Abstract
Phosphatidylinositol 3-phosphate (PI3P) plays essential roles in vesicular trafficking, organelle biogenesis and autophagy. Two class III phosphatidylinositol 3-kinase (PI3KC3) complexes have been identified in mammals, the ATG14L complex (PI3KC3-C1) and the UVRAG complex (PI3KC3-C2). PI3KC3-C1 is crucial for autophagosome biogenesis, and PI3KC3-C2 is involved in various membrane trafficking events. Here we report the cryo-EM structures of human PI3KC3-C1 and PI3KC3-C2 at sub-nanometer resolution. The two structures share a common L-shaped overall architecture with distinct features. EM examination revealed that PI3KC3-C1 “stands up” on lipid monolayers, with the ATG14L BATs domain and the VPS34 C-terminal domain (CTD) directly contacting the membrane. Biochemical dissection indicated that the ATG14L BATs domain is responsible for membrane anchoring, whereas the CTD of VPS34 determines the orientation. Furthermore, PI3KC3-C2 binds much more weakly than PI3KC3-C1 to both PI-containing liposomes and purified endoplasmic reticulum (ER) vesicles, a property that is specifically determined by the ATG14L BATs domain. The in vivo ER localization analysis indicated that the BATs domain was required for ER localization of PI3KC3. We propose that the different lipid binding capacity is the key factor that differentiates the functions of PI3KC3-C1 and PI3KC3-C2 in autophagy.
Keywords: PI3KC3, ATG14L, UVRAG, autophagy, PI, cryo-EM
Introduction
Autophagy is an evolutionarily conserved, lysosome-based degradation pathway that plays essential roles in maintaining cellular homeostasis1. During autophagy, double-membrane vesicles named autophagosomes are formed and later fuse with lysosomes to generate autolysosomes1. As a crucial step in the initiation of autophagy, phosphatidylinositol 3-phosphate (PI3P) is generated on specialized subdomains of the endoplasmic reticulum (ER) named omegasomes2. Omegasomes give rise to intermediate structures named isolation membranes (or phagophores), which then mature into autophagosomes3. It is well established that class III phosphatidylinositol-3-kinase (PI3KC3) complexes are responsible for generating PI3P4,5,6.
So far, two types of PI3KC3 complex have been identified, PI3KC3-C1 and PI3KC3-C2. They share three common components (P150, VPS34 and Beclin1) but are distinguished from each other by a fourth mutually exclusive component, ATG14L for PI3KC3-C1 or UVRAG for PI3KC3-C2 (Figure 1A)7. The ATG14L-containing PI3KC3-C1 (referred to as C1 hereafter) is essential for autophagy8,9, whereas the UVRAG-containing PI3KC3-C2 (referred to as C2 hereafter) is involved in multiple cellular processes including autophagy, endocytic trafficking, cytokinesis and Golgi-ER retrograde transport10,11,12.
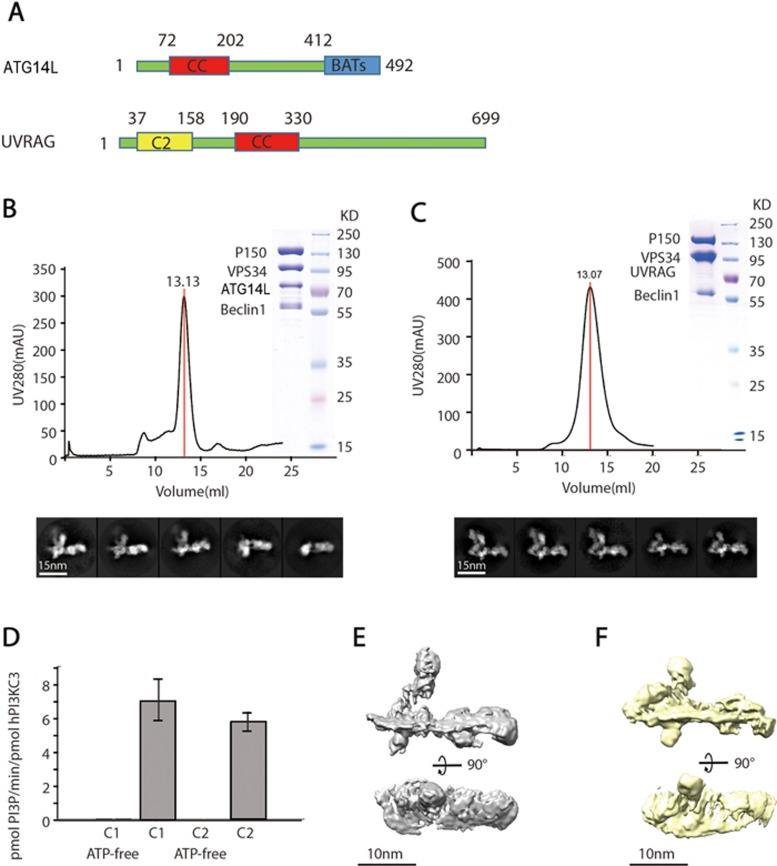
Figure 1.
Purification and reconstruction of C1 and C2. (A) Domain organization of ATG14L and UVRAG. (B, C) Chromatography of C1 and C2 on Superose 6 10/300 GL columns. The dark curves show the UV absorbance at 280 nm, and the peaks containing C1 and C2 are marked by red lines along with the elution volume. SDS-PAGE analyses of the complexes in the peaks are shown at the right of the top panels in B and C. The corresponding proteins are labeled on the left of the PAGE images. The molecular weights of the standard markers are labeled on the right. Classical 2D class averages of cryo-EM images of C1 and C2 are shown in the bottom panels. (D) In vitro lipid kinase assay of C1 and C2. Data are represented as mean ± SD (n = 3). (E) 3D reconstruction volume of C1 (grey) with a threshold of 3.9 σ. The model in the bottom panel is rotated 90 degrees along the main axis compared to the model in the top panel. (F) 3D reconstruction volume of C2 (yellow) with a threshold of 3.8 σ. The model in the bottom panel is rotated 90 degrees along the main axis compared to the model in the top panel.
During the initiation of autophagy, C1 is recruited to ER/mitochondria contact sites13, where it phosphorylates PI, a relatively abundant phospholipid in the ER14, to generate PI3P, thus giving rise to omegasomes. The BATs domain in the C-terminal portion of ATG14L is capable of binding PI3P15. However, it is unlikely that ATG14L/C1 is recruited to the ER through interaction with PI3P, as starvation significantly enhances PI3P generation on the ER, whereas the recruitment of C1 to the ER is largely unchanged in response to starvation16. So far, it is not clear how C1 binds to its substrate PI, or whether this interaction is required to recruit C1 to membranes.
