ABSTRACT
The general secretory pathway (Sec) and twin-arginine translocase (Tat) operate in parallel to export proteins across the cytoplasmic membrane of prokaryotes and the thylakoid membrane of plant chloroplasts. Substrates are targeted to their respective machineries by N-terminal signal peptides that share a tripartite organization; however, Tat signal peptides harbor a conserved and almost invariant arginine pair that is critical for efficient targeting to the Tat machinery. Tat signal peptides interact with a membrane-bound receptor complex comprised of TatB and TatC components, with TatC containing the twin-arginine recognition site. Here, we isolated suppressors in the signal peptide of the Tat substrate, SufI, that restored Tat transport in the presence of inactivating substitutions in the TatC twin-arginine binding site. These suppressors increased signal peptide hydrophobicity, and copurification experiments indicated that they restored binding to the variant TatBC complex. The hydrophobic suppressors could also act in cis to suppress substitutions at the signal peptide twin-arginine motif that normally prevent targeting to the Tat pathway. Highly hydrophobic variants of the SufI signal peptide containing four leucine substitutions retained the ability to interact with the Tat system. The hydrophobic signal peptides of two Sec substrates, DsbA and OmpA, containing twin lysine residues, were shown to mediate export by the Tat pathway and to copurify with TatBC. These findings indicate that there is unprecedented overlap between Sec and Tat signal peptides and that neither the signal peptide twin-arginine motif nor the TatC twin-arginine recognition site is an essential mechanistic feature for operation of the Tat pathway.
KEYWORDS: Sec pathway, Tat pathway, protein secretion, signal peptide, suppressor genetics
IMPORTANCE
Protein export is an essential process in all prokaryotes. The Sec and Tat export pathways operate in parallel, with the Sec machinery transporting unstructured precursors and the Tat pathway transporting folded proteins. Proteins are targeted to the Tat pathway by N-terminal signal peptides that contain an almost invariant twin-arginine motif. Here, we make the surprising discovery that the twin arginines are not essential for recognition of substrates by the Tat machinery and that this requirement can be bypassed by increasing the signal peptide hydrophobicity. We further show that signal peptides of bona fide Sec substrates can also mediate transport by the Tat pathway. Our findings suggest that key features of the Tat targeting mechanism have evolved to prevent mistargeting of substrates to the Sec pathway rather than being a critical requirement for function of the Tat pathway.
INTRODUCTION
The general secretory (Sec) and twin-arginine translocation (Tat) pathways operate in parallel to transport proteins across the cytoplasmic membranes of prokaryotes and the thylakoid membranes of plant chloroplasts. The Sec pathway translocates substrates in an unfolded conformation, whereas the Tat system transports folded proteins. Many Tat substrates contain redox cofactors that are noncovalently associated, and the Tat system is essential for photosynthesis and some modes of respiratory growth (reviewed in reference 1).
Targeting of substrates to the Sec and Tat pathways is mediated by the presence of N-terminal signal peptides. Sec and Tat targeting sequences each have a recognizable tripartite structure with a positively charged n-region, a hydrophobic h-region, and a polar c-region that usually contains a cleavage site for leader peptidase (2, 3) (Fig. 1A). One of the primary differences between them is the presence of an almost invariant arginine pair in the n-region of Tat signal peptides. These consecutive arginines are reported to be mechanistically essential for substrate translocation by the Tat pathway, and even conservative alterations to lysine are poorly tolerated (e.g., references 4 and 5). Twin arginines, however, are also compatible with the Sec pathway, and some Sec signal peptides have paired arginines in their n-regions. A second key difference is the relative hydrophobicity of the two types of signal peptide. Tat targeting sequences are notably less hydrophobic than Sec signal peptides, and increasing the hydrophobicity of the TorA signal peptide reroutes a passenger protein from Tat to Sec (6, 7). Finally, one or more positive charges are frequently found in the c-region of Tat signals that are not mechanistically required for Tat transport but serve to block interaction of the signal peptide with the Sec pathway (6–9). Nonetheless, despite these differences, over half of the Escherichia coli Tat signal peptides that were tested showed some level of engagement with the Sec pathway when fused to a Sec-compatible reporter protein (10).
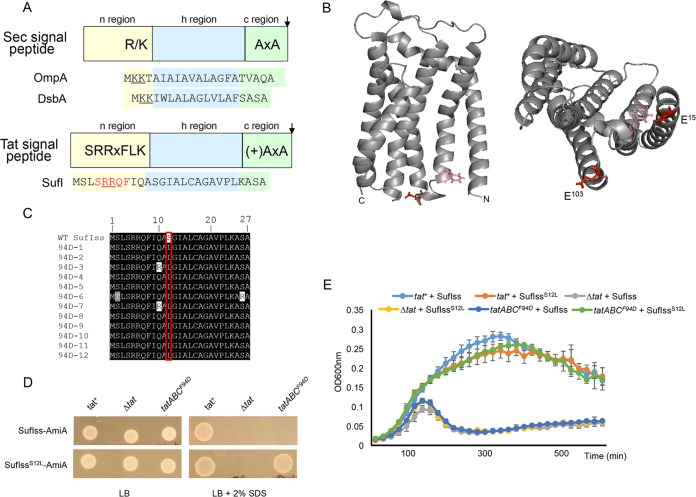
FIG 1 .
(A) Schematic representation of Sec and Tat signal peptides. The sequences of the OmpA and DsbA Sec-targeting signals and the SufI Tat-targeting signal peptide are shown. Positive charges in the signal peptide n-regions are shown underlined, and the amino acids of the SufI Tat consensus motif are shown in red. (B) Models of E. coli TatC. (Left) Side view, with F94 and E103 residues that are located in the signal peptide binding site given in pink and red, respectively. (Right) View of the cytoplasmic face, with E15 additionally shown. (C) Alignment of the amino acid sequence of 12 suppressors in the SufI signal peptide that compensate for the TatC F94D substitution. WT, wild type. (D and E) Cells of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring pTH19kr (empty vector; annotated Δtat) or pTAT101 producing wild-type TatAB along with either wild-type TatC (tat+) or TatCF94D (tatABCF94D) and a compatible plasmid (either pSUSufIss-mAmiA or pSUSufIS12Lss-mAmiA, as indicated) were subcultured at 1:100 into fresh LB medium following overnight growth. (D) Cells were incubated for 3 h at 37°C with shaking. Cells were pelleted and resuspended in sterile phosphate-buffered saline (PBS) supplemented with appropriate antibiotics to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. (E) Alternatively, cultures were supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +standard deviations (n = 3 biological replicates).
Tat signal peptides interact with the membrane-bound Tat receptor complex. In E. coli, the receptor contains TatA, TatB, and TatC proteins, most likely in a 1:1:1 stoichiometry (11–13). The receptor is multivalent (14–16) and contains multiple copies of the TatABC heterotrimer (e.g., references 17 and 18). The primary recognition site for the Tat signal peptide is TatC (e.g., references 19 to 23), with two conserved glutamates on the cytoplasmic face of TatC forming a patch that interacts with the signal peptide twin arginines (24) (Fig. 1B). The signal peptide can also transition to a deep binding mode where it is inserted into the receptor complex, forming cross-links to the transmembrane helix (TM) of TatB and TM5 of TatC (17, 19, 25, 26). Signal peptide insertion into the receptor drives structural reorganization of the complex (13, 18, 26, 27) and the recruitment of further TatA molecules (19, 28–30). The assembled TatA oligomer mediates the transport of folded substrates across the membrane in an unknown manner, potentially by forming a translocation channel or by facilitating a localized weakening and transient disruption of the bilayer (31–33).
A recent study isolated genetic suppressors that restored transport activity to a Tat system that harbored an inactivating substitution in the TatC signal peptide binding site (27). These suppressing substitutions, located primarily in the TM of TatB, could also separately restore Tat transport to a substrate with a defective Tat signal peptide. Biochemical analysis revealed that these substitutions did not act to restore detectable signal peptide binding to the receptor complex, but instead, at least some of them induced conformational changes that apparently mimicked the substrate-activated state (27). In this work, we have taken a complementary approach by searching for signal peptide suppressors able to restore Tat transport when the TatC signal peptide binding site was inactivated. We show that two separate inactive TatC variants, F94D and E103K, can be suppressed by single substitutions that increase the hydrophobicity of a Tat signal peptide. Remarkably, the same hydrophobic substitutions can suppress in cis by restoring Tat transport to a twin-arginine-mutated signal peptide. Our results show that neither the twin-arginine motif nor its cognate recognition site on TatC is required for Tat transport activity. We further show that hydrophobic Sec signal peptides containing paired lysines can also mediate export by the Tat pathway, pointing to an unexpected degree of overlap between Sec and Tat targeting requirements.
RESULTS
Isolation of suppressors of the inactivating TatC F94D substitution.
A series of cross-linking studies, along with direct binding assays using purified TatC variants, have identified that the cytoplasmic N-terminal region and the cytoplasmic loop between TM2 and TM3 form a binding site for the twin-arginine motif of Tat signal peptides (23, 24, 34, 35). Amino acid substitutions in the TM2-TM3 loop in particular are associated with loss of Tat activity, and residues F94 and E103 are almost completely invariant among TatC sequences from all three domains of life (34, 36, 37). Along with E15, E103 has been implicated in coordinating the positively charged twin arginines of the signal peptide (24, 38) (Fig. 1B).
The twin arginines are part of a larger consensus motif, S-R-R-x-F-L-K (Fig. 1A), where the other amino acids are semiconserved (2). The consensus phenylalanine is frequently present, particularly in bacterial Tat signal peptides and, for example, is found in approximately two-thirds of E. coli Tat targeting sequences (39). It has been proposed through modeling studies that if the signal peptide n-region is in an extended conformation, TatCF94 may stack against this consensus F residue (40). We initially sought to test this hypothesis genetically.
It has been shown previously that a TatCF94D substitution inactivates Tat transport and that strains harboring this substitution are unable to grow on media containing the detergent SDS (27) (Fig. 1D). This phenotype arises due to an inability to export two Tat substrates, AmiA and AmiC, which remodel the cell wall during growth (41, 42). We used a fusion protein whereby the signal peptide of SufI, which has the consensus F residue (Fig. 1A), was fused to the mature region of AmiA (27) and constructed a random library of codon substitutions at F8. We then screened this library against a strain lacking native amiA/amiC and harboring TatCF94D, plating onto LB medium containing 2% SDS to select for suppressors of this inactivating substitution. However, after screening more than 1,000 clones we failed to isolate any suppressors of TatCF94D from this library.
