Abstract
Adult mammalian hearts cannot repair by themselves after injury due to limited proliferation of cardiomyocytes; removal of cell cycle blocker and/or addition of drugs that boost proliferation of cardiomyocytes provide potential means to cardiac regeneration. Three publications that appeared recently in Nature and Cell Research now provide new hope to the treatment of heart injuries.
Myocardial infarction (MI) is a worldwide disease that becomes the leading cause of death. Currently, close to 40 million patients worldwide suffer from heart failure due to loss of cardiomyocytes and excessive fibrotic tissue replacement1. In acute severe MI, a huge number of cardiomyocytes die due to loss of sufficient blood supply. The remaining cardiomyocytes have very limited regenerative capacity to restore the damaged tissue, ultimately leading to cardiac fibrosis and heart failure. How to enable an adult mammalian heart to regenerate after injury remains a fundamental question that is of utmost significance to the cardiovascular field.
Currently there are two main paradigms pursued to generate new cardiomyocytes for cardiac regeneration. The first is through activation of resident cardiac stem cells that have been reported to give rise to new cardiomyocytes after injury2. However, recently several independent studies cast doubt on the authenticity of c-kit+ stem cells in vivo3,4, raising controversies on stem cell-based regenerative medicine in the cardiovascular field. The second paradigm focuses on the reactivation of pre-existing cardiomyocytes for expansion after cardiac injury. The mammalian heart is a terminally differentiated organ without noticeable cardiomyocyte proliferation. However, a recent study on human heart showed evidence of cardiomyocyte turnover, ranging from 0.3% to 1% per year5. However, this low cell turnover rate is usually not sufficient to produce a meaningful number of cardiomyocytes that is of clinical significance after cardiac injury. Indeed, lower vertebrates such as zebrafish could regenerate the heart after apex resection6. Mammalian mouse heart also possesses this remarkable regenerative capacity after heart apex resection at early neonatal stages such as postnatal day 1 (P1), but loses this ability soon after P77. In these regenerative models, new cardiomyocytes are derived from pre-existing ones through proliferation, dedifferentiation and redifferentiation, in which cytoskeleton and myofibers are disassembled and reassembled to facilitate these processes7,8. Elucidating the molecular mechanisms especially the key barriers of cell cycle regulation will provide new means for cardiac regeneration. Inspired by the idea of helping the heart to repair itself at early but not late neonatal stage, researchers begin to focus more on the molecular components that govern cardiomyocyte proliferation at different stages. The difference between P1 and P7 poses some intriguing questions: what is the key component that causes regenerative difference between P1 and P7 hearts? What are the roadblocks that prevent cardiomyocytes from entering the cell cycle?
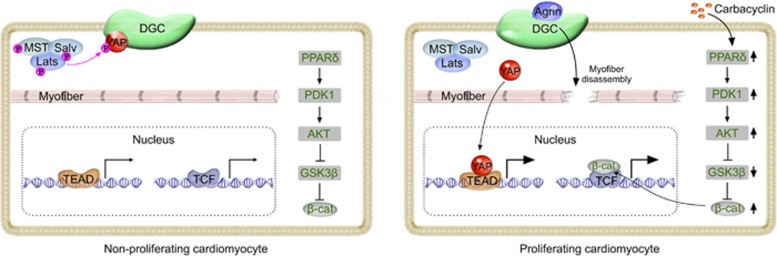
Bassat et al.9 reported in Nature that extracellular matrices (ECM) from P1 and P7 hearts differ significantly in stimulating cardiomyocyte proliferation. Through mass spectrometry analysis, Bassat et al. identified the ECM-associated molecule agrin. This matrix glycoprotein agrin promotes cardiomyocyte proliferation in vitro and is largely detected in coronary endothelial cells in the heart tissue. Genetic ablation of agrin in cardiac lineages leads to impaired regeneration after apex resection at P19. Amazingly, a single myocardial injection of agrin significantly induces cardiomyocyte proliferation, reduces cardiac fibrosis and improves heart function in both juvenile and adult hearts. Agrin also promotes proliferation and attenuates maturation of human cardiomyocytes. Mechanistically, agrin binds its membrane-bound receptor dystroglycan 1 (DAG1), which promotes disassembly of Dystrophin-glycoprotein complex (DGC). This results in reduced integrity of the cytoskeleton and myofibers, which is required for cardiomyocyte dedifferentiation. Interestingly, agrin-DAG1 interaction also facilitated dissociation of Yap protein from DGC and its translocation to the nucleus (Figure 1). Yap activates the transcription of genes involved in cardiomyocyte proliferation, and is negatively regulated by the Hippo signaling pathway that controls heart size10. Another study led by Martin group11 concurrently published in Nature provided an independent line of evidence that DGC sequesters YAP to inhibit cardiomyocyte proliferation. By simultaneous deletion of Hippo pathway component Salvador (Salv) and Mdx (loss of dystrophin function), an excessive myocardial growth is observed after apex resection, which strongly argues for the convergence of two signaling pathways (DGC and Hippo) in regulating cardiomyocyte proliferation. Mechanistically, DAG1 directly binds YAP to inhibit cardiomyocyte proliferation, and their interaction was enhanced by Hippo-induced Yap phosphorylation. Simultaneous removal of DAG1 and Hippo release YAP into the nucleus, promoting cardiomyocyte over-proliferation (Figure 1). These two back-to-back Nature papers employ elegant genetic methods to elucidate the in vivo function of key components of two pathways. Additionally, the powerful single magic injection of agrin represents a novel therapeutic treatment for cardiac repair and regeneration in this field. The potential application of disruption of DAG1-YAP interaction not only helps adult cardiomyocyte to proliferate after heart injuries, but also may improve the poor cardiac function in children with muscular dystrophy. Undoubtedly, these two studies provide a new research beacon that a latent regeneration machinery in adult mammalian heart can be awoken by removal of two or more (rather than one) roadblocks of cardiomyocyte proliferation.