Recent single-particle reconstructions of human C1 and C2 using negative-stain EM showed that they have a similar overall shape and architecture at low resolution17. The structure of yeast C2, which consists of VPS34, VPS15 (yeast homolog of P150), ATG6 (yeast homolog of Beclin1) and VPS38 (yeast homolog of UVRAG), was determined by X-ray crystallography at 4.4 Å resolution18. Yeast C2 has a similar L-shape architecture as the human PI3KC3 complexes. Despite this structural information, the molecular basis of the functional difference between the human C1 and C2 complexes is still unclear.
In this study, we obtained 3D reconstructions of the human C1 and C2 complexes using single-particle cryo-EM at ∼9 Å resolution. These allowed us to compare the structural similarities and differences between the two complexes in more detail. Furthermore, we studied the interaction of the two complexes with membranes of various phospholipid compositions and directly visualized the orientation of the molecules on membranes by EM. In combination with biochemical assays, these results provide structural and mechanistic insights into the functional difference between the two types of PI3KC3 complex.
Results
Protein purification and architecture definition
We used a transient transfection system in the 293F cell line to co-express the C1 and C2 complexes with strep-tagged ATG14L or strep-tagged UVRAG, respectively, and purified the complexes using Strep-Tactin affinity resin and size exclusion chromatography on Superose 6 columns (see Materials and Methods). Both complexes appeared as well-behaved molecules in a prominent peak with similar elution volumes (Figure 1B and 1C). SDS-PAGE verified the stoichiometric subunit composition of each complex (Figure 1B and 1C). The purified complexes effectively phosphorylated PI to PI3P in in vitro assays (Figure 1D).
We performed negative-staining EM and single-particle analysis of the two complexes. 2D and 3D reconstructions of C1 revealed L-shaped architectures (Supplementary information, Figure S1) similar to those previously reported17. Using various MBP-tagged or domain-deleted constructs, we defined the locations of different domains in the C1 complex (Supplementary information, Figure S1B). 2D class averages of the C2 complex exhibited a very similar L-shaped structure to that of C1, except that a globular domain on the short arm of the L-shape appeared more flexible in C1 than in C2 (Supplementary information, Figure S1A and S1C). It is tempting to hypothesize that this globular structural element, which is clearly distinct in the two complexes (red arrows in Supplementary information, Figure S1A and S1C), is due to the presence of ATG14L and UVRAG, which are exclusive components of C1 and C2, respectively. Such a possibility, however, was ruled out by the fact that the globular domain was completely lost in a C1 complex containing VPS34 with a C-terminal truncation (Supplementary information, Figure S1B). Furthermore, the C-terminus of UVRAG is located at the distal end of the long arm of the L-shape (Supplementary information, Figure S1D), in a similar location to the C-terminus of ATG14L in the C1 complex (Supplementary information, Figure S1B). Using random conical tilt reconstruction, we obtained 3D reconstructions of both C1 and C2 from their negatively stained specimens. These reconstructions share a highly homologous architecture, which allows us to roughly define the subunit locations in the C1 and C2 3D models (Supplementary information, Figure S1E-S1G).
Cryo-EM reconstructions of the C1 and C2 complexes
Despite the similar overall shape of the C1 and C2 complexes, it is critical to unveil their structures in more detail in order to understand their different functions. We therefore performed cryo-EM of the two complexes. From electron micrographs of complexes embedded in vitreous ice, we were able to pick particle images and perform single-particle analysis of the C1 and C2 complexes (Supplementary information, Figure S1H-S1K). 2D classification of the images generated class averages with fine details, presumably showing different views of the L-shaped structure (Figure 1B and 1C; Supplementary information, Figure S1I and S1K). It is noteworthy that the 2D class averages of C1 are of relatively poor quality compared to those of C2. In particular, the density corresponding to the C-terminal domain (CTD) of VPS34 is more blurred in C1. In contrast, the CTD of VPS34 in C2 is more rigid in a location adjacent to the CTD of P150. We performed 3D classification and refinement of the C1 and C2 cryo-EM data set using the procedures shown in Supplementary information, Figure S2. In agreement with the 2D classification results, the VPS34 CTD density was largely missing in the C1 complex due to its high flexibility (Supplementary information, Figure S2A). The same density is also missing in some 3D classes of the C2 complex but is present in others (Supplementary information, Figure S2B). To further improve the resolution of the reconstruction, we used a mask to exclude the VPS34 CTD and refined the complexes to 8.5 Å for C1 and 8.6 Å for C2 (Figure 1E and 1F; Supplementary information, Figure S3A and S3B). The most rigid portion of the long arm of C1 was further refined to a resolution of 5.9 Å (Supplementary information, Figure S3C).
Structural comparison of C1 and C2
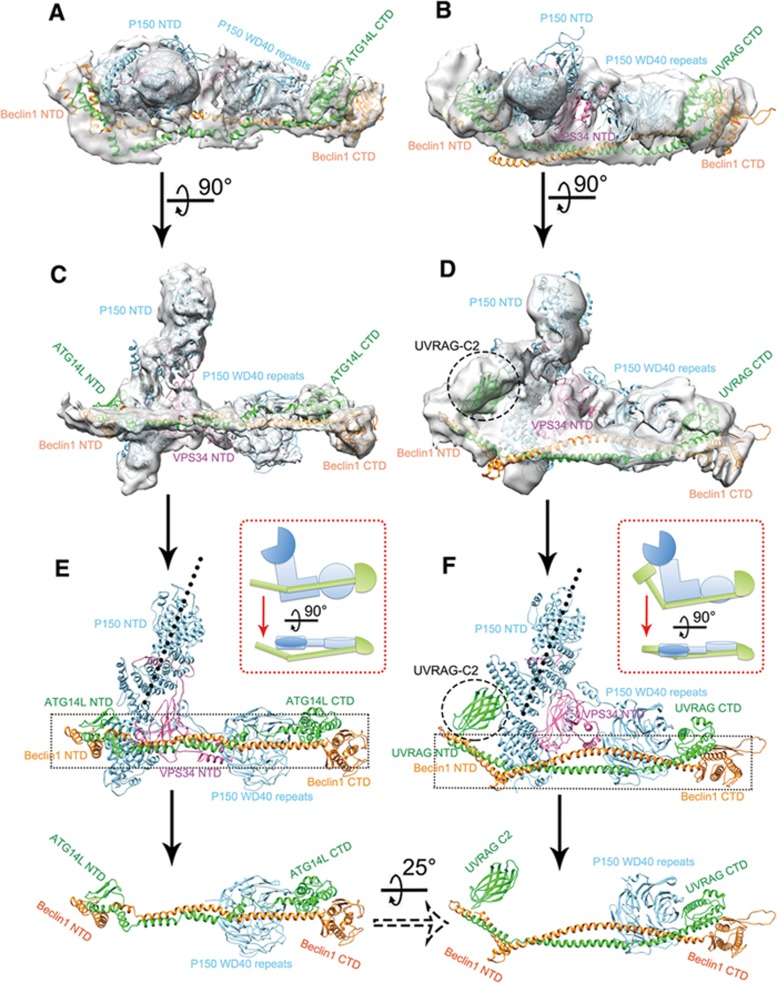
The 3D reconstructions of C1 and C2 by cryo-EM showed a similar L shape overall, with distinct differences. We used homologous structures to build atomic models in the 3D map of C1 and further refined the model using molecular dynamics flexible fitting (MDFF) in the region of relatively high-resolution density in the long arm (see Materials and Methods). The cross correlation coefficient between the final atomic model and the EM density of C1 is about 0.92 (Figure 2A and 2C). For C2, we found that the atomic model of the yeast C2 solved by X-ray crystallography (PDB code: 5DFZ) can be docked into our EM map as a rigid body with a cross correlation coefficient of 0.83 (Figure 2B and 2D). The model and the map agree in most of the features. Therefore, we directly used the crystal structure of yeast C2 as a hypothetical model of human C2 for structural comparison with human C1.
