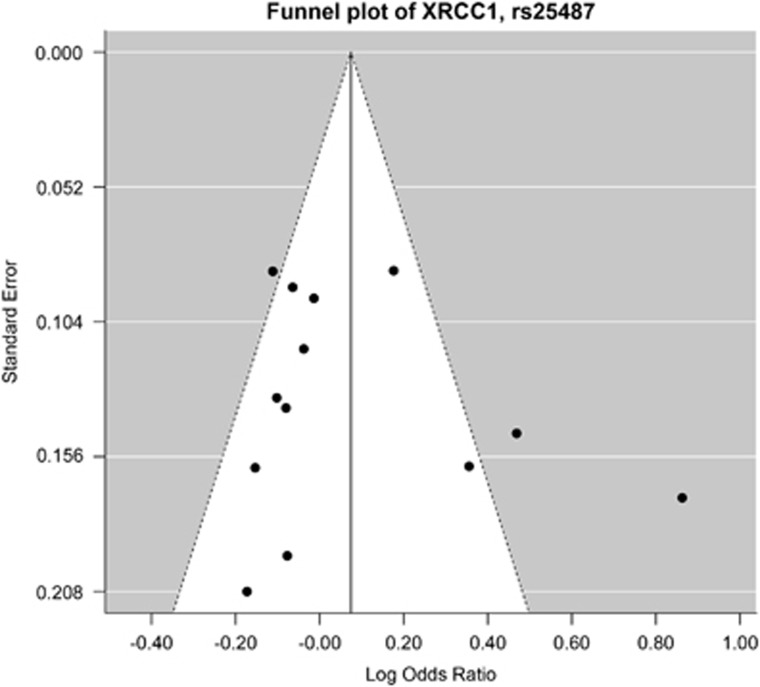
The majority of funnel plots were symmetrical and, therefore, did not demonstrate publication bias. Exceptions include XRCC1 rs25487 and XRCC1 rs1799782 (Figure 3).
Figure 3.
Funnel plot for XRCC1, rs25487. Studies represented as dots are plotted against log OR on the x axis and standard error on the y axis. Unequal number of studies on each side of the vertical line indicates publication bias is present. Nevertheless, Egger’s test reveals no significant funnel plot asymmetry (Table 5). OR, odds ratio.
Despite this, Egger’s test indicated no significant funnel plot asymmetry for any polymorphism, supporting the absence of publication bias (Table 5).
Table 5. Egger’s test for funnel plot asymmetry.
| Pathway | Gene | rs number | Egger’s t-value | P value |
|---|---|---|---|---|
| BER | OGG1 | rs1052133 | −0.2892 | 0.778 |
| XRCC1 | rs25487 | 0.9108 | 0.3819 | |
| rs1799782 | 0.7366 | 0.4801 | ||
| NER | TP53 | rs104522 | 0.6763 | 0.5771 |
| XPD | rs13181 | 0.2142 | 0.8358 |
Meta-analysis could not be conducted for 39 polymorphisms because of insufficient numbers of reported studies (Supplementary Table 2 online). Of these, a statistically significant association with CRC risk was identified for six polymorphisms in individual published reports.
Discussion
This study investigated the association between low-penetrance polymorphisms in DNA repair genes and CRC. As carcinogenesis is associated with an accumulation of acquired somatic mutations, it follows that changes in DNA repair genes would lead to genomic instability and influence an individual’s susceptibility to cancer.6 Six polymorphisms within six genes in the BER, HRR, NER, and DRR pathways were found to be significantly associated with the risk of CRC. The importance of DNA repair genes is indicated and polymorphisms in critical genes can, therefore, indeed alter the susceptibility to CRC.
Base excision repair
BER involves correcting everyday oxidative and alkylative modifications to bases or the sugar phosphate backbone together with most forms of spontaneous hydrolytic decay.19
This study found no association between the polymorphisms rs3219489 and rs1052133 of the two DNA glycosylase genes MUTYH and OGG1, respectively, corroborating with studies by Piceli et al.8 and Zhang et al.20 However, as there were only three eligible studies for MUTYH rs3219489, sensitivity analysis was not possible. Interestingly, subgroup analysis revealed that the association between rectum cancer and OGG1 rs1052133 was discernible. Therefore, one can conclude that there is no association between OGG1 rs1052133 and colon cancer, but further studies with large population groups and varying ethnicities, enabling subgroup analysis, are warranted to assess risk of MUTYH rs3219489 with CRC and risk of OGG1 rs1052133 with rectum cancer.