We therefore addressed whether it was possible to isolate substitutions elsewhere in the SufI signal peptide that would suppress TatCF94D. To this end, we constructed a random library of mutations throughout the SufI signal peptide coding region of the SufIss-AmiA fusion that had some 13,000 members and an error rate of approximately 2%. After screening more than 20,000 individual transformants for the ability to grow in the presence of 2% SDS, we isolated 12 suppressors that supported growth on the detergent. Sequence analysis indicated that each of the suppressors shared a common alteration of serine at position 12 of the signal peptide to leucine (Fig. 1C), and indeed, this single S12L substitution was sufficient to support growth of a strain producing TatCF94D on LB agar containing SDS (Fig. 1D). Since the phenotypic growth test is largely qualitative, we also undertook a more quantitative assessment of growth of the strain coproducing TatCF94D and SufIS12Lss-AmiA by measuring growth curves in the presence of SDS. Figure 1E shows that the strain producing TatCF94D and SufIS12Lss-AmiA grew identically to the same strain producing wild-type TatC and SufIS12Lss-AmiA.
The SufI S12L substitution restores transport activity to a different substitution in the TatC signal peptide binding site.
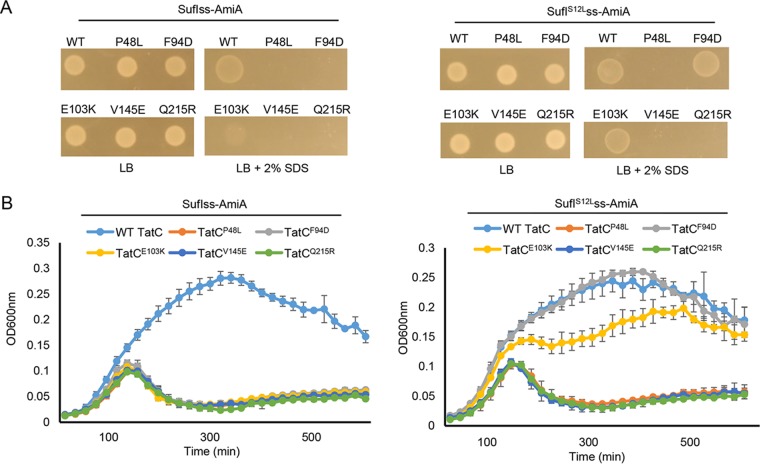
To determine whether the suppressor activity of the signal peptide S12L substitution was specific for TatCF94D, we tested whether this substitution could restore Tat transport to other TatC-inactivating substitutions, including P48L, V145E, and Q215R, located in consecutive periplasmic loops, or E103K, located in the signal peptide binding site (37). Figure 2 shows that the inactivating TatCE103K substitution could also be suppressed by the SufIS12L variant, but transport activity was not restored to any of the substitutions in the periplasmic loops. We conclude that the S12L substitution specifically restores Tat transport to substitutions in the TatC signal peptide binding site.
FIG 2 .
The SufIS12L substitution can restore Tat transport to TatCE103K. Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring either pSUSufIss-mAmiA or pSUSufIS12Lss-mAmiA alongside plasmid pTAT101 producing wild-type (WT) TatAB along with the indicated substitution of TatC, as indicated, were subcultured at a 1:100 dilution. (A) Cultures were grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. (B) Alternatively, cultures were supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +standard deviations (n = 3 biological replicates).
The S12L substitution can restore transport activity to signal peptides that contain inactivating twin-arginine substitutions.
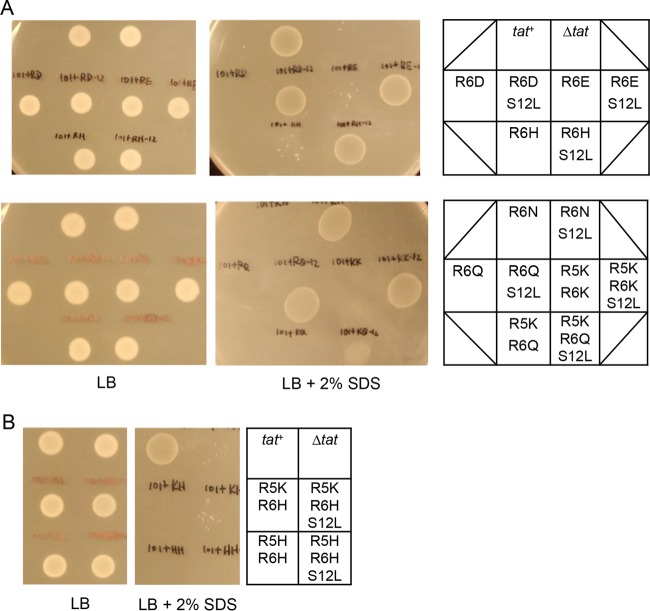
Since the signal peptide S12L substitution can act in trans to suppress inactivating substitutions in the TatC signal peptide binding site, we next asked whether it could act in cis to rescue inactivating substitutions at the twin-arginine motif. Previously, it has been shown that substitutions for one or both consensus arginines of the SufI signal peptide are poorly tolerated (4), and indeed, single mutations of R6 to D, E, H, N, or Q or of R5R6 to KK, KH, KQ, or HH in the SufIss-AmiA fusion are sufficient to prevent growth of cells in the presence of SDS (27) (Fig. 3; also see Fig. S1 in the supplemental material). Interestingly, however, introduction of the S12L substitution alongside R6D, R6E, R6H, R6N, R6Q, or R5K/R6K restored strong growth of cells producing these fusion proteins in the presence of SDS (Fig. 3A and S1B to G). The S12L substitution could also partially compensate for the R5K,R6Q substitution (Fig. 3A and S1H) but could not rescue transport activity of the R5K,R6H or R5H,R6H variants (Fig. 3B and S1I and J). For each of these variant signal peptides, we confirmed that the transport of the AmiA substrate that we observed was dependent on the Tat pathway, since growth on SDS was not observed when the Tat system was absent (Fig. S2). We conclude that the SufIS12L signal peptide substitution can at least partially compensate for substitutions at the twin-arginine motif.
FIG 3 .
The SufIS12L substitution can act in cis to suppress inactivating substitutions in the SufI signal peptide twin-arginine motif. Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring either pTH19kr alongside pSUSufIss-mAmiA (Δtat) or pTAT101 (producing wild-type TatABC) alongside either unsubstituted pSUSufIss-mAmiA (tat+) or pSUSufIss-mAmiA encoding the (A) R6D, R6D-S12L, R6E, R6E-S12L, R6H, R6H-S12L, R6N, R6N-S12L, R6Q, R6Q-S12L, R5KR6K, R5KR6K-S12L, R5KR6Q, and R5KR6Q-S12L, or the (B) R5KR6H, R5KR6H-S12L, R5HR6H, and R5HR6H-S12L substitutions in the SufI signal peptide were subcultured at a 1:100 dilution, grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h.
The SufIS12L substitution can act in cis to suppress inactivating substitutions in the SufI signal peptide twin-arginine motif. Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring pTH19kr alongside pSUSufIss-mAmiA (Δtat) or pTAT101 (producing wild-type TatABC) alongside unsubstituted pSUSufIss-mAmiA (tat+) (A) or pTAT101 (producing wild-type TatABC) alongside pSUSufIss-mAmiA encoding the indicated substitutions in the SufI signal peptide (B to J) were subcultured at a 1:100 dilution into LB supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +SD (n = 3 biological replicates). Download FIG S1, PDF file, 0.1 MB (137KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The twin-arginine substituted SufIS12L signal peptide variants mediate strict Tat-dependent transport of AmiA. (A and B) Overnight cultures of the tat− strain MC4100 ΔamiA ΔamiC ΔtatABC harboring either pTAT101 (producing wild-type TatABC) alongside unsubstituted pSUSufIss-mAmiA (tat+) or pTH19kr alongside either unsubstituted pSUSufIss-mAmiA (Δtat) or pSUSufIss-mAmiA encoding the indicated substitutions in the SufI signal peptide as indicated were subcultured at 1:100 dilution. (A) Cultures were grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. (B) Alternatively, cultures were supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +SD (n = 3 biological replicates). Download FIG S2, PDF file, 0.1 MB (114.5KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Single hydrophobic substitutions along the length of the SufI signal peptide h-region can also suppress inactivating TatC substitutions in the signal peptide binding site.
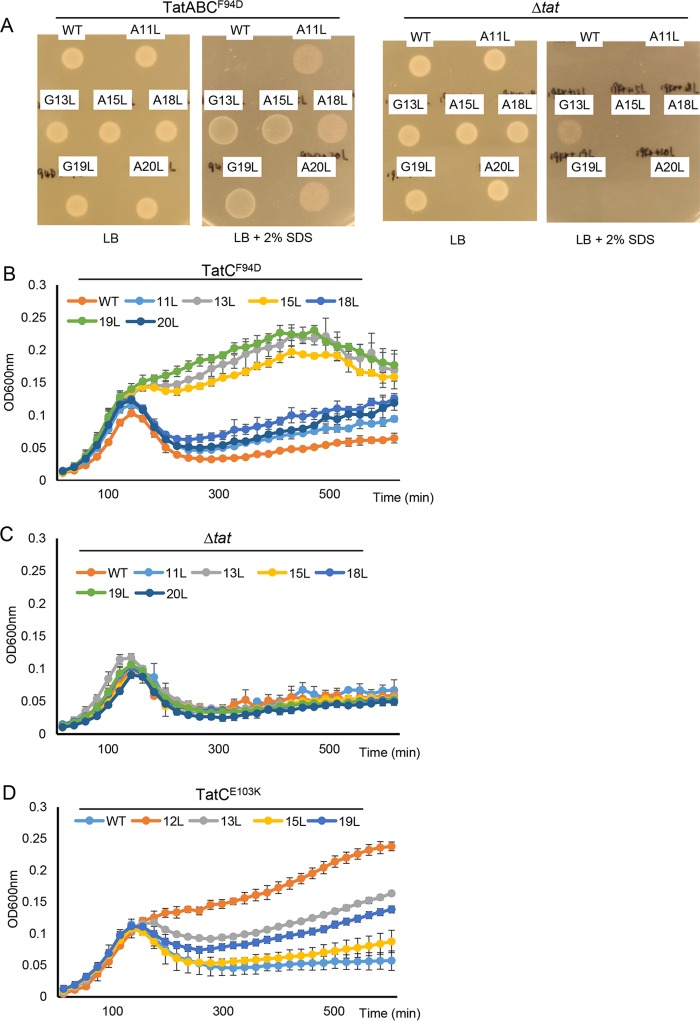
The h-regions of Tat signal peptides are less hydrophobic than Sec signal sequences, containing significantly more glycine and fewer leucine residues (6). The S12L substitution replaces a polar residue near the start of the SufI signal peptide h-region with a highly hydrophobic amino acid, markedly increasing its hydrophobicity score (Table 1). To test whether single hydrophobic substitutions elsewhere in the SufI signal peptide h-region could also suppress Tat transport defects, we increased hydrophobicity of the h-region by constructing individual A11L, G13L, A15L, A18L, G19L, and A20L variants. Figure 4A shows that when each of these individual substitutions was introduced into the SufIss-AmiA construct and produced in a strain harboring tatCF94D, growth on SDS was restored. We assume that we did not obtain any of these substitutions in our original screen because, unlike the S12L substitution, which needed only one nucleotide change, these other substitutions require at least two nucleotide changes.