Figure 2.
Atomic model building and analysis of C1 and C2. (A, C) The chimeric EM volume of C1 docked with the built atomic model (VPS34-CTD excluded). Within the atomic model, the Beclin1, ATG14L, VPS34 and P150 protein subunits are colored orange, light green, hot pink and cyan, respectively. The chimeric EM volume is presented as a translucent grey surface. A shows the top view and C represents the side view. (B, D) The refined EM volume of C2 docked with the hypothetical atomic model of C2 (VPS34-CTD excluded), which was directly extracted from the model of yeast PI3K complex 2 (PDB code: 5DFZ). Within the atomic model, the Beclin1, UVRAG, VPS34 and P150 subunits are colored orange, light green, hot pink and cyan, respectively. The chimeric EM volume is presented as a translucent grey surface. B shows the top view and D represents the side view. The dashed circle in D indicates the electron density from the β-barrel insertion within the NTD of UVRAG. (E, F) The built atomic model of C1 and the docked model of C2. The dashed rectangles indicate Beclin1, ATG14L and the P150 WD domain in C1 and Beclin1, UVRAG and the P150 WD domain in C2, which were extracted from the overall model and are presented in the bottom panels with the same orientation. The relative orientation of these two extracted parts is marked with the rotation axis and angle. The top right panels in E and F show cartoon models of the side and top views of C1 and C2. The green shapes indicate the Beclin1/ATG14L subunits in C1 and the Beclin1/UVRAG subunits in C2. The blue shapes indicate the P150/VPS34-NTD subunits in both complexes.
Besides the different flexibilities of the CTD of VPS34 in C1 and C2, the structures of the two complexes have other distinct features related to their unique subunits ATG14L or UVRAG. Sequence alignment of ATG14L with UVRAG suggested that UVRAG contains a longer N-terminal domain (NTD; C2 domain) than ATG14L but lacks a BATs domain at its C-terminal end (Figure 1A). Consistent with the longer UVRAG NTD, the EM map of C2 shows an extra density attached to the NTD of P150 compared to P150 in C1 (Figure 2D). This extra density corresponds to the β-barrel insertion within the UVRAG NTD. It is intriguing to see that ATG14L and UVRAG both form long helix bundles with Beclin1 and span the entire complex. Interestingly, the helix bundle formed by UVRAG and Beclin1 in C2 is rotated downward about 25 degrees around the long arm of the L-shaped complex compared to the C1 counterpart formed by ATG14L and Beclin1 (Figure 2E and 2F). As a consequence, the helix bundle in the C1 complex has a larger interaction interface (4 459 Å2) with the helical and WD40 domains than that in the C2 complex (4 130 Å2). This may cause the C1 complex to adopt a relatively more rigid L-shaped structure than the C2 complex, which might explain the different abilities of the complexes to sense membrane curvature, as has been shown previously15,18.
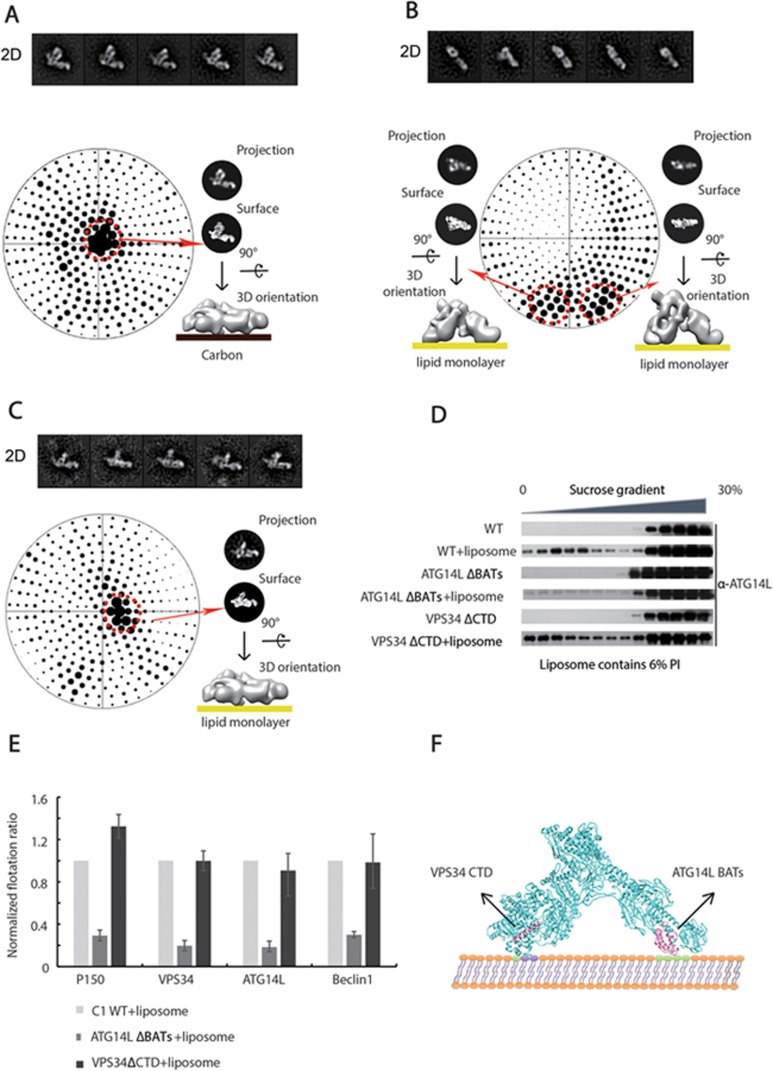
ATG14L-CTD and VPS34-CTD determine the orientation of C1 on membranes
To directly visualize the orientation of the C1 complex on membranes, we developed an EM protocol (see Materials and Methods) to determine the orientation of PI3K complexes on lipid monolayers (Supplementary information, Figure S4A). Interestingly, we found that the majority of C1 particles adopt a “standing up” orientation on lipid monolayers, in dramatic comparison to their mostly L-shaped appearance on hydrophilic carbon films (Figure 3A and 3B). Statistical orientation analysis showed that the majority of C1 particles interact with membranes via the two vertexes (VPS34-CTD and ATG14L-CTD) of the L shape (Figure 3B). To verify that C1 binds to membranes through these two specific domains, we generated mutated complexes with deletion of either VPS34-CTD or ATG14L-BATs, and analyzed their membrane-binding capacity using liposome flotation assays. We found that the ATG14L-BATs deletion strongly impaired the ability of C1 to bind membranes, whereas the VPS34-CTD deletion barely influenced the