After base removal by DNA glycosylase, APE1 catalyzes an acid–base reaction to incise the phosphodiester bond of DNA.19 PARP1 binds to single strand break intermediates formed by APE1 incision. This study found significant associations between APE1 rs1130409 and PARP1 rs1136410 with CRC risk, although Shen et al.21 and Hua et al.22 revealed no such associations. The discrepancy may lie in the different methodologies such as the per-allele model or the more stringent inclusion/exclusion criteria used in the present study. Allowing studies with fewer cases increases the number of included studies but may not reliably reflect the studied population. Future meta-analyses should therefore have similarly rigorous inclusion/exclusion criteria.
XRCC1 is a key effector in the final step of BER. Coupled with DNA ligase IIIα, a covalent phosphodiester bond between 3′-OH end of the upstream nucleotide and the 5′-PO4 is formed. This is termed “nick sealing”.19 Meta-analyses conducted for polymorphisms rs25487, rs25489, and rs1799782 revealed no statistically significant association with CRC. Qin et al.23 concurred for rs25487 and Liu et al.24 also found no association with CRC risk for all three polymorphisms. Sensitivity analysis revealed that removal of the study by Gsur et al.14 would result in a statistically significant association with CRC for the polymorphism rs179978. Asymmetrical funnel plots for rs25487 and rs1799782 revealed possible publication bias, possibly because smaller studies showing no statistically significant effects remain unpublished. Nevertheless, Egger’s test revealed no publication bias for either polymorphism. However, more studies are required before making any conclusive determination regarding the association of each polymorphism with CRC risk.
Double-strand break repair
There are two major conserved pathways for the repair of double-strand breaks: HRR and NHEJ. HRR involves transfer of nucleotide sequence information from the intact DNA double helix to the site of the double-strand break of the broken helix.5 NHEJ involves binding of the Ku heterodimer to the double-strand break to serve as a scaffold allowing other NHEJ factors to be recruited, such as the XRCC4-DNA Ligase IV complex to ligate the ends.25 Because of insufficient number of studies, meta-analyses could not be conducted on polymorphisms of NHEJ genes.
RAD51 catalyzes the strand invasion step allowing homologous pairing. Meta-analyses by Cheng et al.26 and the present study revealed that RAD51 rs1801320 was not associated with CRC risk. Interestingly, only one included study27 suggests significant association between CRC and the polymorphism, with an OR notably lower than the other studies. Nevertheless, sensitivity analysis revealed no marked change. XRCC3 is a paralog of RAD51 and combines with RAD51C, although its mode of action as a mediator is unclear.28 Meta-analyses by Namazi et al.,29 Liu et al.,24 and the present study found no association between XRCC3 rs861539 and CRC risk. Contrary to larger studies, the smaller studies by Nissar et al.30 and Jin et al.31 showed significant association with CRC risk, indicating the importance of a large population number. Although this polymorphism may not be associated with CRC, it is vital to search for other polymorphisms of XRCC3 and to better understand the function of this protein.
RAD18 is integral in the orchestration of HRR and enhances polymerization of RAD51 by transmitting DNA damage signals at sites of DNA breaks.32 To our knowledge, this is the first meta-analysis of any RAD18 polymorphism and risk of CRC. A significant association was found for RAD18 rs373572, but given the low number of studies included, it is vital that more case–control studies are conducted to further assess its association with colorectal carcinogenesis.