TABLE 1 .
Relative hydrophobicities of signal peptide variants used in this worka
| Signal peptide | Sequence | Hydrophobicity |
|---|---|---|
| WT SufI | MSLSRRQFIQASGIALCAGAVPLKASA | 1.36 |
| SufI 11L | MSLSRRQFIQLSGIALCAGAVPLKASA | 1.48 |
| SufI 12L | MSLSRRQFIQALGIALCAGAVPLKASA | 1.63 |
| SufI 13L | MSLSRRQFIQASLIALCAGAVPLKASA | 1.61 |
| SufI 15L | MSLSRRQFIQASGILLCAGAVPLKASA | 1.48 |
| SufI 18L | MSLSRRQFIQASGIALCLGAVPLKASA | 1.48 |
| SufI 19L | MSLSRRQFIQASGIALCALAVPLKASA | 1.61 |
| SufI 20L | MSLSRRQFIQASGIALCAGLVPLKASA | 1.48 |
| SufI 12L,13L | MSLSRRQFIQALLIALCAGAVPLKASA | 1.88 |
| SufI 12L,13L,14L,15L | MSLSRRQFIQALLLLLCAGAVPLKASA | 1.96 |
| SufI 17L,18L,19L,20L | MSLSRRQFIQASGIALLLLLVPLKASA | 1.92 |
| WT OmpA | MKKTAIAIAVALAGFATVAQA | 2.31 |
| OmpA i18K | MKKTAIAIAVALAGFATKVAQA | 2.31 |
| WT DsbA | MKKIWLALAGLVLAFSASA | 2.49 |
| DsbA i16K | MKKIWLALAGLVLAFKSASA | 2.55 |
In each case, the h-region sequence used to calculate the score is shown underlined. For SufI, the signal peptide h-region for each variant was defined using TATFIND 1.4 (62, 63) (http://signalfind.org/tatfind.html). For OmpA and DsbA, the h-region for each variant was defined using Phobius (64) (http://phobius.sbc.su.se/). Hydrophobicity was scored using the grand average of hydropathy (GRAVY) value at http://www.gravy-calculator.de/. WT, wild type. Bold letter K in sequences indicates inserted lysine residue.
FIG 4 .
Single leucine substitutions throughout the SufI signal peptide h-region can suppress the TatC F94D and E103K substitutions. (A to C) Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring either pTH19kr (Δtat) or pTAT101 producing TatABCF94D along with pSUSufIss-mAmiA producing the indicated substitution in the SufI signal peptide were subcultured at a 1:100 dilution. (A) Cultures were grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. (B and C) Alternatively, cultures were supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. (D) MC4100 ΔamiA ΔamiC ΔtatABC harboring pTAT101 producing TatABCE103K along with pSUSufIss-mAmiA producing the indicated substitution in the SufI signal peptide was subcultured into LB containing 0.5% SDS and grown at 37°C without shaking. For all growth curves, the optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +standard deviations (n = 3 biological replicates). WT, wild type.
When we examined growth on SDS semiquantitatively by monitoring growth curves (Fig. 4B), it could be seen that the G13L, A15L, and G19L substitutions suppressed the tatCF94D allele more strongly than A11L, A18L, or A20L. Table 1 shows the hydrophobicity scores for the h-regions of these SufI signal peptide variants. It can be seen that the substitutions that give the biggest increase in hydrophobicity result in the strongest level of suppression. It should be noted that the SufIssG13L substitution appeared to result in a very low level of the fusion protein being routed to the Sec pathway, as weak growth could be detected in a strain lacking the Tat pathway (Fig. 4A and C). None of the other substitutions, however, led to any detectable transport by Sec.
We next tested whether these further hydrophobic substitutions could also suppress a second signal peptide binding site substitution, TatCE103K. It can be seen (Fig. 4D and S3) that these variants could also compensate for loss of Tat activity resulting from this substitution. They could not, however, compensate for any of the TatC P48L, V145E, and Q215R mutations (not shown). As seen for the suppression of TatCF94D, the substitutions giving the biggest increase in hydrophobicity (S12L, G13L, and G19L) restored the highest level of Tat activity in the presence of TatCE103K (Fig. 4D). Finally, we also tested whether the strongest suppressors of TatCE103K could also rescue transport in cis when introduced into the twin-lysine variant of the SufI signal peptide. Figure S4 indicates that, similarly to the S12L substitution, introduction of any of the G13L, A15L, and G19L substitutions into KK-SufIss-AmiA restored strong growth of cells producing these fusion proteins in the presence of SDS. We conclude that increasing h-region hydrophobicity can suppress transport defects associated with either the signal peptide twin-arginine motif or the signal peptide binding site.
Hydrophobic substitutions in the SufI signal peptide can rescue the transport defect of TatCE103K. Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring pTAT101 producing TatABCE103K along with pSUSufIss-mAmiA producing the indicated substitution in the SufI signal peptide were subcultured at a 1:100 dilution, grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. Download FIG S3, PDF file, 0.04 MB (37.5KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hydrophobic substitutions in the SufI signal peptide can act in cis to suppress the signal peptide R5K,R6K substitution. Overnight cultures of MC4100 ΔamiA ΔamiC ΔtatABC harboring pTAT101 (producing wild-type TatABC) alongside either unsubstituted pSUSufIss-mAmiA (WT) or pSUSufIss-mAmiA encoding the indicated substitutions in the SufI signal peptide were subcultured at a 1:100 dilution in the presence of 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Download FIG S4, PDF file, 0.1 MB (84.3KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The h-region suppressors support transport of full-length SufI.
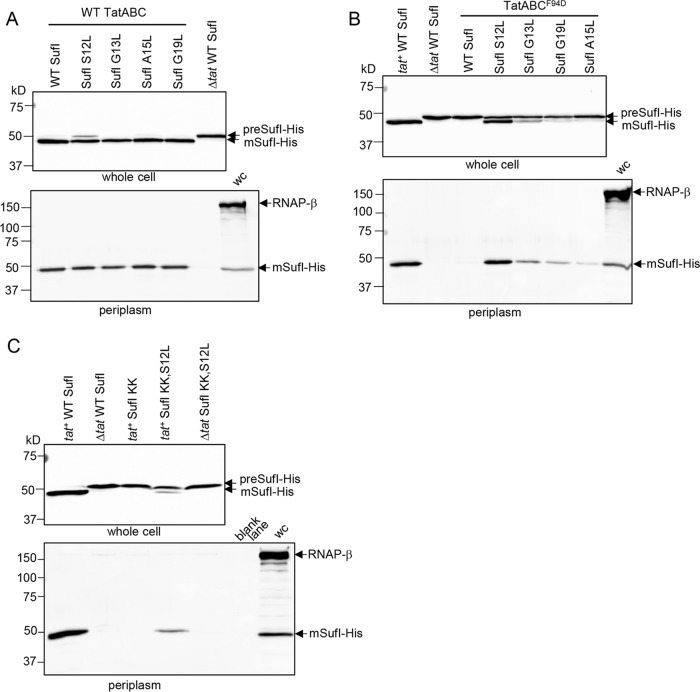
To assess the level of Tat transport mediated by these hydrophobic variants of the SufI signal sequence, we introduced the S12L, G13L, A15L, and G19L substitutions individually into a construct encoding C-terminally His-tagged but otherwise wild-type SufI. We initially expressed these in a strain producing native TatABC and fractionated cells to obtain the periplasm. Figure 5A shows that each of these single hydrophobic variants of the SufI signal peptide supported strong export of SufI-His. Transport of these SufI-His variants was dependent on the Tat pathway since, with the exception of SufI G13L, where a very faint trace of SufI was seen in the periplasm, no periplasmic SufI-His could be detected when the Tat pathway was absent (Fig. S5A).
FIG 5 .
Analysis of SufI export mediated by signal peptide leucine substitutions in the TatCF94D background or when combined with a signal peptide twin-lysine substitution. (A and B) E. coli strain DADE coproducing His-tagged but otherwise native SufI or SufI with the indicated single leucine substitutions in the signal peptide (from a pQE80 plasmid) alongside either wild-type (WT) TatABC (A) or wild-type TatAB and TatCF94D (from pTAT101) (B). Strain DADE coproducing His-tagged but otherwise native SufI alongside an empty vector was used as a negative control (lanes annotated “Δtat WT SufI”). (C) Strain DADE producing His-tagged SufI harboring an R5K,R6K double substitution (SufI KK) and with an additional S12L substitution where indicated, alongside either empty vector pTH19kr (Δtat) or pTAT101 encoding wild-type TatABC (tat+). In each case, strains were grown to mid-log phase and fractionated into whole cell (upper panels) and periplasm (lower panels) and then analyzed by Western blotting with anti-6×His tag or anti-RNA polymerase β-subunit antibodies (cytoplasmic control protein). wc, whole cell. Equivalent volumes of sample were loaded for each of the whole-cell samples and for each of the periplasmic samples.
