Abstract
Membrane transporter proteins are critical for cellular uptake and export of molecules, and are reported to function by a number of different molecular mechanisms. The new occluded state structure of the uracil transporter, UraA, from Escherichia coli, reveals that both coordinated movement of the two domains of a single protomer together with dimer formation are important for transport activity.
The ability to effectively transport nutrients, ions and waste products across the semi-permeable membrane that surrounds cells and organelles is essential for life. In recent years, our understanding of the molecular mechanisms of membrane transport has increased, largely as a result of a number of high-resolution structures of the integral membrane transporter proteins, however, many questions still remain. One key group of transporter proteins, the secondary active transporters, couple the downhill transport of an ion (typically H+ or Na+) to the uphill transport of a substrate. These transporters broadly function via an “alternating access mechanism” whereby the substrate-specific binding site is exposed on one side of the membrane, and upon substrate and ion binding the protein undergoes a significant conformational change to reveal the binding site on the other side of the membrane. There are a number of different models describing the precise conformational rearrangements associated with transporter activity1. One of these, the so-called “elevator model”, was first described for the glutamate transporter from Pyrococcus horikoshi2 and has since been suggested as a mechanism of action for both a range, and rapidly increasing number, of transporters (e.g.,3,4).
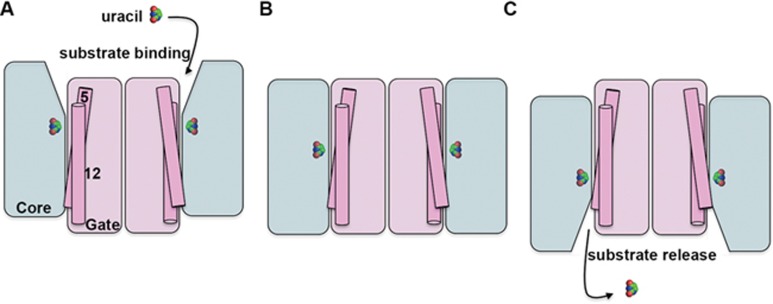
Central to the elevator model of transport activity is the organization of each transporter protomer into two discrete domains, with one domain often referred to as the core and the other as the oligomerization or gate domain. The substrate binds to the core domain, which then undergoes a translation through the membrane moving against the gate domain, thus carrying the substrate from one side to the other like an elevator (Figure 1). The structures obtained to date have indicated substantial conformational change in the core domain relative to the gate, with little or no detectable rearrangement of the gate domain, which is assumed to act rather like a scaffold, supporting movement of the core domain. However, the occluded state structure of the uracil transporter, a nucleobase ascorbate transporter (NAT), from Escherichia coli reported by Yu and colleagues5 reveals conformational changes in the gate domain associated with the transport cycle. Transmembrane domains (TMs) 5 and 12 of UraA form part of the gate domain and sit at the interface between the gate and the core. Comparison with the inward facing conformation of UraA6 indicates that the transition from the occluded to the inward open form appears to involve conformational rearrangement of TMs 5 and 12. This is suggested to contribute to the opening of the translocation channel allowing release of substrate from the binding site into the cytoplasm. This is an indication that the gate domain can act as more than just a static scaffold.
Figure 1.
Schematic illustrating the elevator model of transporter activity. In the case of UraA, the substrate uracil binds to the outward facing conformation (A) and the protein transitions occur via the occluded state (B), to the inward facing conformation (C) where the substrate is released. The conformational changes involved in transport are largely observed in the core domain (blue), which is thought to move against the gate domain (pink). The new occluded state structure of UraA indicates that conformational changes in TMs 5 and 12 (indicated), which sit at the interface between the two domains, might also be involved. Uracil is shown in space filling model.
The earlier inward open structure of UraA although initially described as a monomer is in fact a dimer, albeit a somewhat distorted one6. The current, occluded state structure confirms this oligomeric form which is also seen in related proteins. Intriguingly, dimer formation appears to be essential for transport activity, however, co-expression of wild-type UraA with a non-functional mutant does not affect the activity indicating a lack of functional interdependency of the two protomers. This is in contrast to the related uric acid/xanthine transporter, from Aspergillus nidulans, UapA7. In this case, studies of dominant negative mutants reveal that one UapA protomer affects the overall transport activity of the associated molecule. These findings suggest that the UapA gate domain may also act as more than a static scaffold, since how one protomer affects the activity of the other is not clear from the predicted elevator mechanism.
Clearly, the findings presented by Yu et al. provide novel insights into the mechanism of action of UraA. Importantly, UraA and the other NATs, are structurally homologous to the SLC4 family of anion exchangers8 and the SLC26 family of anion transporters9. Mutations in these transporters are associated with a number of disease states10,11. Thus, the information provided by this new UraA structure may be relevant for both a deeper understanding of how these transporters function and of the molecular basis of the associated diseases, and may ultimately contribute to the design of drugs to treat them.
While several questions still remain, such as how are all the conformational changes required for transport coordinated? And exactly what role does dimer formation have in transport through UraA and other related transporters? This study reveals that the molecular basis of transport via the elevator mechanism is more complicated and potentially more variable than previously thought.
References
- Drew D, Boudker O. Annu Rev Biochem 2015; 85:543–572. [DOI] [PubMed]
- Reyes N, Ginter C, Boudker O. Nature 2009; 462:880–885. [DOI] [PMC free article] [PubMed]
- Lee C, Kang HJ, Ballmoos von C, et al. Nature 2013; 501:573–577.23995679
- Hirschi M, Johnson ZL, Lee SY. Nature 2017; 459:347. [DOI] [PMC free article] [PubMed]
- Yu X, Yang G, Yan C, et al. Cell Res 2017 Jun 16. doi:10.1038/cr.2017.83
- Lu F, Li S, Jiang Y, et al. Nature 2011; 472:243–246. [DOI] [PubMed]
- Alguel Y, Amillis S, Leung J, et al. Nat Commun 2016; 7:11336. [DOI] [PMC free article] [PubMed]
- Arakawa T, Kobayashi-Yurugi T, Alguel Y, et al. Science 2015; 350:680–684. [DOI] [PubMed]
- Geertsma ER, Chang YN, Shaik FR, et al. Nat Struct Mol Biol 2015; 22:803–808. [DOI] [PubMed]
- Alper SL, Sharma AK. Mol Aspects Med 2013; 34:494–515. [DOI] [PMC free article] [PubMed]
- Reithmeier RA, Casey JR, Kalli AC, et al. Biochim Biophys Acta 2016; 1858:1507–1532. [DOI] [PubMed]