Mismatch repair
MMR corrects mismatches created largely during DNA replication.33 MMR increases the accuracy of DNA 20–400-fold, and so any defects raise the spontaneous mutation rate.33
Meta-analyses were conducted for MLH1 rs1799977, MSH2 rs2303425, and MSH6 rs1042821. Results revealed no significant association with CRC risk. Chen at al.34 also found no association between MLH1 rs1799977 and CRC risk but, notably, included studies that did not meet the inclusion criteria of the present study. Given the small number of eligible studies, sensitivity analysis could only be conducted on MLH1 rs1799977, revealing no significant difference. Many studies were excluded based on the inclusion of those with hereditary syndromes or no histological confirmation of cases. It is imperative that further studies assess patients with MMR gene polymorphisms and risk of sporadic CRC in particular.
Nucleotide excision repair
NER has two pathways: the global genome NER sub-pathway and transcription-coupled NER. The genome NER pathway repairs lesions throughout the genome and is triggered by helix distortion, whereas transcription-coupled NER is confined to the repair of transcription blocking DNA lesions triggered by the stalling of RNA polymerase III.35 In the next steps, the two sub-pathways converge and form a transcription initiation factor IIH complex. The core proteins remain the same except XPC, which is specific to genome NER.
XPC is the first factor to detect lesions and recruit other repair machinery to form a transcription initiation factor IIH complex.35 Meta-analyses conducted by Wang et al.36 and the present study for the polymorphism rs2228000 revealed no significant association with risk of CRC. Nevertheless, given the small number of eligible studies, further case–control studies must be conducted to better understand the relationship this polymorphism may have with CRC risk. Meta-analysis for the polymorphism rs2228001 revealed a significant association with CRC. This concurs with Peng et al.37 and Liu et al,38 although both included studies ineligible for this meta-analysis. This result should be treated cautiously as it was only just statistically significant and sensitivity analysis revealed removal of studies by Wu et al.,16 Liu et al.,17 or Aizat et al.39 would leave an insignificant result showing no association with CRC risk.
Ultraviolet damage repair has been shown to be dependent on p53 action.40 Nevertheless, it is not known whether p53 directly participates or whether the response is mediated through p53-regulated gene products.40 Meta-analyses by Economopoulos et al.,41 Liu et al.,42 and this meta-analysis found no association with the polymorphism TP53 rs1042522 and risk of CRC. However, due to insufficient data, subgroup analyses were not possible in the present study but Liu et al.42 and Economopoulos et al.41 concluded that race-specific effects may feature. Additional studies providing sufficient data for subgroup analyses are desirable.
Mutations in XPD prevent it from interacting with the transcription initiation factor IIH complex, hence reducing its helicase activity and increasing the chance of repair defects.15 Zhang et al.43 and the present meta-analysis revealed no significant association for both rs13181 and rs1799793 with CRC risk. However, sensitivity analysis revealed that removal of the study by Sliwinski et al.15 would show a statistically significant result for rs13181, albeit marginally. Given that subgroup analysis was not possible for both polymorphisms and the small number of qualifying studies for rs1799793, more studies are imperative to better understand the role of XPD polymorphisms and risk of CRC.
Lesion excision is the point at which the reaction must be completed to avoid leaving dangerous intermediates. Structure-specific endonucleases, XPF-ERCC1 complex, and XPG (encoded by ERCC5) incise 5′ and 3′ from the lesion, respectively.35 To our knowledge, this is the first meta-analysis conducted on a polymorphism of ERCC1 and CRC risk. Results revealed no association between ERCC1 rs11615 and risk of CRC. Furthermore, Zeng et al.44 and this meta-analysis found a significant association between ERCC5 rs17655 and CRC risk. However, a lack of sufficient data prohibited subgroup analyses. This study allows insight into the role these polymorphisms may have in CRC carcinogenesis but further research is required to conclusively determine it.
Direct reversal repair
DRR involves removal of alkylating groups in O6-methylguanine base residues, produced by endogenous and exogenous alkylating agents. Therefore, MGMT has an important role in preventing carcinogenesis.45
Meta-analyses by Chen et al.,46 Lu et al.,47 and the present study found no association for the polymorphisms MGMT rs12917 and rs2308321 with CRC risk. Subgroup analyses were not possible due to insufficient data. Given the small number of eligible studies, more case–control studies are warranted to clarify whether these two polymorphisms have a role in the development of CRC.