Analysis of SufI export mediated by signal peptide leucine substitutions in the TatCE103K background and in a tat deletion strain. E. coli strain DADE coproducing His-tagged but otherwise native SufI or SufI with the indicated single leucine substitutions in the signal peptide (from a pQE80 plasmid) alongside either an empty plasmid vector (Δtat) (A) or wild-type TatAB and TatCE103K (from pTAT101) (B) was grown to mid-log phase, fractionated into whole-cell (upper panels) and periplasm (lower panels) samples, and then analyzed by Western blotting with anti-6×His tag or anti-RNA polymerase β-subunit antibodies (cytoplasmic control protein). Strain DADE coproducing His-tagged but otherwise native SufI alongside either wild-type TatABC (tat+ WT SufI) or an empty vector (Δtat WT SufI) was used as a positive or negative control, respectively. wc, whole cell. Equivalent volumes of sample were loaded for each of the whole-cell samples and for each of the periplasmic samples. Download FIG S5, PDF file, 0.1 MB (136.2KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we assessed the degree of transport mediated by these variant signal peptides in cells producing TatABCF94D. It can be seen (Fig. 5B) that although wild-type SufI-His was not exported in the presence of TatCF94D, transport was detected when any of the single hydrophobic substitutions was present in the signal peptide. The S12L substitution in particular could strongly suppress TatCF94D, with high levels of SufI-His detected in the periplasm when the signal peptide harbored this mutation. These same signal peptide substitutions could also restore good transport of SufI-His in the presence of the inactivating TatCE103K substitution (Fig. S5B).
Since the hydrophobic substitutions can act in cis to restore transport activity to a twin-lysine variant of the SufI signal peptide twin-arginine motif, we assessed the export of the KK variant of SufI-His harboring the S12L substitution. Figure 5C shows that there was clear Tat transport activity conferred on the twin-lysine signal peptide variant by the presence of the S12L suppressor. Taken together, the results presented so far indicate that the signal peptide consecutive arginines and the TatC twin-arginine recognition site are not essential mechanistic features for operation of the Tat pathway, and substitutions in either of these can be at least partially compensated for by an increase in signal peptide hydrophobicity.
The h-region suppressors restore signal peptide binding to TatBC.
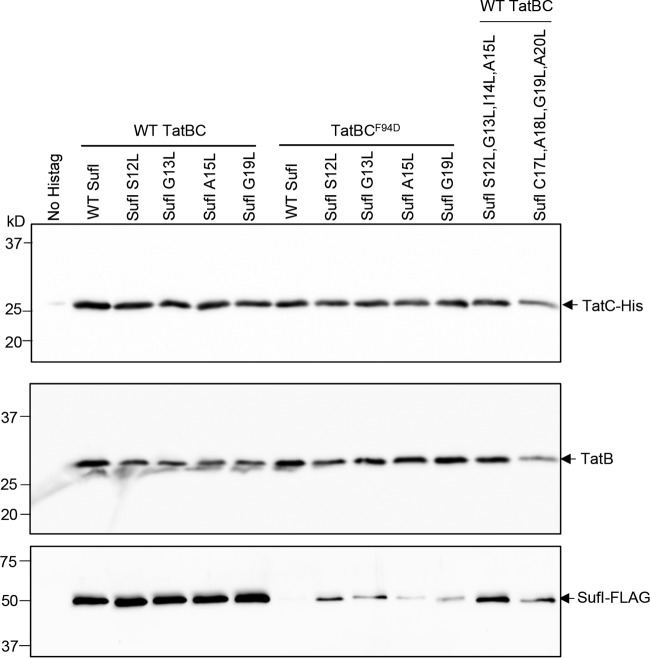
A previous study identified suppressors in the TatB component that could also restore transport activity to substitutions in the TatC twin-arginine binding site. It was shown that substrate precursors could be copurified with wild-type TatBC complexes but did not copurify when the signal peptide binding site was mutated, even in the presence of the TatB suppressors. Thus, it was concluded that the TatB suppressors did not detectably restore binding of signal peptides to the TatBC complex (27). To determine whether the suppressors that we identified here that increase signal peptide hydrophobicity could restore binding to TatBC complexes harboring the TatCF94D substitution, we coproduced FLAG-tagged variants of SufI with these suppressors alongside TatB and His-tagged TatC. After purification of TatBC complexes from digitonin-treated cell lysates, we assessed the level of copurifying SufI by immunoblotting. As shown in Fig. 6, the single substitutions S12L, G13L, A15L, or G19L in the SufI signal peptide did not detectably affect interaction of SufI with wild-type TatBC, since qualitatively similar levels of FLAG-tagged SufI were seen to copurify with TatBC-His. When the F94D substitution was present in His-tagged TatC, no SufI-FLAG was copurified with the variant TatBC-His complex (Fig. 6), even though SufI was clearly detected in the cell lysate (Fig. S6). However, when the S12L, G13L, A15L, or G19L substitution was introduced into SufI, it could now be detected in the fractions containing purified TatBCF94D-His. Qualitatively, it appeared that more SufI-FLAG copurified with TatBCF94D-His when a stronger suppressing substitution (S12L or G13L) was present than when a weaker suppressing substitution (A15 or G19) was present, suggesting that an increased degree of binding was associated with better suppression. Taken together, these observations indicate that the SufI h-region suppressors restore some degree of signal peptide binding to the TatBCF94D complex.
FIG 6 .
Hydrophobic variants of the SufI signal peptide mediate binding to TatBC and TatBCF94D. Cells of strain DADE-P coproducing C-terminally FLAG-tagged SufI with its native signal peptide (WT SufI) or harboring the indicated leucine substitutions, alongside TatB and C-terminally His-tagged TatC or TatCF94D, were lysed and incubated with digitonin, and His-tagged TatC was isolated with Ni-charged beads. Following elution of bound TatC-His, equivalent volumes of the eluate from each sample were analyzed by Western blotting with anti-His, anti-TatB, or anti-FLAG antibodies as indicated. DADE-P coproducing C-terminally FLAG-tagged SufI with its native signal peptide alongside TatB and nontagged TatC (lane annotated “No Histag”) was used as a negative control. Equivalent volumes of sample were loaded in each lane. WT, wild type.
Detection of SufI-FLAG variants in input samples prior to Ni affinity purification. Cells of strain DADE-P coproducing C-terminally FLAG-tagged SufI with its native signal peptide (WT SufI) or harboring the indicated leucine substitutions, alongside TatB and C-terminally His-tagged TatC or TatCF94D, were lysed and incubated with digitonin. Thirty microliters of the digitonin-solubilized fraction was mixed with an equal volume of 2× Laemmli buffer, and 10 μl of each sample was subjected to SDS-PAGE (12% acrylamide) followed by Western blotting with anti-FLAG antibody. Download FIG S6, PDF file, 0.1 MB (78KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Highly hydrophobic signal peptides are compatible with the Tat pathway.
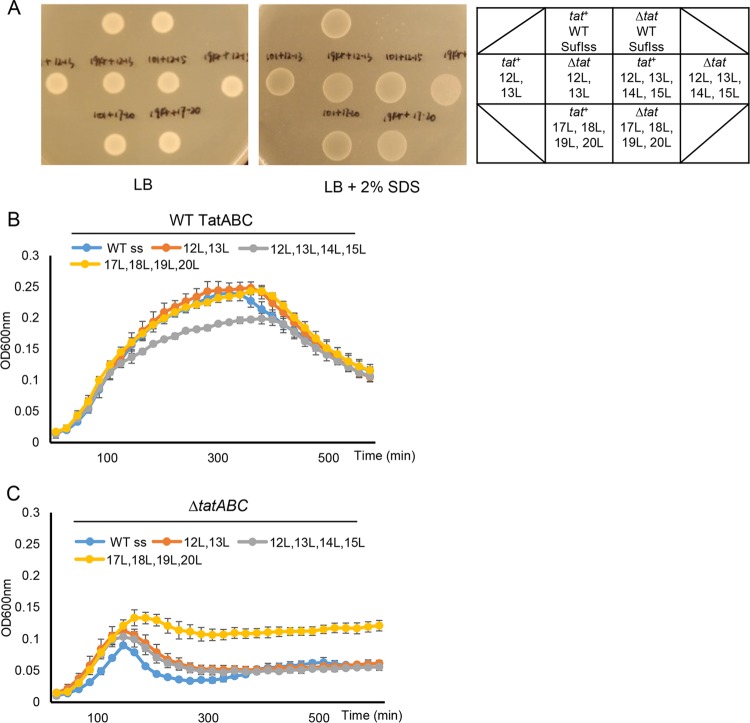
It has previously been reported that the relatively low hydrophobicity of Tat signal peptides partially prevents their routing to the Sec pathway (6). It is not clear, however, whether low h-region hydrophobicity is a mechanistic requirement for engagement with Tat. To explore this in more detail, we investigated the effect of further increasing the hydrophobicity of the SufI signal peptide on transport of the SufIss-AmiA fusion. To this end, we introduced an S12L/G13L double substitution and two quadruple substitutions, S12L/G13L/I14L/A15L and C17L/A18LG19L/A20L, into the SufI signal sequence. These substitutions markedly increase the signal sequence hydrophobicity score, bringing it into the range of the Sec signal sequences of OmpA and DsbA (Table 1).
Figure 7A shows that each of these SufIss-AmiA fusion proteins was able to support growth on solid medium in the presence of SDS, although growth was also seen in a strain lacking the Tat pathway, indicating that there is some export of these more hydrophobic SufIss-AmiA fusion proteins by Sec. These findings were confirmed by monitoring growth of these strains in liquid culture (Fig. 7B and C). However, it is clear that in the absence of a functional Tat system, growth in the presence of SDS was much poorer than when the Tat system was present. This observation suggests that there must be some recognition of these hydrophobic signal peptide variants by the Tat pathway. To confirm this, we coproduced FLAG-tagged S12L/G13L/I14L/A15L and C17L/A18LG19L/A20L SufI variants alongside TatBC-His. When His-tagged TatC was purified from digitonin-solubilized cell lysates, each of these hydrophobic SufI-FLAG variants was copurified (Fig. 6), indicating that they retained the ability to interact with the TatBC complex. Taken together, these results show that highly hydrophobic signal peptides are mechanistically compatible with the Tat pathway.
FIG 7 .
Multiple leucine substitutions in SufI signal peptide h-region partially reroute AmiA to the Sec pathway. Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring either pTH19kr (Δtat) or pTAT101 producing wild-type (WT) TatABC (tat+) along with pSUSufIss-mAmiA producing the indicated substitutions in the SufI signal peptide were subcultured at a 1:100 dilution. (A) Cultures were grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. (B and C) Alternatively, cultures were supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +standard deviations (n = 3 biological replicates).
The OmpA and DsbA signal peptides functionally interact with the Tat pathway.