membrane-binding capacity (Figure 3D and 3E; Supplementary information, Figure S4E). Next, we examined the orientation of these mutants on membranes. We found that for the complex with the ATG14L-BATs deletion, almost no particles were observed on lipid monolayers, which is consistent with the fact that the ATG14L-CTD is essential for membrane binding of C1. Interestingly, for the C1 complex with the VPS34-CTD deletion, most particles presented an L-shaped side view, which is similar to the orientation of C1 on hydrophilic carbon films (Figure 3C). From these data, we concluded that the BATs domain of ATG14L is responsible for anchoring C1 on membranes, whereas the CTD of VPS34 determines the orientation of C1 on membranes (Figure 3F).
Figure 3.
Membrane binding orientation of C1. (A) Orientation of C1 on carbon films. The upper panel presents the five top 2D class averages of WT C1 on carbon. The lower panel presents the Euler angle distribution of all the particles generated by SPIDER. The sizes of the dark dots correlate with the number of particles in a certain orientation. A classical surface view coupled with the projection for the preferred orientation is shown on the right. The cartoon model indicates the general appearance of the particles on carbon film in their preferred orientation. (B) Orientation of C1 on lipid monolayers. The upper panel presents the five top 2D class averages of WT C1 bound to lipid monolayers. The lower panel presents the Euler angle distribution of all the particles generated by SPIDER. The sizes of the dark dots correlate with the number of particles in a certain orientation. Two classical surface views corresponding to the projections of the preferred orientations are shown on the left and right. The cartoon models indicate the general appearance of WT C1 particles on lipid monolayers in their preferred orientations. (C) Orientation of truncated C1 with VPS34 CTD deletion. The descriptions of the figure panels are the same as in A. (D) Flotation assay to examine the binding of C1 WT, C1 VPS34 ΔCTD and C1 ATG14L ΔBATs to liposomes containing 6% PI in a sucrose gradient (from top to bottom: 0%, 20%, 25%, 30%). All 14 fractions from top to bottom were immunoblotted using an antibody against ATG14L. (E) The flotation ratio from D for ATG14L and Supplementary information, Figure S4E for P150, VPS34 and Beclin1 was calculated by dividing the total intensity of the bands in all the fractions with the intensity of the bands in the top 8 fractions, then normalizing to the value for C1 WT. Data are represented as mean ± SD (n = 3). (F) Membrane binding model of C1. The structural areas responsible for membrane binding (ATG14L BATs domain) and the orientation of the complex (VPS34 CTD) are colored in hot pink.
The BATs domain determines the ER-binding capacity of PI3KC3 complexes
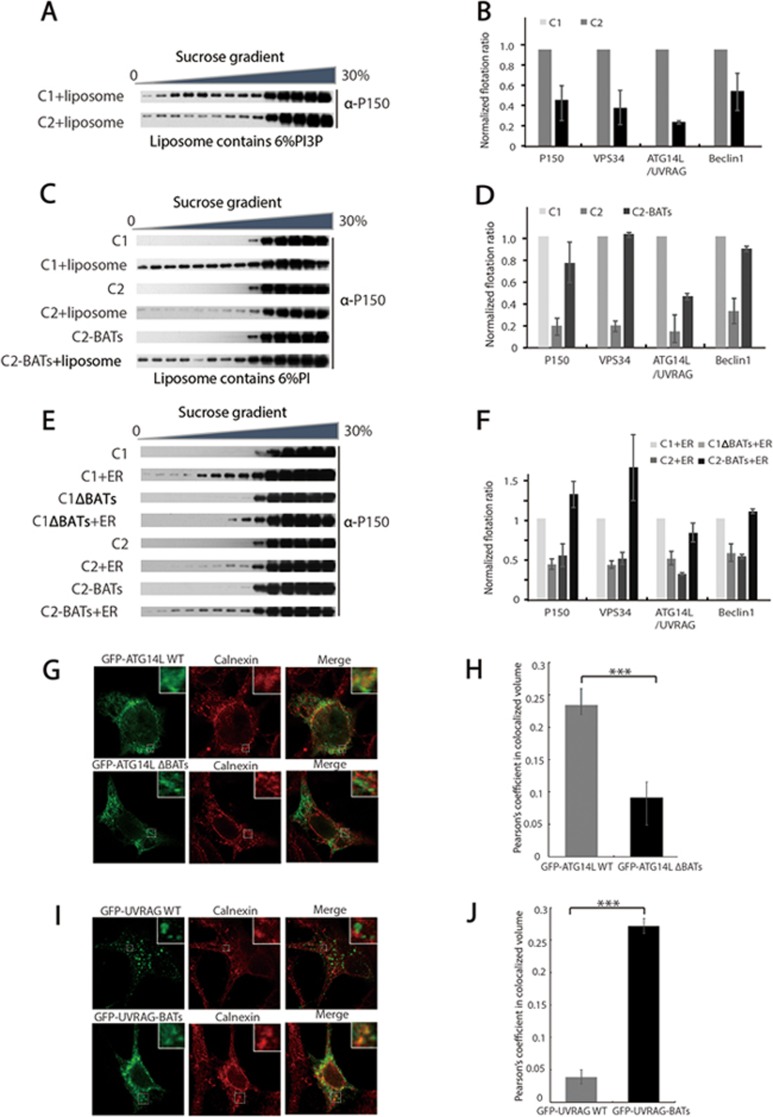
Despite the fact that C1 and C2 have very similar architectures, they are involved in different membrane trafficking processes. To investigate the molecular mechanism underlying the functional difference between C1 and C2, we performed liposome flotation assays to assess the membrane-binding properties of the two complexes. We found that both C1 and C2 can bind to liposomes containing PI3P, PI(4,5)P2 or PI(3,4,5)P3, although the binding of C2 is weaker than C1 (Figure 4A and 4B; Supplementary information, Figures S5C, S6C and S6E). To our surprise, C2 showed no or very weak binding affinity to liposomes containing PI but not PI3P (Figure 4C and 4D; Supplementary information, Figure S6B and S6D). In contrast, C1 strongly bound to the PI-only liposomes (Figure 4C and 4D; Supplementary information, Figure S5B). Next, we generated a chimeric protein by fusing the ATG14L-BATs domain to the C-terminus of UVRAG (UVRAG-BATs) and purified the C2 complex containing this recombinant protein. Interestingly, we found that the chimeric C2 complex was able to bind to the PI-only liposomes as effectively as the wild-type C1 (Figure 4C and 4D; Supplementary information, Figure S5B).