Limitations
Many polymorphisms could not be investigated due to a low number of eligible studies or due to insufficient population size. Patients should be advised about the importance and possible beneficial effects of enrollment in such research studies. Increased patient numbers would allow greater reliability and statistical power when detecting an association.
Data such as genotype frequencies or patient demographics were often missing from studies, limiting subgroup analyses to tumor location and European/Chinese ethnicities. It would benefit the scientific community if publications would provide these data in the form of Supplementary Tables online or facilitate requests to allow extensive and complete analyses. In particular, study designs should allow investigation of whether variants differ significantly by other ethnicities and by gender.
The bulk of published literature reviewed in this meta-analysis subdivides tumors by rectum/colon location rather than by right and left sides. However, the differentiation of CRC by left and right sides is more biologically and clinically meaningful, and future study design should follow the right/left-sided paradigm. An example of this is the prevalence of microsatellite-unstable tumors on the right side of the colon.
Cases were often not histologically confirmed and therefore not included. It is vital histology is performed to ensure correct diagnoses of cases and to avoid misinterpretation of results.
The role of DNA repair gene polymorphisms and colorectal carcinogenesis
This comprehensive analysis demonstrates the association of DNA repair gene polymorphisms with CRC risk. However, given that the ORs calculated are very small and often only just significant, it is conceivable that the polymorphisms act synergistically to affect risk of CRC. It was not possible to assess this in the present study, and should be investigated in future larger studies.
Sixty-one polymorphisms were investigated and twenty-one were meta-analyzed. However, these polymorphisms account for a small proportion of all DNA repair gene polymorphisms, and it is likely there are many more, which are associated with risk of CRC, which requires a systematic whole-genomic approach.
This study synthesizes current published information regarding which DNA repair gene polymorphisms are associated with risk of CRC. This knowledge will allow physicians to risk assess patients more comprehensively in the future, enabling them to diagnose earlier and provide advice accordingly. It enhances our understanding of the pathophysiology of CRC carcinogenesis and can allow the production of better treatments, which may be tailored to meet the needs of each individual patient.
Although the functional role of these pathways is clearly very important in carcinogenesis, the functional role of the polymorphisms identified is less clear, and such studies, although required, are technically difficult, and need to be robust in their design. DNA repair and genome integrity SNPs are remarkable by their absence from the list of common CRC predisposition variants—no known DNA repair gene is tagged by the known predisposition SNPs for CRC in GWAS. One possible explanation is that these highly conserved pathways are so important to the cell that any alleles with more than very small functional effects are strongly selected against. Nevertheless, common DNA repair alleles with very small effects on cancer risk may still exist, yet their effects may be too small (with OR<1.4 per allele) to be detected individually even in the large sample size required for GWAS.
Conclusion
This study identified a significant association between six DNA repair gene polymorphisms and risk of CRC. This knowledge allows a better understanding of global risk and the role of DNA repair genes in colorectal carcinogenesis. The results indicate that further studies are warranted and should focus on assessing whole-genomic polymorphism risk and the synergistic effects of this variation on CRC risk. This study will enable inclusion of these DNA repair gene polymorphisms in future CRC risk assessments, and may ultimately contribute to global CRC risk assessment and personalized care pathways pending further data.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Guarantor of the article: Kevin J. Monahan, FRCP, PhD.
Specific author contributions: Study design: N.A., S.M., M.M., and K.M. Data search and collection: N.A., S.S., and KM. Data analysis and interpretation: N.A., N.D., and S.M. Drafting of manuscript: N.A. Critical revision of manuscript: N.A., N.D., S.M., S.S., M.M., and K.M.
Financial support: None.
Potential competing interests: None.
Supplementary Material
References
- Fletcher O, Houlston RS. Architecture of inherited susceptibility to common cancer. Nat Rev Cancer 2010; 10: 353–361. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000; 343: 78–85. [DOI] [PubMed] [Google Scholar]
- De la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer 2004; 4: 769–780. [DOI] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 2008; 40: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J DNA Repair. Available at http://www.ncbi.nlm.nih.gov/books/NBK26879/ (2002). Accessed 3 April 2016.
