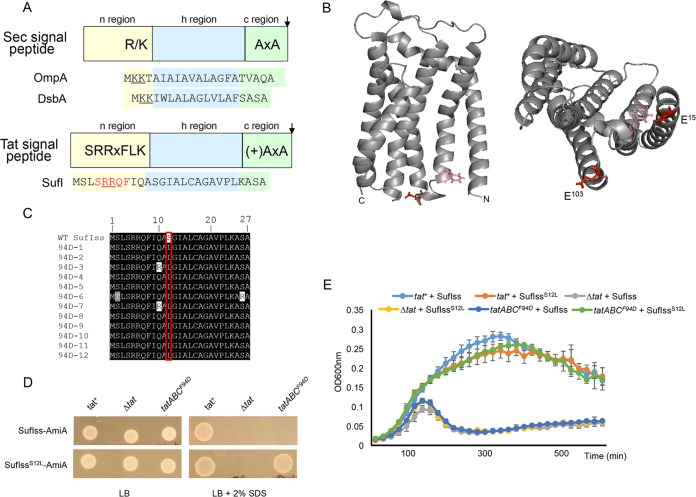
FIG 1 .
(A) Schematic representation of Sec and Tat signal peptides. The sequences of the OmpA and DsbA Sec-targeting signals and the SufI Tat-targeting signal peptide are shown. Positive charges in the signal peptide n-regions are shown underlined, and the amino acids of the SufI Tat consensus motif are shown in red. (B) Models of E. coli TatC. (Left) Side view, with F94 and E103 residues that are located in the signal peptide binding site given in pink and red, respectively. (Right) View of the cytoplasmic face, with E15 additionally shown. (C) Alignment of the amino acid sequence of 12 suppressors in the SufI signal peptide that compensate for the TatC F94D substitution. WT, wild type. (D and E) Cells of strain MC4100 ΔamiA ΔamiC ΔtatABC harboring pTH19kr (empty vector; annotated Δtat) or pTAT101 producing wild-type TatAB along with either wild-type TatC (tat+) or TatCF94D (tatABCF94D) and a compatible plasmid (either pSUSufIss-mAmiA or pSUSufIS12Lss-mAmiA, as indicated) were subcultured at 1:100 into fresh LB medium following overnight growth. (D) Cells were incubated for 3 h at 37°C with shaking. Cells were pelleted and resuspended in sterile phosphate-buffered saline (PBS) supplemented with appropriate antibiotics to an OD600 of 0.1, and 8 μl of sample was spotted onto LB agar or LB agar containing 2% SDS. Plates were incubated at 37°C for 16 h. (E) Alternatively, cultures were supplemented with 0.5% SDS (final concentration) and grown at 37°C without shaking. The optical density at 600 nm was monitored every 20 min using a plate reader. Error bars are +standard deviations (n = 3 biological replicates).