Figure 4.
The BATs domain determines the ER-binding capacity of PI3KC3 complexes in vitro. (A) Flotation assay to examine the interaction of C1 and C2 with liposomes containing 6% PI3P in a sucrose gradient (from top to bottom: 0%, 20%, 25%, 30%). All 14 fractions from top to bottom were immunoblotted using an antibody against P150. (B) The flotation ratio from C for P150 and Supplementary information, Figure S5C for VPS34, ATG14L/UVRAG and Beclin1 was calculated by dividing the band intensities from all the fractions with the band intensities of the top 8 fractions, then normalizing to the value for C1. Data are represented as mean ± SD (n = 2 or 3). (C) Flotation assay to examine the binding of C1, C2 and C2-BATs to liposomes containing 6% PI in a sucrose gradient (from top to bottom: 0%, 20%, 25%, 30%). All 14 fractions from top to bottom were analyzed by immunoblotting using an antibody against P150. (D) The flotation ratio from A for P150 and Supplementary information, Figure S5B for VPS34, ATG14L/UVRAG and Beclin1 was calculated by dividing the band intensities of all the fractions with the band intensities of the top 8 fractions, then normalizing to the value for C1. Data are represented as mean ± SD (n = 2 or 3). (E) Flotation assay to examine the interaction of C1, C1 ΔBATs, C2 and C2-BATs with the ER fraction in a sucrose gradient (from top to bottom: 0%, 20%, 25%, 30%). All 14 fractions from top to bottom were analyzed using an antibody against P150. (F) The flotation ratio from E for P150 and Supplementary information, Figure S5F for VPS34, ATG14L/UVRAG and Beclin1 was calculated by dividing the input with the top 8 fractions, then normalizing to the value for C1. Data are represented as mean ± SD (n = 2 or 3). (G) Confocal analyses of the subcellular co-localization of GFP-ATG14L WT or GFP-ATG14L ΔBATs with the ER marker calnexin in HEK293 cells. The insets in the top right corner show a high magnification of the selected areas. (H) Quantification of co-localization of GFP-ATG14L WT or GFP-ATG14L ΔBATs with calnexin (data are shown as mean ± SD, n = 30 cells obtained by gathering data from three independent experiments). ***P < 0.001. (I) Confocal analyses of the subcellular co-localization of GFP-UVRAG WT or GFP-UVRAG-BATs with the ER marker calnexin in HEK293 cells. The insets in the top right corner show a high magnification of the selected areas. (J) Quantification of co-localization of GFP-UVRAG WT or GFP-UVRAG-BATs with calnexin (data are shown as mean ± SD, n = 30 cells obtained by gathering data from three independent experiments). ***P < 0.001.
Because the ER contains significantly more PI than PI3P, we hypothesized that the presence of the BATs domain in C1 should enable it to interact more strongly with the ER than C2. To test this hypothesis, we purified microsomes from 293F cells and performed flotation assays (Figure 4E and 4F; Supplementary information, Figure S5F). Similar to the reconstituted liposomes, we found that C2 bound more weakly to the ER than C1. Deletion of the BATs domain significantly decreased the ability of C1 to bind to the ER. The chimeric C2 complex containing recombinant UVRAG-BATs was able to interact with the ER to a similar extent as the wild-type C1.
Furthermore, we found that the BATs domain was also crucial for localization of PI3KC3 to the ER in vivo. We deleted the BATs domain from ATG14L and found that GFP-ATG14L ΔBATs was separated from the ER (Figure 4G and 4H). The presence of the BATs domain significantly enhanced the localization of GFP-UVRAG-BATs to the ER compared to GFP-UVRAG WT (Figure 4I and 4J). These results indicated that the BATs domain determines the ER-binding capacity of PI3KC3 complexes.
Discussion
In this study, we determined the cryo-EM structures of the human C1 and C2 complexes. The overall architecture of these two complexes is very similar; however, they differ in the arrangement of the Beclin1-ATG14L and Beclin1-UVRAG sub-complexes. The conformation of C2 is more rigid than C1 in the VPS34 CTD region. Furthermore, we determined the orientation of C1 by direct EM visualization and observed that ATG14L-BATs and VPS34-CTD, which are localized on the tips of opposite arms of the L-shaped complex, directly contact membranes. The ATG14L-BATs domain is responsible for anchoring C1 on membranes, whereas the VPS34-CTD determines the orientation of the complex. Finally, we showed that C1 and C2 differ in their ability to bind PI, and this difference is due to the BATs domain of the C1 complex.