- Tomlinson IP, Houlston RS, Montgomery GW et al. Investigation of the effects of DNA repair gene polymorphisms on the risk of colorectal cancer. Mutagenesis 2012; 27: 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulogiannis G, Frayling IM, Arends MJ. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 2010; 56: 167–179. [DOI] [PubMed] [Google Scholar]
- Picelli S, Bermejo JL, Chang-Claude J et al. Meta-analysis of mismatch repair polymorphisms within the cogent consortium for colorectal cancer susceptibility. PLoS ONE 2013; 8: e72091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseer L, Moher D, Clarke M et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. Br Med J 2015; 349: g7647. [DOI] [PubMed] [Google Scholar]
- Kontopantelis E, Springate DA, Reeves D. A re-analysis of the Cochrane Library data: the dangers of unobserved heterogeneity in meta-analyses. PLoS ONE 2013; 8: e69930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne JA, Sutton AJ, Ioannidis JP et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. Br Med J 2011; 343: d4002. [DOI] [PubMed] [Google Scholar]
- Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Software 2010; 36: 1–48. [Google Scholar]
- 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsur A, Bernhart K, Baierl A et al. No association of XRCC1 polymorphisms Arg194Trp and Arg399Gln with colorectal cancer risk. Cancer Epidemiol 2011; 35: e38–e41. [DOI] [PubMed] [Google Scholar]
- Sliwinski T, Krupa R, Wisniewska-Jarosinska M et al. Common polymorphisms in the XPD and hOGG1 genes are not associated with the risk of colorectal cancer in a Polish population. Tohoku J Exp Med 2009; 218: 185–191. [DOI] [PubMed] [Google Scholar]
- Wu Y, Jin M, Liu B et al. The association of XPC polymorphisms and tea drinking with colorectal cancer risk in a Chinese population. Mol Carcinog 2011; 50: 189–198. [DOI] [PubMed] [Google Scholar]
- Liu D, Wu HZ, Zhang YN et al. DNA repair genes XPC, XPG polymorphisms: relation to the risk of colorectal carcinoma and therapeutic outcome with Oxaliplatin-based adjuvant chemotherapy. Mol Carcinog 2012; 51 (Suppl 1): E83–E93. [DOI] [PubMed] [Google Scholar]
- Aizat AA, Shahpudin SN, Mustapha MA et al. Association of Arg72Pro of P53 polymorphism with colorectal cancer susceptibility risk in Malaysian population. Asian Pac J Cancer Prev 2011; 12: 2909–2913. [PubMed] [Google Scholar]
- Kim YJ, Wilson DM3rd. Overview of base excision repair biochemistry. Curr Mol Pharmacol 2012; 5: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, He B, Pan Y et al. Association of OGG1 Ser326Cys polymorphism with colorectal cancer risk: a meta-analysis. Int J Colorectal Dis 2011; 26: 1525–1530. [DOI] [PubMed] [Google Scholar]
- Shen E, Liu C, Wei L et al. The APE1 Asp148Glu polymorphism and colorectal cancer susceptibility: a meta-analysis. Tumor Biol 2014; 35: 2529–2535. [DOI] [PubMed] [Google Scholar]
- Hua R, Li H, Liang Y et al. Association between the PARP1 Val762Ala polymorphism and cancer risk: evidence from 43 studies. PLoS ONE 2014; 9: e87057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Xu K, Chen Z et al. XRCC1 R399Q polymorphism and colorectal cancer risk in the Chinese Han population: a meta-analysis. Tumor Biol 2015; 36: 461–466. [DOI] [PubMed] [Google Scholar]
- Liu L, Miao L, Ji G et al. Association between XRCC1 and XRCC3 polymorphisms and colorectal cancer risk: a meta-analysis of 23 case–control studies. Mol Biol Rep 2013; 40: 3943–3952. [DOI] [PubMed] [Google Scholar]
- Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2013; 2: 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Shi H, Zhang K et al. RAD51 Gene 135G/C polymorphism and the risk of four types of common cancers: a meta-analysis. Diagn Pathol 2014; 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa R, Sliwinski T, Wisniewska-Jarosinska M et al. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer—a case control study. Mol Biol Rep 2011; 38: 2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer W. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 2008; 18: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namazi A, Abedinzadeh M, Nourbaksh P et al. Association between the XRCC3 Thr241Met polymorphism and risk of colorectal cancer: a meta analysis of 5,193 cases and 6,645 controls. Asian Pac J Cancer Prev 2015; 16: 2263–2268. [DOI] [PubMed] [Google Scholar]