Figure 1.
Signaling pathways governing cardiomyocyte proliferation. Left: YAP phosphorylated by Hippo signaling is sequestered by DGC. Myofiber is mature with intact sarcomeric structure. PPARδ/PDK1/AKT/GSK3β/β-cat activity is maintained at basal level. Right: ECM protein agrin binds DGC, which promotes the disassembly of DGC and subsequently myofiber disassembly and YAP activation9,11. Chemical compound carbacyclin increases PPARδ activity, which activates β-cat via PDK1/AKT/GSK3β pathway12. YAP-TEAD and β-cat-TCF promote transcriptional regulation of cardiomyocyte proliferation.
Almost concurrently, Magadum et al.12 reported in Cell Research a new way to induce mammalian cardiomyocyte proliferation. By a modified Fucci-based cell cycle stage screening assay for chemical compound libraries, Engel and his colleagues identified carbacyclin, an agonist of PPARδ, as a potential inducer of postnatal cardiomyocyte proliferation. Carbacyclin also increased proliferation of human induced pluripotent stem cell-derived cardiomyocytes12. Mechanistically, carbacyclin promotes cardiomyocyte proliferation via PPARδ/PDK1/Akt pathway, upregulating the expression of positive regulators of cell cycle progression and downregulating the cell cycle inhibitors (Figure 1). Adult zebrafish heart has regeneration capacity via cardiomyocyte proliferation8, but when the zebrafish was treated with a PPARδ inhibitor after heart injury, its regenerative ability was inhibited, indicating that PPARδ signaling is required for adult cardiomyocyte cell cycle activity. Given a significant difference in regenerative potential between fish and mouse hearts, Engel and colleagues also overexpressed PPARδ in adult mouse heart to test its potential efficacy in treatment of MI. Notably, overexpression of active PPARδ in cardiomyocytes as well as PPARδ agonist treatment significantly induced cell cycle progression, leading to reduced infarct scars and improved heart function12. This impressive piece of work advances the cardiovascular field by presenting a new pathway that could be explored to promote heart muscle regeneration after injury. The elegant live cardiomyocyte screening platform represents a useful and efficient way to identify new chemical compounds that accelerate cardiomyocyte cell cycle. This study also raises more intriguing questions that remain to be addressed. Whether treatment with a single chemical compound alone could stimulate cardiomyocyte proliferation in vivo and protect myocardium from excessive fibrosis and heart failure? Do PPARδ/PDK1/Akt pathway and DAG1-Yap converge at some point to regulate cardiomyocyte proliferation? What is the common downstream effector pathway by which carbacyclin, agrin and Hippo pathway function to release cardiomyocytes from the inhibitory checkpoint, e.g., β-catenin? It would be interesting to hypothesize that combination of addition of cell cycle stimulators and removal of brakes would further spur the regenerative potential of adult mammalian heart after injuries.
References
- Tzahor E, Poss KD. Science 2017; 356:1035–1039. [DOI] [PMC free article] [PubMed]
- Ellison GM, Vicinanza C, Smith AJ, et al. Cell 2013; 154:827–842. [DOI] [PubMed]
- van Berlo JH, Kanisicak O, Maillet M, et al. Nature 2014; 509:337–341. [DOI] [PMC free article] [PubMed]
- Liu Q, Yang R, Huang X, et al. Cell Res 2016; 26:119–130. [DOI] [PMC free article] [PubMed]
- Bergmann O, Bhardwaj RD, Bernard S, et al. Science 2009; 324:98–102. [DOI] [PMC free article] [PubMed]
- Poss KD, Wilson LG, Keating MT. Science 2002; 298:2188–2190. [DOI] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Science 2011; 331:1078–1080. [DOI] [PMC free article] [PubMed]
- Kikuchi K, Holdway JE, Werdich AA, et al. Nature 2010; 464:601–605. [DOI] [PMC free article] [PubMed]
- Bassat E, Mutlak YE, Genzelinakh A, et al. Nature 2017; 547:179–184. [DOI] [PMC free article] [PubMed]
- Heallen T, Zhang M, Wang J, et al. Science 2011; 332:458–461. [DOI] [PMC free article] [PubMed]
- Morikawa Y, Heallen T, Leach J, et al. Nature 2017; 547:227–231. [DOI] [PMC free article] [PubMed]
- Magadum A, Ding Y, He L, et al. Cell Res 2017. Jun 16. doi:10.1038/cr.2017.84