Our results collectively show that the hallmark twin arginines of Tat signal peptides are not a mechanistic requirement for Tat-dependent transport and that a single arginine or twin lysines in the n-region are compatible with the Tat pathway if compensatory mutations are introduced that increase the hydrophobicity of the signal peptide. Interestingly, many Sec-dependent signal peptides share these parameters (Table S1), raising the possibility that bona fide Sec signal peptides may be able to interact with the Tat pathway. To explore this, we selected two well-studied Sec signal peptides—those of OmpA, which is a posttranslational Sec substrate, and of DsbA, which directs cotranslational translocation (43, 44) (Table 1)—and fused their signal peptides to the mature portion of AmiA. We also made two additional constructs where we introduced a “Sec-avoidance” lysine residue into the signal peptide c-regions, to reduce interaction with the Sec pathway (8).
E. coli proteins with known or predicted signal peptides. Protein sequences encoded by E. coli MG1655 were retrieved from UniProt (http://www.uniprot.org) and analyzed for the presence of a signal peptide using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP). The n-regions were analyzed manually; twin arginines are colored red, twin lysines are colored green, and Lys-Arg or Arg-Lys is shown in blue. Pairings of Arg-Asn, Arg-Asp, Arg-Gln, Arg-Glu, and Arg-His (which, from this study, can be suppressed by increased signal peptide hydrophobicity) are shown in orange. For those signal peptides harboring any of these dipeptides in their n-regions, the approximate location of the h-region is shown underlined. The start of the h-region was defined by Phobius (http://phobius.sbc.su.se/), and the end of the h-region and the cleavage site was defined by SignalP. Pairings within five residues of the start of the h-region were considered likely to interact with the Tat pathway. Those marked as N? have one of the indicated pairings 5 or more residues away from the h-region and are considered unlikely to interact with the Tat pathway. Known or probable Tat substrates are shown in yellow highlight; EfeO is predicted to be a Tat substrate, but this has not been demonstrated experimentally. Download TABLE S1, DOCX file, 0.04 MB (41.9KB, docx) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
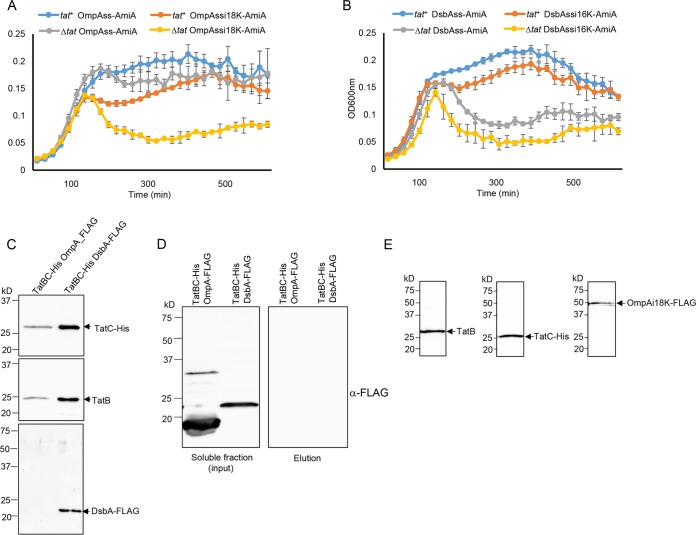
Figure 8A shows that there is Sec-dependent transport of AmiA mediated by the OmpA signal peptide, as there is strong growth of the Δtat strain producing OmpAss-AmiA in the presence of SDS. Introduction of a lysine at position 18 of the OmpA signal peptide clearly reduces interaction with the Sec pathway, as growth of the Δtat strain producing this variant is significantly reduced. However, there is good growth of the tat+ strain producing this variant fusion protein, indicating that some of this fusion must be interacting with the Tat pathway. Similarly, Fig. 8B shows that there is some low-level growth of the Δtat strain producing DsbAss-AmiA in SDS-containing medium, which is reduced by inclusion of a Sec-avoidance lysine in the c-region of the DsbA signal peptide. In contrast, the tat+ strain harboring either of these fusion proteins shows markedly stronger growth in the presence of SDS, indicating that the DsbA signal peptide is productively engaging with the Tat machinery.
FIG 8 .
The OmpA and DsbA signal peptides are able to functionally engage with the Tat machinery. (A and B) Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring either pTH19kr (Δtat) or pTAT101 producing wild-type TatABC (tat+) along with a plasmid encoding the indicated signal peptide fusion to AmiA were subcultured at a 1:100 dilution, supplemented with 0.5% SDS (final concentration), and grown at 37°C without shaking. The optical density at 600 nm (y axis of both panels) was monitored every 20 min using a plate reader. Error bars are +standard deviations (n = 3 biological replicates). (C) Cells of strain DADE coproducing TatB, C-terminally His-tagged TatC, and either C-terminally FLAG-tagged DsbA or OmpA, as indicated, were lysed and incubated with digitonin, and His-tagged TatC was isolated using Ni-charged beads. Following elution of bound TatC-His, equivalent volumes of the eluate from each sample were analyzed by Western blotting with anti-His, anti-TatB, or anti-FLAG antibodies. (D) An aliquot cell lysate prior to digitonin treatment from the experiment shown in panel C was ultracentrifuged to remove the cell membranes. A small amount of the supernatant was retained as the input fraction, and the remainder was incubated with Ni-charged beads. The beads were washed three times with wash buffer, and aliquots of the input and eluate samples were analyzed by Western blotting using an anti-FLAG antibody. (E) Cells of DADE-P coproducing C-terminally FLAG-tagged SufI fused to the OmpA signal peptide harboring a lysine insertion after codon 17, alongside TatB and C-terminally His-tagged TatC, were treated as described in the legend to Fig. 6, and equivalent volumes of the elution fraction were analyzed by Western blotting with anti-His, anti-TatB, or anti-FLAG antibodies as indicated.
To confirm that these signal peptides are able to interact with Tat, we coproduced C-terminally FLAG-tagged variants of full-length OmpA or full-length DsbA alongside TatBC-His. When His-tagged TatC was purified from digitonin-solubilized cell lysates, DsbA-FLAG, which migrated very close to the expected mass of 24.1 kDa, was seen to copurify (Fig. 8C). This copurification was clearly dependent on the presence of the Tat proteins, since when membranes were removed by an ultracentrifugation step, the cytoplasmic form of DsbA-FLAG was no longer isolated by Ni-affinity purification (Fig. 8D). We conclude that FLAG-tagged but otherwise native DsbA can interact with TatBC. In contrast, we were not able to detect copurification of FLAG-tagged OmpA with TatBC under these conditions (Fig. 8C). However, in these experiments we noted that OmpA-FLAG migrated at a lower mass than the predicted size of the tagged protein (38.2 kDa) or of folded OmpA (which migrates at an estimated mass of approximately 30 kDa [45]), raising the possibility that it may have been subjected to proteolysis. We therefore took a second approach to assessing whether the OmpA signal peptide could interact with TatBC by fusing the OmpA signal peptide variant containing the K18 insertion to the N terminus of mature SufI and coproducing it with TatBC-His. Figure 8E indicates that this fusion protein could indeed be copurified alongside TatB and His-tagged TatC, indicating that the OmpA signal peptide is able to interact with the TatBC complex.
DISCUSSION
In this study, we have sought to identify SufI signal sequence variants that restore Tat transport activity in the presence of substitutions that inactivate the twin-arginine recognition site on TatC. Our results have shown that an increase in signal peptide hydrophobicity can overcome two different inactivating substitutions, TatCF94D and TatCE103K, and that these suppressors act to restore detectable binding of the SufI signal sequence to the variant TatBCF94D complex. We further showed that the same hydrophobic substitutions can act in cis to compensate for a range of inactivating substitutions at the SufI signal peptide twin-arginine motif. These results demonstrate that neither the consecutive arginines of the signal peptide nor the conserved recognition site on the cytoplasmic surface of TatC is mechanistically essential for operation of the Tat pathway and that they can be bypassed if the signal peptide hydrophobicity is increased. Taken together, our findings indicate that the signal peptide features that can facilitate interaction with the Tat pathway are remarkably similar to those that facilitate interaction with Sec, namely, the presence of at least one basic charge in the n-region and a relatively hydrophobic h-region. Indeed, we show that even a highly hydrophobic signal peptide that naturally directs its passenger into the cotranslocational Sec pathway can functionally engage with the Tat system.
If the Tat pathway can interact with hydrophobic signal peptides lacking the twin-arginine motif, why then do almost all Tat substrates that have been identified contain paired arginine residues and only moderately hydrophobic h-regions? In prokaryotes and plant chloroplasts, the Tat system always coexists with the Sec pathway. In bacteria, ribosome-associated signal recognition particle (SRP) and cytosolic or ribosome-bound SecA capture Sec substrates at an early stage of biogenesis, at least partially through interaction with their signal sequences (46, 47). Signal sequence hydrophobicity is a key sorting feature for Sec substrates; highly hydrophobic signals generally interact with SRP, whereas those with lower hydrophobicity bind to SecA (46). Photo-cross-linking and/or genetic studies have indicated that Tat signal peptides interact with ribosomally bound trigger factor and with general cytoplasmic chaperones, including DnaK (48–51), but no cross-links to SecA have been reported, and in vitro analysis indicates that Tat signal peptides do not productively engage with SecA to the same extent as a Sec signal peptide (52). It is therefore likely that Tat signal peptides evolved lower hydrophobicity to avoid the targeting pathways that feed into the Sec translocon and that the paired arginines and the twin-arginine binding site are necessary features to strengthen recognition of these weakly hydrophobic signal peptides by the Tat machinery. In this context, it is interesting that although paired arginines in the signal peptide n-region are compatible with the Sec pathway, this pairing is relatively rare in Sec signal peptides, at least in E. coli, being found in only five of the 244 probable Sec signal peptides listed in Table S1 in the supplemental material (compared with 53 that have paired lysines). If lysine and arginine are equivalent in the amino-terminal region of a Sec signal peptide, as implied by kinetic analysis (53), this might suggest that there is selection pressure against the presence of paired arginines in Sec signals.
The presence of one or more positively charged amino acids in the c-region of Tat signal peptides is a further feature that has no mechanistic requirement for Tat translocation but leads to rejection of these signal sequences by the Sec pathway (6, 8, 9). C-terminal positive charges may act at a late stage during Sec translocation when the signal peptide is already engaged with the Sec translocon, and Sec avoidance motifs are particularly abundant in membrane proteins that require the dual action of the Sec and Tat pathways for their assembly (7, 54). Here, the Tat-dependent signal sequence (which is internal to the protein and follows a series of Sec-dependent transmembrane domains) has several c-region positive charges that result in abortive interaction with the Sec pathway, freeing up the sequence to be recognized by Tat (7). Taken together, it is clear that there is strong selective pressure, particularly at the level of Tat signal peptides, to refine features that minimize mistargeting to the Sec pathway.