The membrane binding of C1 is significantly reduced when the BATs domain of ATG14L is absent, suggesting that the BATs domain is essential for the interaction of C1 with membranes. On lipid monolayers, wild-type C1 adopted an orientation that may represent a “standing up” conformation or a flipped inverted triangle conformation. Although the CTD domain of VPS34 is not essential for membrane binding of C1, deletion of CTD changed the orientation of C1 on lipid monolayers, suggesting that it directly interacts with the membrane. Collectively, our results showed that the BATs domain is mainly responsible for membrane binding of C1, whereas the VPS34 CTD domain determines the orientation on the membrane. This indicated that C1 does not adopt an inverted triangle conformation, but instead “stands up” on the membrane, with the BATs domain and VPS34 CTD directly interacting with the membrane.
Autophagosomes emerge from omegasomes, which come from specialized subdomains of the ER. The formation of omegasomes depends on the phosphorylation of ER-enriched PI to PI3P by PI3KC3 kinases. Until now, it is unclear how the kinases were recruited to the ER. Our observations that ATG14L-BATs can bind PI and that ATG14L-BATs is necessary and sufficient to mediate the binding of PI3KC3 to PI-containing membranes suggests an appealing hypothesis that C1 directly interacts with PI and is therefore recruited to the ER. In this scenario, an autophagic stimulus triggers the interaction of ATG14L-BATs with PI and mediates enrichment of C1 on the ER. C1 “stands up” on the ER membrane, allowing the PI3K domain to be in close proximity to its substrate. The ER-localized C1 then phosphorylates PI to PI3P, which subsequently recruits other PI3P effectors, thus ensuring that autophagy proceeds smoothly.
It is well known that the C1 and C2 are involved in various membrane trafficking processes. C1 is essential for autophagy, whereas C2 mainly plays an important role in endosome-to-Golgi retrograde trafficking. It is interesting to note that PI3P is highly enriched in endosomes but not in ER19, whereas PI accounts for about 10% of the total ER lipid content14. These membrane systems are also different in their shapes and curvatures. The long helical bundles formed by Beclin1 and ATG14L in the C1 complex and by Beclin1 and UVRAG in the C2 complex adopt different configurations, creating different interaction interfaces between the helical bundles and the rest of the complex. This may account for the different rigidity of two arms of the L-shaped molecule upon binding to lipid membranes, C1 having more rigid L-shaped arms than C2. We speculate that the membrane binding selectivity of the two complexes is determined by the different lipid binding domains (BATs) and the rigidity of the molecules on membranes. Future studies of the detailed structures of C1 and C2 on their target membranes will illuminate the precise molecular features that underlie the functional differences between these two complexes.
Materials and Methods
Constructs and antibodies
The plasmids pCAG-OSF-ATG14L and pCAG-NT-VPS34 were gifts from Yoshimori's lab. The coding sequences of Beclin1, P150, UVRAG and the mutants ATG14L-ΔBATs (AA1-412) and VPS34-ΔCTD (AA1-421) were amplified by PCR and inserted into the KpnI/XhoI sites of pCAG-NT, pCAG-OSF or pEGFP-C3. The chimeric construct pCAG-OSF-UVRAG-BATs was obtained by inserting the BATs domain of ATG14L (AA413-492) into pCAG-OSF-UVRAG. The chimeric construct pEGFP-UVRAG-BATs was obtained by inserting the BATs domain of ATG14L (AA413-492) into pEGFP-UVRAG. Mouse anti-P150 and rabbit anti-VPS34 were purchased from Sigma. Mouse anti-ATG14L, rabbit anti-Beclin1 and mouse anti-UVRAG were purchased from MBL. Rabbit anti-Calnexin was purchased from Abcam. Mouse anti-GFP was purchased from Roche.
Protein expression and purification
For C1, the four plasmids pCAG-OSF-ATG14L, pCAG-NT-P150, pCAG-NT-VPS34 and pCAG-NT-Beclin1 were transfected into HEK293F cells. The cells were harvested and lysed in buffer containing 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM DTT, 1% Triton X-100 and protease inhibitor cocktail (Roche). After centrifugation at 15 000 rpm for 1 h at 4 °C, the supernatant was loaded onto a column containing Strep-Tactin resin (IBA) and incubated for 1 h at 4 °C. The resin was then washed 3 times. The protein was eluted with 10 mM desthiobiotin (Sigma) and further purified by gel filtration on Superose 6 10/300. For C2 (pCAG-OSF-UVRAG, pCAG-NT-P150, pCAG-NT-VPS34, pCAG-NT-Beclin1) and C2-BATs (pCAG-OSF-UVRAG-BATs, pCAG-NT-P150, pCAG-NT-VPS34, pCAG-NT-Beclin1), purification was carried out in the same way as for C1.
Negative-staining EM
Samples of the PI3KC3 complexes were diluted to a final concentration of 50-80 nM and negatively stained in a 2% (w/v) solution of uranyl acetate (Electron Microscopy Sciences) following the standard deep-staining procedure on glow-discharged holey carbon-coated EM copper grids covered with a thin layer of continuous carbon. The negatively stained specimens were then mounted onto a transmission electron microscope holder and examined by an FEI Tecnai Spirit electron microscope operated at 120-kV. Magnified digital micrographs of the specimen were taken at a nominal magnification of 49 000× on a Gatan Ultrascan 4000 CCD camera with a pixel size of 2.29 Å at the specimen level. The defocus values used were about −1.0 to −2.5 μm, and the total accumulated dose at the specimen was about 70 electrons per Å2.
Lipid monolayer sample preparation for EM analysis
A clean, dry Teflon plate with 3 × 3 wells was fixed on a flat table free from vibration. About 36 μl protein solution at a concentration of ∼100 nM was loaded into each well. The system was incubated at room temperature (RT) for 5 min to ensure that the protein solution was warmed to RT. Subsequently, about 0.6–0.8 μl of lipid mixture with a concentration of 1 mM (in chloroform solution) was gently loaded onto the surface of the protein solution using a micro-injector. The reaction system was kept static at RT for 20 min. The lipid monolayer on the top surface of the protein solution was then transferred to a hydrophobic carbon-coated Cu EM gird using vertical transfer. The sample on the Cu grid was then lightly washed once with ∼40 μl dilution buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM DTT) and twice with ∼40 μl 2% (w/v) uranyl acetate solution. The solution was blotted off each time with filter paper. 4 μl of stain solution was left on the grid to stain the specimen for ∼40 s before it was blotted off by a piece of filter paper and the specimen was allowed to air-dry. The grids were screened and imaged by TEM within 12 h of preparation.