- Nissar S, Sameer AS, Lone TA et al. XRCC3 Thr241Met gene polymorphism and risk of colorectal cancer in Kashmir: a case control study. Asian Pac J Cancer Prev 2014; 15: 9621–9625. [DOI] [PubMed] [Google Scholar]
- Jin M, Chen K, Song L et al. The association of the DNA repair gene XRCC3 Thr241Met polymorphism with susceptibility to colorectal cancer in a Chinese population. Cancer Genet Cytogenet 2005; 163: 38–43. [DOI] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 2009; 11: 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K. DNA mismatch repair in eukaryotes and bacteria. J Nucleic Acids 2010; doi:10.4061/2010/260512. [DOI] [PMC free article] [PubMed]
- Chen H, Shen Z, Hu Y et al. Association between MutL homolog 1 polymorphisms and the risk of colorectal cancer: a meta-analysis. J Cancer Res Clin Oncol 2015; 141: 2147–2158. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W et al. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 2014; 15: 465–481. [DOI] [PubMed] [Google Scholar]
- Wang G, Sun H, Liu Z et al. Lack of associations between XPC polymorphisms and colorectal cancer: a meta-analysis. J BUON 2015;20:770–774. [PubMed] [Google Scholar]
- Peng Q, Lao X, Tang W et al. XPC Lys939Gln polymorphism contributes to colorectal cancer susceptibility: evidence from a meta-analysis. Diagn Pathol 2014; 9: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yin Q, Ying M et al. XPC Lys939Gln and Ala499Val polymorphisms in colorectal cancer susceptibility: a meta-analysis of case–control studies. Mol Biol Rep 2014; 41: 1171–1178. [DOI] [PubMed] [Google Scholar]
- Aizat AAA, Nurfatimah MSS, Aminudin MM et al. XPC Lys939Gln polymorphism, smoking and risk of sporadic colorectal cancer among Malaysians. World J Gastroenterol 2013; 19: 3623–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Seo YR. p53 regulation of DNA excision repair pathways. Mutagenesis 2002; 17: 149–156. [DOI] [PubMed] [Google Scholar]
- Economopoulos KP, Sergentanis TN, Zagouri F et al. Association between p53 Arg72Pro polymorphism and colorectal cancer risk: a meta-analysis. Onkologie 2010; 33: 666–674. [DOI] [PubMed] [Google Scholar]
- Liu Y, Qin H, Zhang Y et al. P53 codon 72 polymorphism and colorectal cancer: a meta-analysis of epidemiological studies. Hepatogastroenterology 2011; 58: 1926–1929. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ding D, Wang X et al. Lack of association between XPD Lys751Gln and Asp312Asn polymorphisms and colorectal cancer risk: a meta-analysis of case–control studies. Int J Colorectal Dis 2011; 26: 1257–1264. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wei L, Wang Y et al. Genetic association between ERCC5 rs17655 polymorphism and colorectal cancer risk: evidence based on a meta-analysis. Asian Pac J Cancer Prev 2015; 16: 5565. [DOI] [PubMed] [Google Scholar]
- Khatami F, Noorinayer B, Mohebi SR et al. Effects of amino acid substitution polymorphisms of two DNA methyltransferases on susceptibility to sporadic colorectal cancer. Asian Pac J Cancer Prev 2009; 10: 1183–1188. [PubMed] [Google Scholar]
- Chen C, Wang L, Liao Q et al. Association between six genetic polymorphisms and colorectal cancer: a meta-analysis. Genet Test Mol Biomarkers 2014; 18: 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Cao M, Gao K et al. The role of O(6)-methylguanine-DNA methyltransferase polymorphisms in colorectal cancer susceptibility: a meta analysis. Int J Clin Exp Med 2015; 8: 791–799. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.