Our findings show that signal peptides with either twin lysines or an unpaired arginine, coupled with a moderately hydrophobic h-region, can functionally interact with the Tat pathway. Inspection of all of the signal peptides present at the N termini of E. coli MG1655 proteins identified using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP [55]) indicates that some 44% of Sec signal peptides contain either KR, RK, KK, RD, RE, RH, RN, or RQ adjacent to their h-regions (Table S1) and therefore potentially have the capability of engaging with the Tat pathway. Whether any of these would ever target to the Tat pathway in vivo is not clear, since presumably under standard conditions E. coli synthesizes sufficient targeting factors to ensure that Sec substrates are efficiently channeled into the Sec pathway. This is particularly difficult to envisage for substrates that are sequestered by SRP at the ribosome and cotranslationally targeted to the Sec translocon. However, there may be exceptional situations when SRP is out-titrated, resulting in SRP-free ribosomes, and it would presumably be under these circumstances where a substrate might escape targeting to Sec and engage with the Tat pathway. Interestingly, it should be noted that in Bacillus subtilis, hyperproduction of a normally Sec-dependent lipase results in overflow into the Tat pathway (56), raising the possibility that transient rerouting of substrates to the Tat pathway may occur on occasions where cells undergo secretion stress.
MATERIALS AND METHODS
Strain and plasmid construction.
Strains used in this study are MC4100 derivatives (57). Strain MC4100 ΔamiA ΔamiC ΔtatABC (F− ΔlacU169 araD139 rpsL150 relA1 ptsF rbs flbB5301 ΔamiA ΔamiC ΔtatABC) was used for signal peptide library screening and for SDS growth tests with signal peptide-AmiA fusion proteins (27). Strain DADE (as MC4100, ΔtatABCD ΔtatE [58]) was used for SufI transport assays, and DADE-P [as DADE, pcnB1 zad-981::Tn10d (Kanr) (59)] was used for copurification experiments.
All plasmids used and constructed in this study are given in Table S2 in the supplemental material (65, 66). Point mutations in plasmids were introduced by QuikChange site-directed mutagenesis (Stratagene) using the primers listed in Table S3. Plasmid pTAT101 was used for low-level production of TatA, TatB, and TatC (37). Plasmid pSUSufIss-mAmiA was used to produce SufIss-AmiA, where the SufI signal peptide is fused to the mature portion of AmiA (27). Plasmid pQE80-SufIhis was used to produce His-tagged SufI (27).
Plasmids used and constructed in this study. Download TABLE S2, DOCX file, 0.02 MB (22.2KB, docx) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download TABLE S3, DOCX file, 0.02 MB (19.4KB, docx) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids pSUDsbAss-mAmiA, pSUDsbAssi16K-mAmiA (with an additional lysine codon inserted after codon 15 of the DbsA signal peptide), pSUOmpAss-mAmiA, and pSUOmpAssi18K-mAmiA (with an additional lysine codon inserted after codon 17 of the OmpA signal peptide) were constructed according to the method in reference 27. Briefly, DNA fragments encoding DsbAss, DsbAssi16K, OmpAss, and OmpAssi18K were amplified by PCR using MC4100 genomic DNA as the template, using primer pairs DsbAss-FE/DsbAss-R, DsbAss-FE/DsbAss16inK-R, OmpA-FE/OmpAss-R, and OmpA-FE/OmpA18inK-R, respectively. DNA fragments encoding the corresponding mature domain of AmiA were amplified by PCR using MC4100 genomic DNA as the template with primer pair OmpA-mAmiA-F/amiA-mRX, or DsbA-mAmiA-F/amiA-mRX. The DNA fragments encoding the signal peptides and the mature domain of AmiA were fused by overlap extension PCR, giving DNA fragments DsbAss-mAmiA, DsbAssi16K-mAmiA, OmpAss-mAmiA, and OmpAi18Kss-mAmiA, which were finally cloned into the pSU18 vector following digestion with EcoRI and XbaI.
Plasmid pFAT75BC-SufIFLAG was modified from pFAT75ΔA-SufIhis (18) via QuikChange using primers FAT75SufIFLAG-1/FAT75SufIFLAG-2. Plasmid pFATBChis-SufIFLAG was modified from pFAT75BC-SufIFLAG via QuikChange using primers FAT75TatChis-1/FAT75TatChis-2. Plasmid pFATBChis-OmpAssi18KSufIFLAG has the SufI signal peptide coding region substituted for DNA encoding OmpAssi18K and was constructed using a restriction enzyme-free cloning method according to the method in reference 60. Briefly, a DNA fragment covering OmpAssi18K was PCR amplified using pSUOmpAssi18K-mAmiA as the template, with primer pair FATHF-OmpA-F/FATHF-OmpA18K-R. The resultant DNA fragment was used as a primer to amplify the whole pFATBChis-SufIFLAG plasmid using a PCR program of 95°C for 2 min followed by 15 cycles of 95°C for 30 s, 48°C for 1.5 min, and 68°C for 15 min and a final extension at 68°C for 10 min. The PCR product was subjected to DpnI digestion and introduced into E. coli JM109 competent cells by transformation. The resultant plasmid was verified by DNA sequencing. Plasmids pQEBChis-OmpAFLAG and pQEBChis-DsbAFLAG were used for coproduction of TatB, His-tagged TatC, and FLAG-tagged OmpA or DsbA, respectively, and were constructed as follows. A DNA fragment encoding TatB and His-tagged TatC was amplified using pFATBChis-SufIFLAG as the template with primer pair QEF/FAT75TatChis-2 and was ligated, via an ApaI restriction site, to a DNA fragment encoding FLAG-tagged OmpA (which was amplified using MC4100 genomic DNA as the template with primer pair FATHF-OmpA-F/OmpAFLAG-SR) or FLAG-tagged DsbA (amplified similarly using primer pair FATHF-OmpA-F/DsbAFLAG-SR). The ligated fragment was enriched by using the ligation mixture as a template in a PCR with primer pair QEF/OmpAFLAG-SR or QEF/DsbAAFLAG-SR, respectively. The amplified fragment was gel purified, digested with EcoRI and SalI, and cloned into similarly digested pQE80. Constructs were verified by DNA sequencing.
Mutant library construction and screening.
To construct a random library of substitutions at codon 8 of the SufI signal peptide, site-directed mutagenesis was carried out via QuikChange using a pair of random primers, SufIF8X1 and SufIF8X2 (Table S3), and pSUSufIss-mAmiA as the template. The PCR product was subsequently introduced into XL1-Gold ultracompetent cells (Agilent). Transformants were scraped from plates, resuspended in LB, pooled, and cultured overnight, after which plasmid DNA was isolated and taken as the F8X random library. The library contained approximately 5,000 clones, and random sequencing of eight of them revealed mutations of the TTC codon to CTT, ATC GGT, GTG, GGT, CAA, AGA, and GGG.
The signal peptide mutagenesis library in plasmid pSUSufIss-mAmiA was constructed as described previously (27). Briefly, an error-containing DNA fragment covering the sufI signal sequence was amplified by error-prone PCR using primers SufIF and SufIR and pSUSufIss-mAmiA as the template. This fragment was used as a megaprimer to amplify the whole pSUSufIss-mAmiA plasmid. The amplified plasmid was introduced into XL1-Gold ultracompetent cells following nick repair using T4 polynucleotide kinase and T4 ligase. Transformants were scraped from plates, resuspended in LB, pooled, and used to inoculate fresh LB to an initial optical density at 600 nm (OD600) of 0.2. Cells were grown at 37°C until OD600 reached 2, after which plasmid DNA was prepared and taken as the signal peptide mutagenesis library.
For library screening, plasmid pTAT101 harboring the tatC point substitution of interest (along with wild-type tatAB) was introduced into MC4100 ΔamiA ΔamiC ΔtatABC. Subsequently, the mutant library was introduced, and cells were plated onto LB agar containing 2% SDS. Plasmids were isolated from colonies growing on this selective medium, and mutations were identified by sequencing.
Protein methods.
Copurification of TatBC-substrate complexes was carried out as described previously (27). Briefly, an overnight culture of DADE-P harboring plasmid pFATBChis-SufIFLAG or its derivatives was subcultured in LB supplemented with 0.5% glycerol and appropriate antibiotics for 2.5 h at 37°C with shaking. Following supplementation with 0.4 mM isopropyl-β-d-galactopyranoside (IPTG), cells were incubated overnight at 30°C. The following morning, cells were harvested, resuspended in 200 μl of 2× lysis buffer (100 mM NaH2PO4, pH 8.0, 600 mM NaCl, 40 mM imidazole, 50 mg lysozyme, DNase I, and protease inhibitor), and mixed gently at room temperature for 1 h. Cells were then frozen at −80°C for 1 h and thawed at room temperature. An equal volume of 2.5% digitonin was added to the cells, and the samples were solubilized for 1 h at 4°C. The insoluble material was pelleted by centrifugation at 4°C. A 30-μl amount of the supernatant was mixed with 2× Laemmli buffer, which was taken as the input sample, and the remaining supernatant was mixed with 50 μl wash buffer-equilibrated nickel beads (Profinity IMAC Ni-charged resin; Bio-Rad,; catalog number 156-0131) for 1 h. The nickel beads were pelleted, washed three times with 1 ml wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 40 mM imidazole, 0.03% digitonin), and then mixed with 100 μl elution buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 700 mM imidazole, 0.03% digitonin). The beads were incubated for 10 min with shaking and then pelleted. The supernatant (elution fraction) was taken and mixed with an equal volume of 2× Laemmli buffer, and 20 μl of the sample was subjected to SDS-PAGE followed by Western blotting with anti-His (anti-6×His tag antibody [GT359] [horseradish peroxidase {HRP}]; Abcam, Inc.; catalog number ab184607), anti-TatB (61), or anti-FLAG antibodies (monoclonal anti-FLAG M2 antibody produced in mouse; Sigma; catalog number F1804). Secondary antibody was goat anti-rabbit IgG (HRP conjugate; Bio-Rad; catalog number 170-6515) or goat anti-mouse IgG (HRP conjugate; Bio-Rad; catalog number 1706516).