Imaging processing for negative-staining EM
For the samples on carbon, the particles were semi-automatically picked in EMAN222, CTF corrected and 2D-classified without reference in IMAGIC-4D24. Random conical tilt reconstruction was performed using tilted pairs of negatively stained specimens using SPIDER as described23. 3D refinement of C1 was performed in SPIDER with the initial model obtained from the random conical tilt reconstruction. For the samples on lipid monolayers, the particles were manually picked in EMAN224 and 2D-classfied in IMAGIC-4D24. Particles were aligned and refined based on the projections of the refined PI3K 3D model. The Euler angle was statistically defined and the distribution map for all the particles was generated with a scaling factor of 10 in SPIDER.
Cryo-EM analysis
3.5 μl of sample with a particle concentration of ∼0.5 μM was applied to a glow-discharged holey carbon grid (Quantifoil Cu R1.2/1.3). The grid was then blotted for 1.5 s with Whatman 55-mm filter paper and flash-frozen in liquid ethane slush cooled at liquid nitrogen temperature in a FEI Mark IV Vitrobot. The prepared grids were transferred to an FEI Titan Krios electron microscope operated at 300-kV acceleration voltage and equipped with a Gatan K2 Summit direct electron counting camera. Micrographs were recorded in super-resolution mode using the semi-automated low-dose acquisition program UCSF-Image4 at a nominal magnification of 22 500×, corresponding to a pixel size of 1.3 Å at the specimen level. The total exposure time of each image was 8 s which was fractionated into 32 sub-frames. The total accumulated dose on the specimen was about 50 electrons per Å2. The defocus values used for the image recording were about −2.4 to −3.5 μm. The 32 frames of each image stack were aligned, decimated and summed by a frame-based motion correction algorithm. Particle picking and coordinate exporting were performed in EMAN2. CTF values of the micrographs were determined by the CTFFIND3 program25. 2D reference-free classification and 3D classification procedures were performed in RELION 1.426. The auto-refinement, post-processing and auto-b-factor correction processes were all performed in RELION 1.4. The soft masks used in the refinement were generated by a home-made script written by Dr Xueming Li of Tsinghua University.
Model building for human C1
The initial atomic models of human ATG14L, P150, VPS34 and Beclin1 were built by homology modeling based on the yeast VPS38, VPS15, VPS30 and VPS15 structures (chains A, B, C and D in PDB: 5DFZ) via Swiss-model software27, respectively. For these four initial models, only the homologous regions with atomic structural information were modeled and superimposed to generate a starting model of human C1 based on the atomic structure of yeast PI3K complex 2. The starting model was rigid body docked into the EM density of human C2 using UCSF Chimera28, and then refined by MDFF29 which incorporated the EM density gradient as an external potential into MD simulation. All MDFF simulations were performed in NAMD 2.9 using CHARMM 22 all-atom force field with CMAP correction. The simulations were carried out at options of T = 300K and 1 bar, employing the Langevin algorithm30 with an integration time step of 1 fs and a scaling factor ζ = 0.3 kcal/mol for 10 ns.
Liposome preparation
POPC, DOPE and LiPI were purchased from Avanti Polar Lipids. PI3P, PI(4,5)P2 and PI(3,4,5)P3 were purchased from Echelon Biosciences. Lipid mixtures (4 mM) were prepared containing POPC:DOPE (2.6:1) with 2% PI, 6% PI, 10% PI, 15% PI, 6% PI3P, 6% PI(4,5)P2 or 6% PI(3,4,5)P3. The mixtures were dried with a nitrogen stream and further dried overnight under vacuum. The lipid film was then hydrated completely with HKN buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM DTT) and subjected to 10 cycles of freezing in liquid nitrogen and thawing in a 42 °C water bath. Finally, the liposomes were extruded 21 times through a 100 nm pore size polycarbonate film.
ER (microsome) isolation
Microsomes were isolated as described previously20. 50 ml of a suspension of cultured 293F cells were collected and resuspended in 8 ml MTE buffer (270 mM 𝒟-mannitol, 10 mM Tris-HCl, 0.1 mM EDTA and protease inhibitor cocktail). The cell suspension was homogenized by 50 strokes in a Dounce homogenizer and centrifuged at 1 400× g for 10 min (4 °C) to remove nuclear and cell debris. The post-nuclear supernatant (PNC) was centrifuged for 10 min at 15 000× g (4 °C) to remove mitochondria. The post-mitochondrial supernatant (PMF) was then layered onto a sucrose density gradient containing 2.0-, 1.5- and 1.3-M discontinuous sucrose gradient in an ultracentrifuge tube and ultracentrifuged for 70 min at 152 000× g (4 °C). The microsome fraction was collected at the interface of the 1.3-M sucrose gradient layer and centrifuged at 100 000× g for 30 min. Finally, the microsome pellet was resuspended in 400 μl HKN (10 mM HEPES, pH 7.4, 150 mM NaCl) buffer and sonicated for 1 min. An equivalent amount of each fraction was subjected to immunoblotting. The band intensity was quantified from three independent experiments using Image J.
Liposome and microsome flotation assays
The flotation assay was described previously21. Briefly, 0.5 μM proteins and 0.8 mM liposomes (or 60 μl microsomes) were incubated at RT for 1 h in a 100 μl reaction system. The mixture was added to 60% (W/V) sucrose to generate 400 μl protein-liposome solution with a final concentration of 30% sucrose and loaded into a centrifuge tube. Then 400 μl 25% sucrose, 400 μl 20% sucrose and 200 μl HKN buffer were sequentially overlaid onto the protein-liposome/microsome mixture. After centrifugation in a TLS-55 rotor (Beckmen) at 35 000 rpm for 2 h, a total of 14 fractions (100 μl/fraction) were collected from one tube and run on a 10% SDS-PAGE gel followed by western blotting. All the components were detected with the corresponding antibodies.
Lipid kinase assays
Lipid kinase assays were carried out using a K-3000 kit from Echelon according to the manufacturer's instructions. 10 μM proteins and 80 μM PI substrate were incubated for 0.5 h at 37 °C in a 25 μl kinase reaction system containing 50 mM HEPES, pH 7.4, 2 mM DTT, 150 mM NaCl, 5 mM MnCl2, 1 mM CHAPS and 100 μM ATP. Control reactions without ATP were also set up for all proteins. The kinase reactions were quenched by adding 5 μl of 100 mM EDTA and diluted to 200 μl with 90 μl ddH2O and 80 μl of 5× PI(3)P Detection Buffer (K-3004). 50 μl reaction solution was transferred to the Detection plate (K-3001) and incubated with 50 μl PI3P Detector (K-3305) for exactly 1 h at RT in a plate shaker with the speed at 250 rpm. The wells were washed 3 times with 200 μl TBST, then 100 μl of diluted (1:200) secondary detector (K-SEC) in TBST was added and the plates were further incubated at RT for 30 min in the shaker. The wells were washed 3 times with 200 μl TBST and 100 μl TMB solution (K-TMB1) was added, and allowed to develop at RT for 30 min. Finally, 50 μl of 1 N H2SO4 solution was added to the wells to stop the development and the absorbance was read at 450 nm on a plate reader.
Cell culture and transfection
HEK293 cells were cultured in DMEM (Hyclone) medium supplemented with 1% penicillin-streptomycin, 1% sodium pyruvate and 1% non-essential amino acids. For transfection of each well of 6-well plate, 2 μg plasmid expressing GFP-ATG14L, GFP-ATG14L ΔBATs, GFP-UVRAG or GFP-UVRAG-BATs were used with 3 μg PEI.
Semi-intact immunofluorescence staining
After transfection for 24 h, cells were washed with phosphate buffered saline (PBS) and permeabilized by incubation with semi-intact buffer (25 mM HEPES-KOH, pH 7.2, 125 mM KOAc, 5 mM MgAc2, 1 mM DTT, 1 mg/ml 𝒟-glucose and 25 μg/ml digitonin) for exactly 5 min on ice. Cells were then incubated with semi-intact buffer without digitonin for 20 min on ice. Next, the cells were fixed in 4% paraformaldehyde for 20 min at RT. After three washes in PBS for 5 min each, cells were blocked with 10% FBS and 0.1% saponin in PBS for 60 min, then stained with primary antibody in blocking buffer for 1 h and washed with PBS three times. Cells were then stained with fluorescein isothiocyanate-conjugated secondary antibody in PBS for 1 h and washed with PBS three times.
Confocal microscopy and statistical analysis
Confocal images were acquired using Fluoview1000 confocal microscopy (Olympus), fitted with a 60× objective and Fluoview 1000 software. Images were collected at 1 024 × 1 024 pixel resolution. The fluorescent staining was imaged sequentially to eliminate crosstalk between the channels. All experiments were independently repeated several times. IMARIS software was used for the co-localization analyses. In the analysis, confocal images of double-stained sections were first subjected to background correction. Pearson's coefficients were calculated and used to obtain the co-localization values as percentages of GFP-tagged proteins overlapping with Calnexin for 30 cells. The experimenters were blinded to the sample identity during analyses. Values indicate the mean ± SD of three independent experiments.
Accession codes
The 3D reconstruction maps obtained by cryo-EM have been deposited into the Electron Microscopy Data Bank under accession codes: EMD-6785 (hPI3KC3-C1), EMD-6786 (hPI3KC3-C1 rigid part), EMD-6787 (hPI3KC3-C2).
Author contributions
LY and H-WW conceived and supervised the study. MM took charge of protein preparation and all the biochemical analysis and in vivo assay. J-JL was responsible for structure determination by cryo-EM. YL, HF and MY were involved in EM data collection. YH and NT carried out the biochemical analysis. YC performed the in vivo experiment. JW was involved in the structure determination. WH prepared the protein sample and provided good suggestions. YD and M-QD verified the structure quality. MM, J-JL, LY and H-WW wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Acknowledgments
We are grateful to members of the Yu and Wang groups for helpful discussions. We thank NY (Tsinghua University) for her kind discussions and suggestions about the paper. We acknowledge the China National Center for Protein Sciences Beijing and the “Explorer100” cluster system of Tsinghua National Laboratory for Information Science and Technology for providing the facility support. This study was supported by the Ministry of Science and Technology of China (2016YFA0500202 to LY; 2016YFA0501100 to H-WW), the National Natural Science Foundation of China (31430053 and 31621063 to LY), the Natural Science Foundation of China International Cooperation and Exchange Program (31561143002 to LY), the Beijing Municipal Science & Technology Commission grant (Z161100000116034 to H-WW), and Independent Research of Tsinghua University (20161080135 to LY).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Architectural definition of C1 and C2.
Work-flow of 3D analysis of C1 and C2.
Resolution of 3D reconstructions of C1 and C2.
Membrane-binding orientation of C1.
The BATs domain determines the ER-binding capacity of PI3KC3 complexes in vitro.
The membrane binding ability of C2 is much weaker than C1.
References
- Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861–2873. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett 2007; 581:2156–2161. [DOI] [PubMed] [Google Scholar]
- Volinia S, Dhand R, Vanhaesebroeck B, et al. A human phosphatidylinositol 3-kinase complex related to the yeast VPS34p-VPS15p protein sorting system. EMBO J 1995; 14:3339–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L, Smrcka A, Cooke FT, Jackson TR, Sternweis PC, Hawkins PT. A novel phosphoinositide 3 kinase activity in myeloid-derived cells is activated by G protein beta gamma subunits. Cell 1994; 77:83–93. [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct VPS34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 2008; 105:19211–19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, ATG14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11:385–396. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, et al. Beclin1-binding UVRAG targets the class C VPS complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008; 10:776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 2010; 316:3368–3378. [DOI] [PubMed] [Google Scholar]
- He S, Ni D, Ma B, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the Beclin 1 complex. Nat Cell Biol 2013; 15:1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389–393. [DOI] [PubMed] [Google Scholar]
- Van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008: 9:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci USA 2011; 108:7769–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via ATG14L. J Cell Biol 2010; 190:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran S, Carlson LA, Stjepanovic G, et al. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife 2014; 3:e05115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostislavleva K, Soler N, Ohashi Y, et al. Structure and flexibility of the endosomal VPS34 complex reveals the basis of its function on membranes. Science 2015; 350:aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 2000; 19:4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozidis P, Williamson CD, Colberg-Poley AM. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus infected primary fibroblasts. Curr Protoc Cell Biol 2007 Dec; Chapter 3:Unit 3.27. [DOI] [PubMed] [Google Scholar]
- Wragg RT, Snead D, Dong Y, et al. Synaptic vesicles position complexin to block spontaneous fusion. Neuron 2013; 77:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 2007; 157:38–46. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 2008; 3:1941–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol 1996; 116:17–24. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 2003; 142:334–347. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 2003; 31:3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 2008; 16:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 1995; 103:4613–4621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Architectural definition of C1 and C2.
Work-flow of 3D analysis of C1 and C2.
Resolution of 3D reconstructions of C1 and C2.
Membrane-binding orientation of C1.
The BATs domain determines the ER-binding capacity of PI3KC3 complexes in vitro.
The membrane binding ability of C2 is much weaker than C1.