Subcellular fractionation was carried out as described previously (27). All experiments were carried out with at least three biological replicates, and representative results are shown. Briefly, overnight cultures of strain DADE harboring pTAT101 or the cognate empty vector pTH19kr along with pQE80-SufIhis or its derivatives were subcultured at 1:50 in LB supplemented with 1 mM IPTG and grown at 37°C until OD600 reached 1. Where the RR-KK substitution was present in the SufI, no IPTG was used (as for unknown reasons this substitution results in high-level expression of SufI even in the absence of IPTG). For whole-cell samples, cells were pelleted from 5 ml of the culture, resuspended in 250 μl resuspension buffer (50 mM Tris-HCl, pH 7.6, 2 mM EDTA), and lysed by sonicating for 15 s. The cell lysate was mixed with an equal volume of 2× Laemmli buffer and boiled at 95°C for 10 min. For preparation of periplasm, cells were pelleted from 20 ml of the culture and resuspended in 500 μl fractionation buffer (20 mM Tris-HCl, pH 7.6, 2 mM EDTA, 20% [wt/vol] sucrose). Freshly made lysozyme (0.6 mg/ml) was added, and the cells were incubated at room temperature for 20 min. The cells were then pelleted by centrifugation, and the supernatant was taken and mixed with an equal volume of 2× Laemmli buffer. Aliquots (20 μl) of the whole-cell or periplasmic fraction samples were separated by SDS-PAGE (10% acrylamide) followed by Western blotting with anti-His antibody or anti-His and anti-RNA polymerase β-subunit mixed antibodies.
Prediction of Sec signal peptides.
All of the protein sequences encoded by E. coli MG1655 were analyzed using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP) (55) with parameters “Gram-negative bacteria” and “input sequence do not include TM regions” selected. Inner membrane proteins were removed manually from the output Sec substrate candidates.
ACKNOWLEDGMENTS
This work was supported by the China Scholarship Council (through a studentship to Q.H.). T.P. is a Wellcome Trust Investigator.
Footnotes
Citation Huang Q, Palmer T. 2017. Signal peptide hydrophobicity modulates interaction with the twin-arginine translocase. mBio 8:e00909-17. https://doi.org/10.1128/mBio.00909-17.
REFERENCES
- 1.Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 2.Berks BC. 1996. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 3.Lüke I, Handford JI, Palmer T, Sargent F. 2009. Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch Microbiol 191:919–925. doi: 10.1007/s00203-009-0516-5. [DOI] [PubMed] [Google Scholar]
- 4.Stanley NR, Palmer T, Berks BC. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem 275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 5.DeLisa MP, Samuelson P, Palmer T, Georgiou G. 2002. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J Biol Chem 277:29825–29831. doi: 10.1074/jbc.M201956200. [DOI] [PubMed] [Google Scholar]
- 6.Cristóbal S, de Gier JW, Nielsen H, von Heijne G. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J 18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tooke FJ, Babot M, Chandra G, Buchanan G, Palmer T. 17 May 2017. A unifying mechanism for the biogenesis of membrane proteins co-operatively integrated by the Sec and Tat pathways. Elife 6:e26577. doi: 10.7554/eLife.26577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogsch E, Brink S, Robinson C. 1997. Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a “Sec-incompatible” mature protein. EMBO J 16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaudeck N, Kreutzenbeck P, Freudl R, Sprenger GA. 2003. Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J Bacteriol 185:2811–2819. doi: 10.1128/JB.185.9.2811-2819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tullman-Ercek D, DeLisa MP, Kawarasaki Y, Iranpour P, Ribnicky B, Palmer T, Georgiou G. 2007. Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J Biol Chem 282:8309–8316. doi: 10.1074/jbc.M610507200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C. 2001. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J Biol Chem 276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- 12.Alcock F, Stansfeld PJ, Basit H, Habersetzer J, Baker MA, Palmer T, Wallace MI, Berks BC. 2016. Assembling the Tat protein translocase. eLife 5:e20718. doi: 10.7554/eLife.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habersetzer J, Moore K, Cherry J, Buchanan G, Stansfeld PJ, Palmer T. 2017. Substrate-triggered position-switching of TatA and TatB is an essential step in the Escherichia coli Tat protein export pathway. bioRxiv doi: 10.1101/113985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X, Cline K. 2000. Precursors bind to specific sites on thylakoid membranes prior to transport on the delta pH protein translocation system. J Biol Chem 275:10016–10022. doi: 10.1074/jbc.275.14.10016. [DOI] [PubMed] [Google Scholar]
- 15.Tarry MJ, Schäfer E, Chen S, Buchanan G, Greene NP, Lea SM, Palmer T, Saibil HR, Berks BC. 2009. Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc Natl Acad Sci U S A 106:13284–13289. doi: 10.1073/pnas.0901566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James MJ, Coulthurst SJ, Palmer T, Sargent F. 2013. Signal peptide etiquette during assembly of a complex respiratory enzyme. Mol Microbiol 90:400–414. doi: 10.1111/mmi.12373. [DOI] [PubMed] [Google Scholar]
- 17.Blummel AS, Haag LA, Eimer E, Muller M, Frobel J. 2015. Initial assembly steps of a translocase for folded proteins. Nat Commun 6:7234. doi: 10.1038/ncomms8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cléon F, Habersetzer J, Alcock F, Kneuper H, Stansfeld PJ, Basit H, Wallace MI, Berks BC, Palmer T. 2015. The TatC component of the twin-arginine protein translocase functions as an obligate oligomer. Mol Microbiol 98:111–129. doi: 10.1111/mmi.13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alami M, Lüke I, Deitermann S, Eisner G, Koch HG, Brunner J, Müller M. 2003. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell 12:937–946. doi: 10.1016/S1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 20.Gérard F, Cline K. 2006. Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J Biol Chem 281:6130–6135. doi: 10.1074/jbc.M512733200. [DOI] [PubMed] [Google Scholar]
- 21.Kreutzenbeck P, Kröger C, Lausberg F, Blaudeck N, Sprenger GA, Freudl R. 2007. Escherichia coli twin arginine (Tat) mutant translocases possessing relaxed signal peptide recognition specificities. J Biol Chem 282:7903–7911. doi: 10.1074/jbc.M610126200. [DOI] [PubMed] [Google Scholar]
- 22.Strauch EM, Georgiou G. 2007. Escherichia coli tatC mutations that suppress defective twin-arginine transporter signal peptides. J Mol Biol 374:283–291. doi: 10.1016/j.jmb.2007.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoufaly S, Fröbel J, Rose P, Flecken T, Maurer C, Moser M, Müller M. 2012. Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J Biol Chem 287:13430–13441. doi: 10.1074/jbc.M112.343798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rollauer SE, Tarry MJ, Graham JE, Jääskeläinen M, Jäger F, Johnson S, Krehenbrink M, Liu SM, Lukey MJ, Marcoux J, McDowell MA, Rodriguez F, Roversi P, Stansfeld PJ, Robinson CV, Sansom MS, Palmer T, Högbom M, Berks BC, Lea SM. 2012. Structure of the TatC core of the twin-arginine protein transport system. Nature 492:210–214. doi: 10.1038/nature11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gérard F, Cline K. 2007. The thylakoid proton gradient promotes an advanced stage of signal peptide binding deep within the Tat pathway receptor complex. J Biol Chem 282:5263–5272. doi: 10.1074/jbc.M610337200. [DOI] [PubMed] [Google Scholar]
- 26.Aldridge C, Ma X, Gerard F, Cline K. 2014. Substrate-gated docking of pore subunit Tha4 in the TatC cavity initiates Tat translocase assembly. J Cell Biol 205:51–65. doi: 10.1083/jcb.201311057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Q, Alcock F, Kneuper H, Deme JC, Rollauer SE, Lea SM, Berks BC, Palmer T. 2017. A signal sequence suppressor mutant that stabilizes an assembled state of the twin arginine translocase. Proc Natl Acad Sci U S A 114:E1958–E1967. doi: 10.1073/pnas.1615056114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori H, Cline K. 2002. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid [Delta]pH/Tat translocase. J Cell Biol 157:205–210. doi: 10.1083/jcb.200202048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcock F, Baker MA, Greene NP, Palmer T, Wallace MI, Berks BC. 2013. Live cell imaging shows reversible assembly of the TatA component of the twin-arginine protein transport system. Proc Natl Acad Sci U S A 110:E3650–E3659. doi: 10.1073/pnas.1306738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose P, Fröbel J, Graumann PL, Müller M. 2013. Substrate-dependent assembly of the Tat translocase as observed in live Escherichia coli cells. PLoS One 8:e69488. doi: 10.1371/journal.pone.0069488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brüser T, Sanders C. 2003. An alternative model of the twin arginine translocation system. Microbiol Res 158:7–17. doi: 10.1078/0944-5013-00176. [DOI] [PubMed] [Google Scholar]
- 32.Cline K. 2015. Mechanistic aspects of folded protein transport by the twin arginine translocase (tat). J Biol Chem 290:16530–16538. doi: 10.1074/jbc.R114.626820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berks BC. 2015. The twin-arginine protein translocation pathway. Annu Rev Biochem 84:843–864. doi: 10.1146/annurev-biochem-060614-034251. [DOI] [PubMed] [Google Scholar]
- 34.Holzapfel E, Eisner G, Alami M, Barrett CM, Buchanan G, Lüke I, Betton JM, Robinson C, Palmer T, Moser M, Müller M. 2007. The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46:2892–2898. doi: 10.1021/bi062205b. [DOI] [PubMed] [Google Scholar]
- 35.Ma X, Cline K. 2013. Mapping the signal peptide binding and oligomer contact sites of the core subunit of the pea twin arginine protein translocase. Plant Cell 25:999–1015. doi: 10.1105/tpc.112.107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchanan G, de Leeuw E, Stanley NR, Wexler M, Berks BC, Sargent F, Palmer T. 2002. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol Microbiol 43:1457–1470. doi: 10.1046/j.1365-2958.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- 37.Kneuper H, Maldonado B, Jäger F, Krehenbrink M, Buchanan G, Keller R, Müller M, Berks BC, Palmer T. 2012. Molecular dissection of TatC defines critical regions essential for protein transport and a TatB-TatC contact site. Mol Microbiol 85:945–961. doi: 10.1111/j.1365-2958.2012.08151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramasamy S, Abrol R, Suloway CJ, Clemons WM Jr. 2013. The glove-like structure of the conserved membrane protein TatC provides insight into signal sequence recognition in twin-arginine translocation. Structure 21:777–788. doi: 10.1016/j.str.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer T, Sargent F, Berks BC. 2010. The tat protein export pathway. EcoSal Plus 4:1. doi: 10.1128/ecosalplus.4.3.2. [DOI] [PubMed] [Google Scholar]
- 40.Berks BC, Lea SM, Stansfeld PJ. 2014. Structural biology of Tat protein transport. Curr Opin Struct Biol 27:32–37. doi: 10.1016/j.sbi.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Ize B, Stanley NR, Buchanan G, Palmer T. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol 48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 42.Bernhardt TG, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fekkes P, de Wit JG, van der Wolk JP, Kimsey HH, Kumamoto CA, Driessen AJ. 1998. Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol Microbiol 29:1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- 44.Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol 185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freudl R, Schwarz H, Stierhof YD, Gamon K, Hindennach I, Henning U. 1986. An outer membrane protein (OmpA) of Escherchia coli K-12 undergoes a conformational change during export. J Biol Chem 261:11355–11361. [PubMed] [Google Scholar]
- 46.Tsirigotaki A, De Geyter J, Šoštaric N, Economou A, Karamanou S. 2017. Protein export through the bacterial Sec pathway. Nat Rev Microbiol 15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 47.Huber D, Rajagopalan N, Preissler S, Rocco MA, Merz F, Kramer G, Bukau B. 2011. SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell 41:343–353. doi: 10.1016/j.molcel.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 48.Pérez-Rodríguez R, Fisher AC, Perlmutter JD, Hicks MG, Chanal A, Santini CL, Wu LF, Palmer T, DeLisa MP. 2007. An essential role for the DnaK molecular chaperone in stabilizing over-expressed substrate proteins of the bacterial twin-arginine translocation pathway. J Mol Biol 367:715–730. doi: 10.1016/j.jmb.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 49.Jong WS, ten Hagen-Jongman CM, Genevaux P, Brunner J, Oudega B, Luirink J. 2004. Trigger factor interacts with the signal peptide of nascent Tat substrates but does not play a critical role in Tat-mediated export. Eur J Biochem 271:4779–4787. doi: 10.1111/j.1432-1033.2004.04442.x. [DOI] [PubMed] [Google Scholar]
- 50.Holzapfel E, Moser M, Schiltz E, Ueda T, Betton JM, Müller M. 2009. Twin-arginine-dependent translocation of SufI in the absence of cytosolic helper proteins. Biochemistry 48:5096–5105. doi: 10.1021/bi900520d. [DOI] [PubMed] [Google Scholar]
- 51.Graubner W, Schierhorn A, Brüser T. 2007. DnaK plays a pivotal role in Tat targeting of CueO and functions beside SlyD as a general Tat signal binding chaperone. J Biol Chem 282:7116–7124. doi: 10.1074/jbc.M608235200. [DOI] [PubMed] [Google Scholar]
- 52.Kebir MO, Kendall DA. 2002. SecA specificity for different signal peptides. Biochemistry 41:5573–5580. doi: 10.1021/bi015798t. [DOI] [PubMed] [Google Scholar]
- 53.Sasaki S, Matsuyama S, Mizushima S. 1990. In vitro kinetic analysis of the role of the positive charge at the amino-terminal region of signal peptides in translocation of secretory protein across the cytoplasmic membrane in Escherichia coli. J Biol Chem 265:4358–4363. [PubMed] [Google Scholar]
- 54.Keller R, de Keyzer J, Driessen AJ, Palmer T. 2012. Co-operation between different targeting pathways during integration of a membrane protein. J Cell Biol 199:303–315. doi: 10.1083/jcb.201204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 56.Kouwen TR, van der Ploeg R, Antelmann H, Hecker M, Homuth G, Mäder U, van Dijl JM. 2009. Overflow of a hyper-produced secretory protein from the Bacillus Sec pathway into the Tat pathway for protein secretion as revealed by proteogenomics. Proteomics 9:1018–1032. doi: 10.1002/pmic.200800580. [DOI] [PubMed] [Google Scholar]
- 57.Casadaban MJ, Cohen SN. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A 76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, Berks BC, Palmer T. 2000. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J Biol Chem 275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
- 59.Lee PA, Orriss GL, Buchanan G, Greene NP, Bond PJ, Punginelli C, Jack RL, Sansom MS, Berks BC, Palmer T. 2006. Cysteine-scanning mutagenesis and disulfide mapping studies of the conserved domain of the twin-arginine translocase TatB component. J Biol Chem 281:34072–34085. doi: 10.1074/jbc.M607295200. [DOI] [PubMed] [Google Scholar]
- 60.van den Ent F, Löwe J. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Sargent F, Gohlke U, De Leeuw E, Stanley NR, Palmer T, Saibil HR, Berks BC. 2001. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur J Biochem 268:3361–3367. [DOI] [PubMed] [Google Scholar]
- 62.Rose RW, Brüser T, Kissinger JC, Pohlschröder M. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 45:943–950. [DOI] [PubMed] [Google Scholar]
- 63.Dilks K, Rose RW, Hartmann E, Pohlschröder M. 2003. Prokaryotic use of the twin arginine translocation pathway: a genomic survey. J Bacteriol 185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Käll L, Krogh A, Sonnhammer ELL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 65.Hashimoto-Gotoh T, Yamaguchi M, Yasojima K, Tsujimura A, Wakabayashi Y, Watanabe Y. 2000. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185–191. doi: 10.1016/S0378-1119(99)00434-5. [DOI] [PubMed] [Google Scholar]
- 66.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78. doi: 10.1016/0378-1119(91)90541-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The SufIS12L substitution can act in cis to suppress inactivating substitutions in the SufI signal peptide twin-arginine motif. Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring pTH19kr alongside pSUSufIss-mAmiA (Δtat) or pTAT101 (producing wild-type TatABC) alongside unsubstituted pSUSufIss-mAmiA (tat+) (A) or pTAT101 (producing wild-type TatABC) alongside pSUSufIss-mAmiA encoding the indicated substitutions in the SufI signal peptide (B to J) were subcultured at a 1:100 dilution into LB supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +SD (n = 3 biological replicates). Download FIG S1, PDF file, 0.1 MB (137KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The twin-arginine substituted SufIS12L signal peptide variants mediate strict Tat-dependent transport of AmiA. (A and B) Overnight cultures of the tat− strain MC4100 ΔamiA ΔamiC ΔtatABC harboring either pTAT101 (producing wild-type TatABC) alongside unsubstituted pSUSufIss-mAmiA (tat+) or pTH19kr alongside either unsubstituted pSUSufIss-mAmiA (Δtat) or pSUSufIss-mAmiA encoding the indicated substitutions in the SufI signal peptide as indicated were subcultured at 1:100 dilution. (A) Cultures were grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. (B) Alternatively, cultures were supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +SD (n = 3 biological replicates). Download FIG S2, PDF file, 0.1 MB (114.5KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hydrophobic substitutions in the SufI signal peptide can rescue the transport defect of TatCE103K. Overnight cultures of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring pTAT101 producing TatABCE103K along with pSUSufIss-mAmiA producing the indicated substitution in the SufI signal peptide were subcultured at a 1:100 dilution, grown for a further 3 h at 37°C, pelleted, and resuspended to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. Download FIG S3, PDF file, 0.04 MB (37.5KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hydrophobic substitutions in the SufI signal peptide can act in cis to suppress the signal peptide R5K,R6K substitution. Overnight cultures of MC4100 ΔamiA ΔamiC ΔtatABC harboring pTAT101 (producing wild-type TatABC) alongside either unsubstituted pSUSufIss-mAmiA (WT) or pSUSufIss-mAmiA encoding the indicated substitutions in the SufI signal peptide were subcultured at a 1:100 dilution in the presence of 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Download FIG S4, PDF file, 0.1 MB (84.3KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of SufI export mediated by signal peptide leucine substitutions in the TatCE103K background and in a tat deletion strain. E. coli strain DADE coproducing His-tagged but otherwise native SufI or SufI with the indicated single leucine substitutions in the signal peptide (from a pQE80 plasmid) alongside either an empty plasmid vector (Δtat) (A) or wild-type TatAB and TatCE103K (from pTAT101) (B) was grown to mid-log phase, fractionated into whole-cell (upper panels) and periplasm (lower panels) samples, and then analyzed by Western blotting with anti-6×His tag or anti-RNA polymerase β-subunit antibodies (cytoplasmic control protein). Strain DADE coproducing His-tagged but otherwise native SufI alongside either wild-type TatABC (tat+ WT SufI) or an empty vector (Δtat WT SufI) was used as a positive or negative control, respectively. wc, whole cell. Equivalent volumes of sample were loaded for each of the whole-cell samples and for each of the periplasmic samples. Download FIG S5, PDF file, 0.1 MB (136.2KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detection of SufI-FLAG variants in input samples prior to Ni affinity purification. Cells of strain DADE-P coproducing C-terminally FLAG-tagged SufI with its native signal peptide (WT SufI) or harboring the indicated leucine substitutions, alongside TatB and C-terminally His-tagged TatC or TatCF94D, were lysed and incubated with digitonin. Thirty microliters of the digitonin-solubilized fraction was mixed with an equal volume of 2× Laemmli buffer, and 10 μl of each sample was subjected to SDS-PAGE (12% acrylamide) followed by Western blotting with anti-FLAG antibody. Download FIG S6, PDF file, 0.1 MB (78KB, pdf) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. coli proteins with known or predicted signal peptides. Protein sequences encoded by E. coli MG1655 were retrieved from UniProt (http://www.uniprot.org) and analyzed for the presence of a signal peptide using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP). The n-regions were analyzed manually; twin arginines are colored red, twin lysines are colored green, and Lys-Arg or Arg-Lys is shown in blue. Pairings of Arg-Asn, Arg-Asp, Arg-Gln, Arg-Glu, and Arg-His (which, from this study, can be suppressed by increased signal peptide hydrophobicity) are shown in orange. For those signal peptides harboring any of these dipeptides in their n-regions, the approximate location of the h-region is shown underlined. The start of the h-region was defined by Phobius (http://phobius.sbc.su.se/), and the end of the h-region and the cleavage site was defined by SignalP. Pairings within five residues of the start of the h-region were considered likely to interact with the Tat pathway. Those marked as N? have one of the indicated pairings 5 or more residues away from the h-region and are considered unlikely to interact with the Tat pathway. Known or probable Tat substrates are shown in yellow highlight; EfeO is predicted to be a Tat substrate, but this has not been demonstrated experimentally. Download TABLE S1, DOCX file, 0.04 MB (41.9KB, docx) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used and constructed in this study. Download TABLE S2, DOCX file, 0.02 MB (22.2KB, docx) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download TABLE S3, DOCX file, 0.02 MB (19.4KB, docx) .
Copyright © 2017 Huang and Palmer.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.